Genome-Wide Identification and Expression Analysis of Growth-Regulating Factor Family in Sweet Potato and Its Two Relatives
Abstract
1. Introduction
2. Results
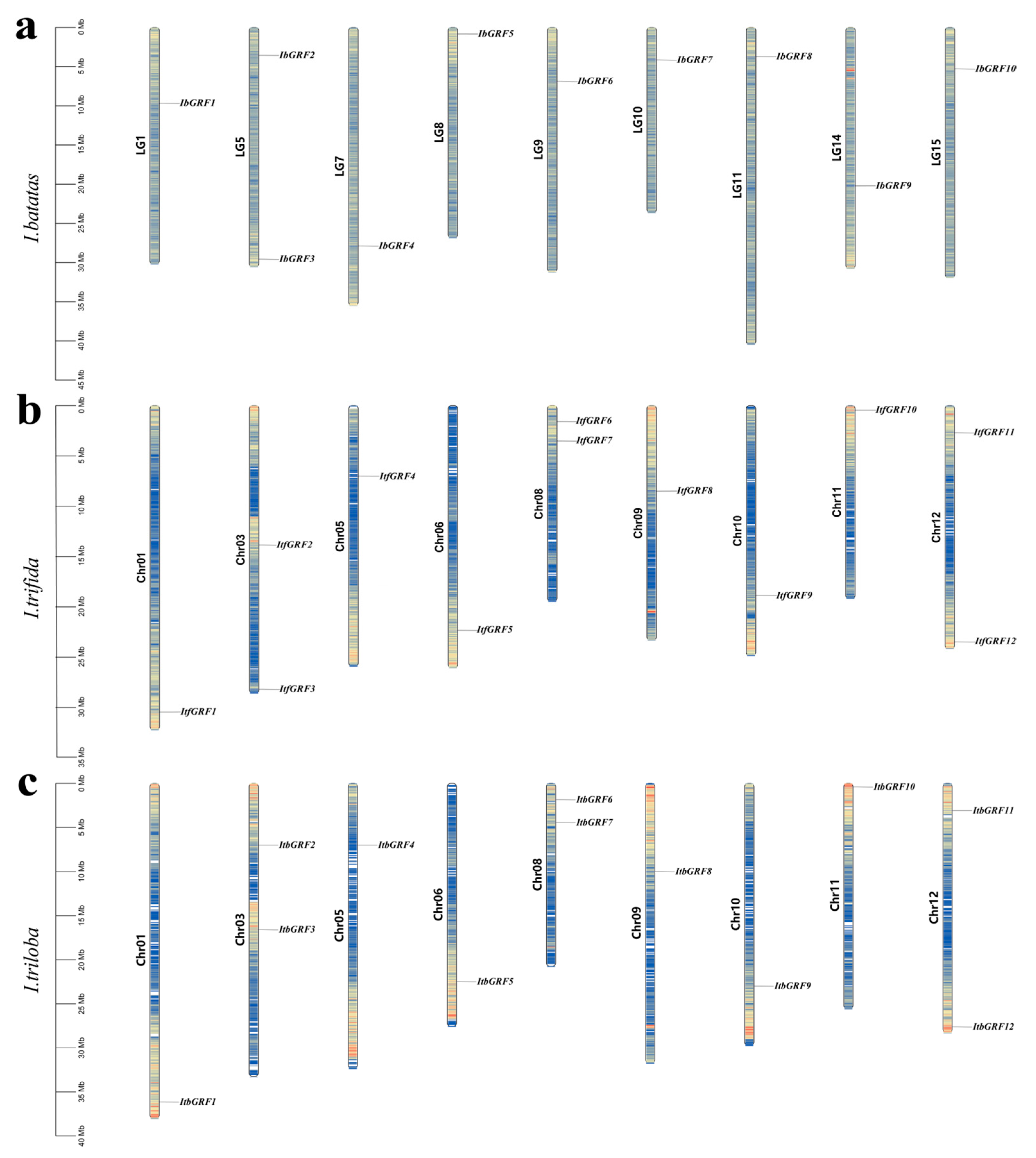
2.1. Identification of GRFs in Sweet Potato and Its Two Diploid Wild Relatives
2.2. Collinearity Analysis of GRF Genes and Ka/Ks Analysis
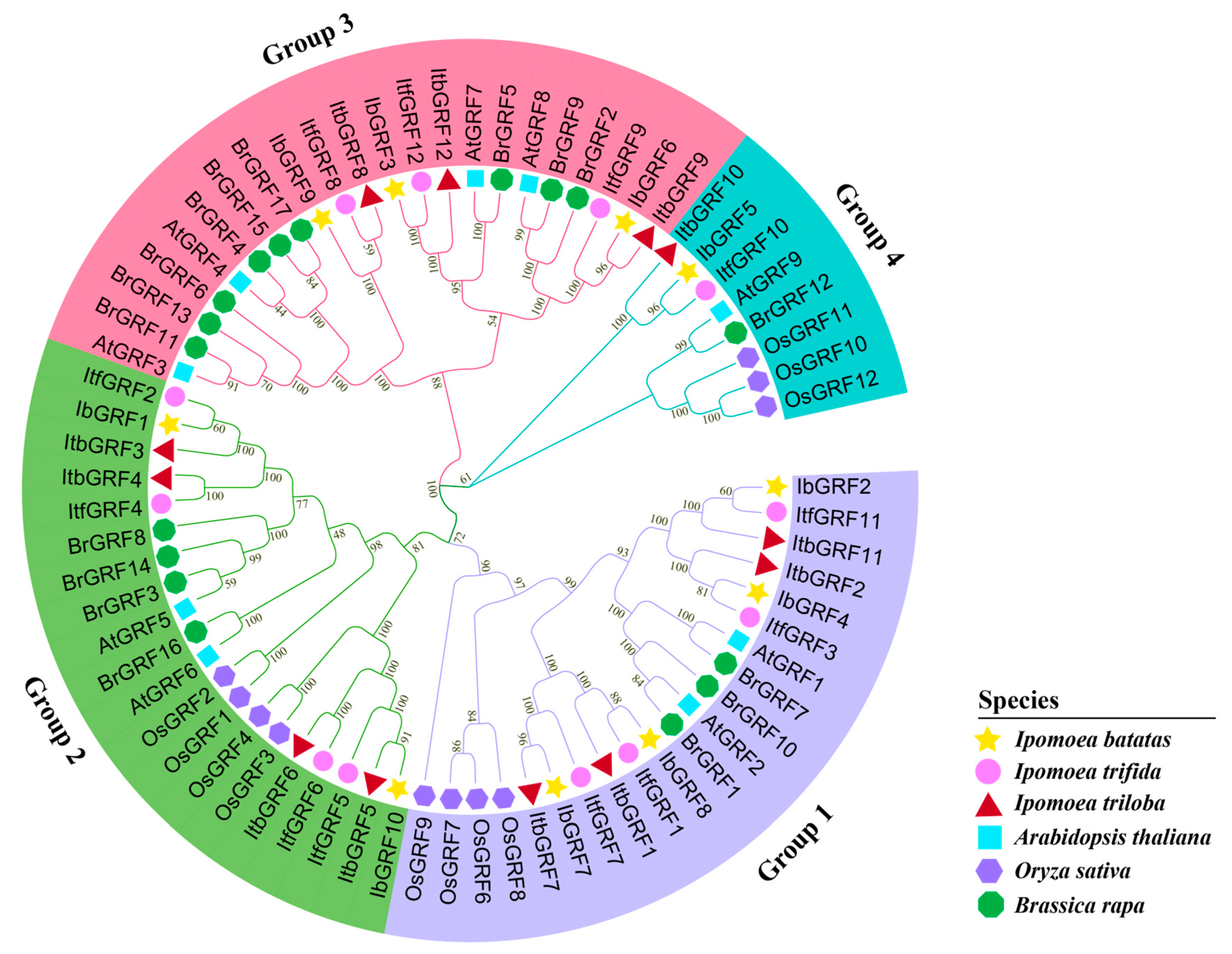
2.3. Phylogenetic Relationship Analysis of GRFs in Sweet Potato and Its Two Diploid Relatives
2.4. Conserved Motifs, Conserved Domain and Exon–Intron Structure Analysis of GRFs from I. batatas and Its Two Relatives I. trifida and I. triloba
2.5. Analysis of Putative Cis-Regulatory Elements of GRFs Promoters from I. batata and Its Two Diploid Relatives I. trifida and I. triloba
2.6. Expression Analysis of GRFs in I. batata, I. trifida and I. triloba
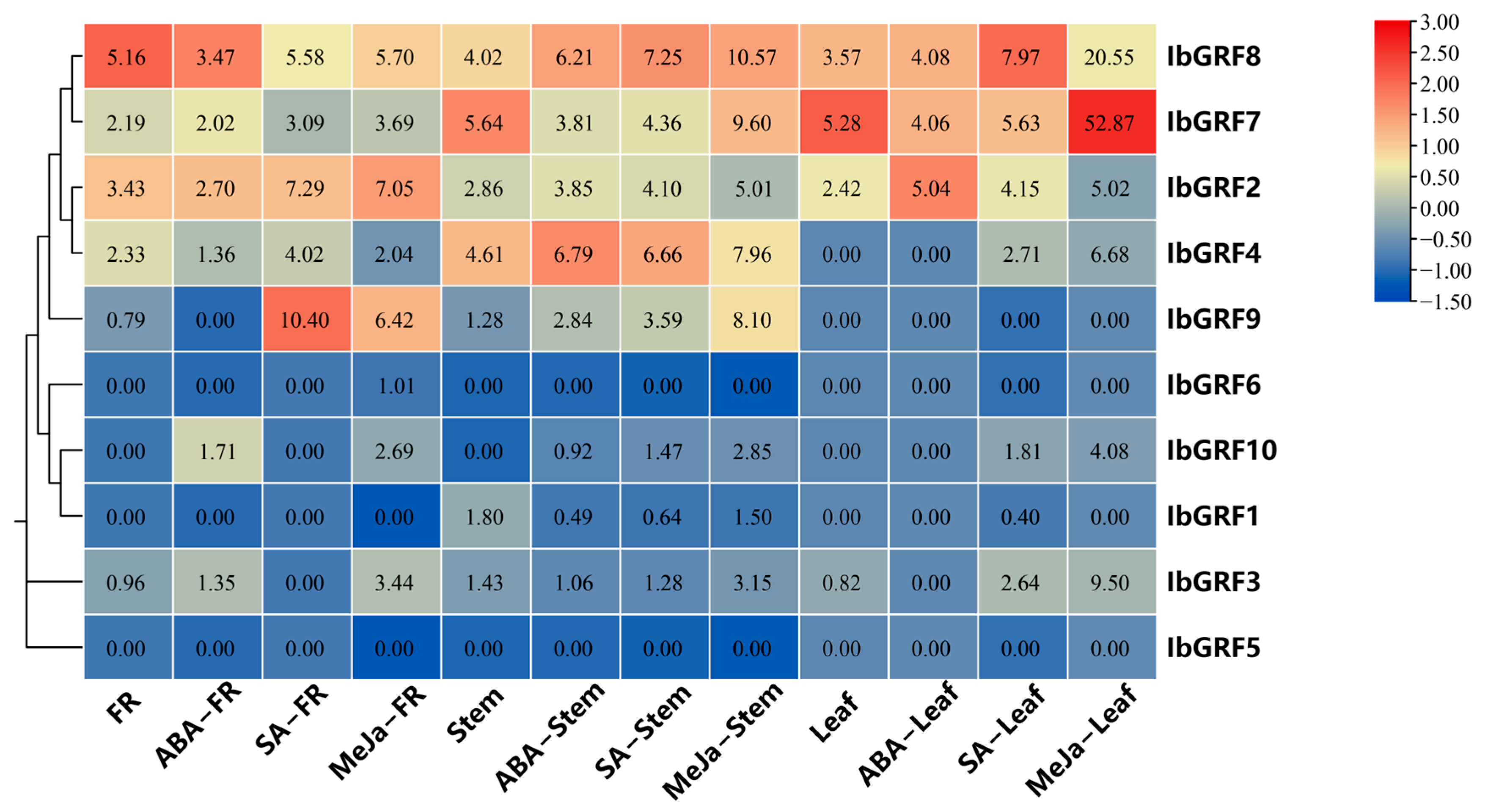
2.6.1. Expression Analysis under Hormone Stress
2.6.2. Expression Analysis under Cold Stress
2.6.3. Expression Analysis under Heat Stress
2.6.4. Expression Analysis under Salt and Drought Stresses
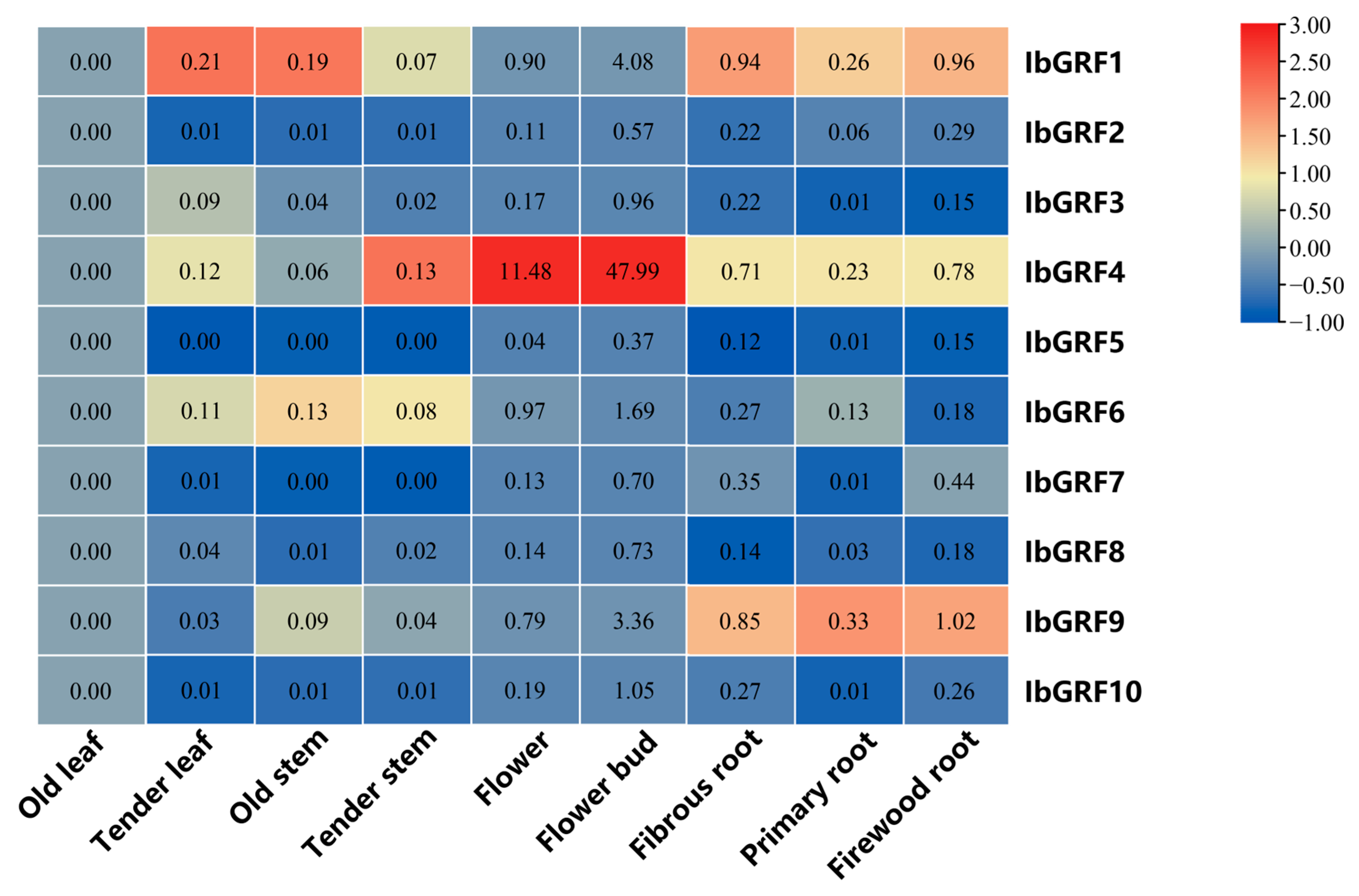
2.6.5. Expression Analysis in Various Tissues
2.7. Protein Interactions Network of GRFs in Sweet Potato
2.8. Transcript Factors Network of Sweet Potato GRF Genes
3. Discussion
3.1. Identification and Evolution of Sweet Potato and Its Two Diploid Relatives I. trifida and I. triloba
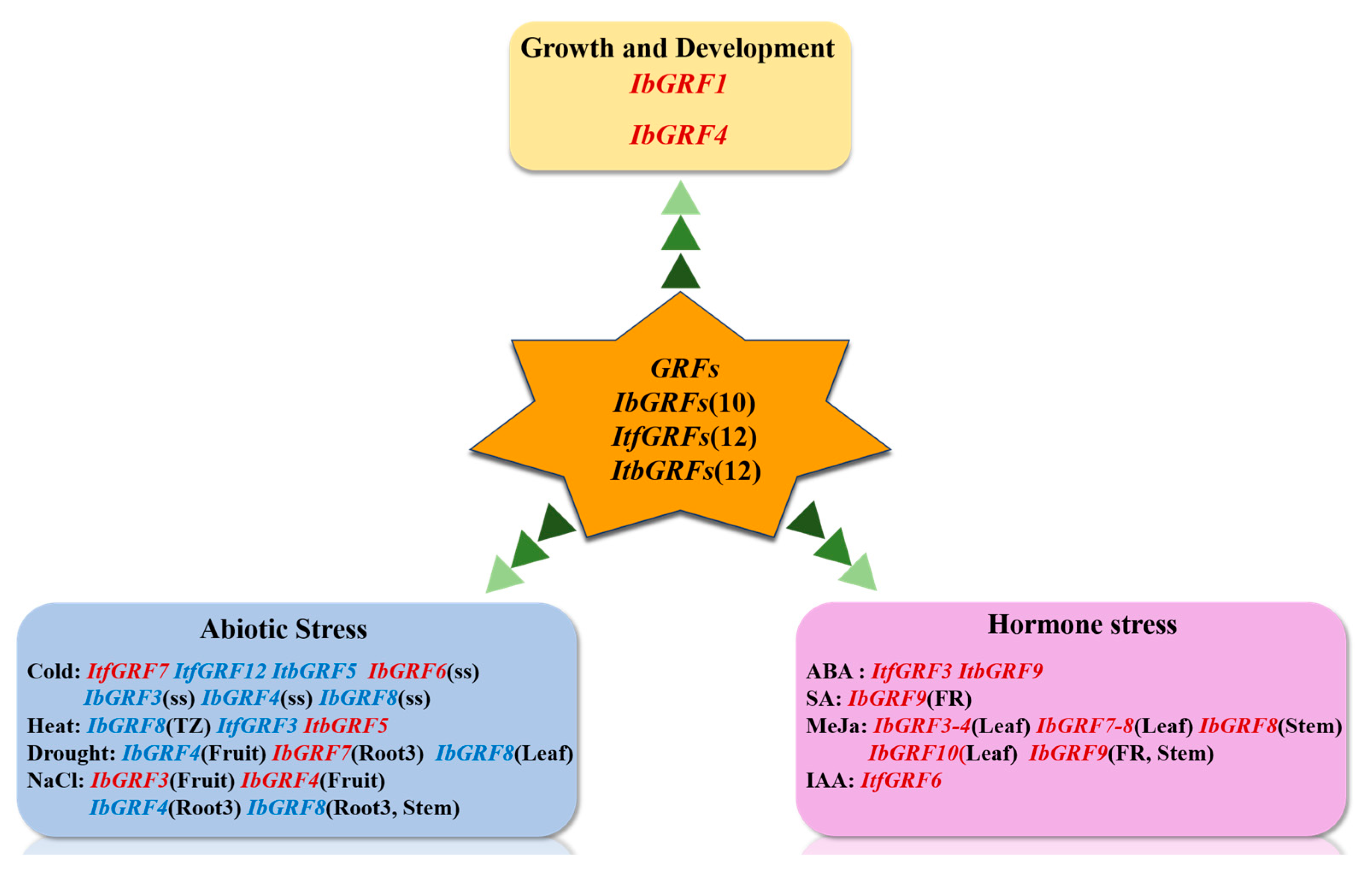
3.2. GRFs Are Involved in Plants Growth and Development Hormonal Regulation
4. Materials and Methods
4.1. Identification and Physicochemical Properties of GRF Family Members
4.2. Collinearity Analysis of GRF Genes and Calculation of Ka/Ks Value
4.3. Phylogenetic Analysis of GRFs
4.4. Conserved Motifs and Gene Structure Analysis of GRFs
4.5. Analysis of Putative Cis-Regulatory Elements of GRFs
4.6. Protein Interactions Analysis of Sweet Potato GRFs
4.7. Transcript Factors Regulatory Network Analysis of Sweet Potato GRFs
4.8. Transcriptome Analysis
4.9. Quantitative Analysis of Candidate IbGRF Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liang, G.; He, H.; Li, Y.; Wang, F.; Yu, D. Molecular Mechanism of microRNA396 Mediating Pistil Development in Arabidopsis. Plant Physiol. 2013, 164, 249–258. [Google Scholar] [CrossRef]
- Rosenquist, M.; Alsterfjord, M.; Larsson, C.; Sommarin, M. Data mining the Arabidopsis genome reveals fifteen 14-3-3 genes. Expression is demonstrated for two out of five novel genes. Plant Physiol. 2001, 127, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Zan, T.; Zhang, L.; Xie, T.; Li, L. Genome-Wide Identification and Analysis of the Growth-Regulating Factor (GRF) Gene Family and GRF-Interacting Factor Family in Triticum aestivum L. Biochem. Genet. 2020, 58, 705–724. [Google Scholar] [CrossRef] [PubMed]
- Carlson, F.D.; Hubbard, J. Physiological and biochemical aspects of nervous integration. Physiol. Zool. 1968, 42, 348–349. [Google Scholar]
- Obsil, T.; Obsilova, V. Structural basis of 14-3-3 protein functions. Semin. Cell Dev. Biol. 2011, 22, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Camoni, L.; Visconti, S.; Aducci, P.; Marra, M. 14-3-3 Proteins in Plant Hormone Signaling: Doing Several Things at Once. Front. Plant Sci. 2018, 9, 297. [Google Scholar] [CrossRef]
- Omidbakhshfard, M.A.; Proost, S.; Fujikura, U.; Mueller-Roeber, B. Growth-Regulating Factors (GRFs): A Small Transcription Factor Family with Important Functions in Plant Biology. Mol. Plant 2015, 8, 998–1010. [Google Scholar] [CrossRef]
- Van der Knaap, E.; Kim, J.H.; Kende, H. A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 2000, 122, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Vasupalli, N.; Cai, O.; Lin, X.; Wu, H. Network of miR396-mRNA in Tissue Differentiation in Moso Bamboo (Phyllostachys edulis). Plants 2023, 12, 1103. [Google Scholar] [CrossRef]
- Zhang, D.-F.; Li, B.; Jia, G.-Q.; Zhang, T.-F.; Dai, J.-R.; Li, J.-S.; Wang, S.-C. Isolation and characterization of genes encoding GRF transcription factors and GIF transcriptional coactivators in Maize (Zea mays L.). Plant Sci. 2008, 175, 809–817. [Google Scholar] [CrossRef]
- Kim, J.H.; Choi, D.; Kende, H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 2003, 36, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.; Kim, J.H.; Kende, H. Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice (Oryza sativa L.). Plant Cell Physiol. 2004, 45, 897–904. [Google Scholar] [CrossRef]
- Wang, F.; Qiu, N.; Ding, Q.; Li, J.; Zhang, Y.; Li, H.; Gao, J. Genome-wide identification and analysis of the growth-regulating factor family in Chinese cabbage (Brassica rapa L. ssp. pekinensis). BMC Genom. 2014, 15, 807. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.Q.; Jian, H.J.; Yang, B.; Lu, K.; Zhang, A.X.; Liu, P.; Li, J.N. Genome-wide analysis and expression profiling of the GRF gene family in oilseed rape (Brassica napus L.). Gene 2017, 620, 36–45. [Google Scholar] [CrossRef] [PubMed]
- FİLİZ, E.; KoÇ, İ.; TombuloĞLu, H. Genome-wide identification and analysis of growth regulating factor genes in Brachypodium distachyon: In silico approaches. Turk. J. Biol. 2014, 38, 296–306. [Google Scholar] [CrossRef]
- Zhu, R.; Cao, B.; Sun, M.; Wu, J.; Li, J. Genome-Wide Identification and Evolution of the GRF Gene Family and Functional Characterization of PbGRF18 in Pear. Int. J. Mol. Sci. 2023, 24, 14690. [Google Scholar] [CrossRef]
- Matías, B.; Florencia, E.M.; Manuel, D.J.; Camila, G.; Rojas, A.M.L.; Florencia, N.; Elena, A.M.; Liesbeth, V.; Dirk, I.; Palatnik, J.F. Robust increase of leaf size by Arabidopsis thaliana GRF3-like transcription factors under different growth conditions. Sci. Rep. 2018, 8, 13447. [Google Scholar]
- Horiguchi, G.; Kim, G.T.; Tsukaya, H. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 2005, 43, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Mizoi, J.; Kidokoro, S.; Maruyama, K.; Nakajima, J.; Nakashima, K.; Mitsuda, N.; Takiguchi, Y.; Ohme-Takagi, M.; Kondou, Y. Arabidopsis growth-regulating factor7 functions as a transcriptional repressor of abscisic acid- and osmotic stress-responsive genes, including DREB2A. Plant Cell 2012, 24, 3393–3405. [Google Scholar] [CrossRef]
- Amin, O.M.; Ushio, F.; Jadwiga, O.J.; Gang-Ping, X.; Salma, B.; Bernd, M.R.; Scott, P.R. GROWTH-REGULATING FACTOR 9 negatively regulates arabidopsis leaf growth by controllingORG3and restricting cell proliferation in leaf primordia. PLoS Genet. 2018, 14, e1007484. [Google Scholar]
- Diao, Z.; Yu, M.; Bu, S.; Duan, Y.; Zhang, L.; Wu, W. Functional characterization of OsmiR396a in rice (Oryza sativa L.). Plant Growth Regul. Int. J. Nat. Synth. Regul. 2018, 85, 351–361. [Google Scholar] [CrossRef]
- Li, S.; Gao, F.; Xie, K.; Zeng, X.; Cao, Y.; Zeng, J.; He, Z.; Ren, Y.; Li, W.; Deng, Q. The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice. Plant Biotechnol. J. 2016, 14, 2134–2146. [Google Scholar] [CrossRef]
- Gao, F.; Wang, K.; Liu, Y.; Chen, Y.; Chen, P.; Shi, Z.; Luo, J.; Jiang, D.; Fan, F.; Zhu, Y.; et al. Blocking miR396 increases rice yield by shaping inflorescence architecture. Nat. Plants 2015, 2, 15196. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Dan, Z.; Li, S. Rice GROWTH-REGULATING FACTOR 7 controls tiller number by regulating strigolactone synthesis. Plant Signal. Behav. 2020, 15, 1804685. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Tan, J.; Zhou, C.; Yang, X.; Shi, Z. The OsmiR396-OsGRF8-OsF3H-flavonoid pathway mediates resistance to the brown planthopper in rice (Oryza sativa). Plant Biotechnol. J. 2019, 17, 1657–1669. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhao, X.; Dai, Z.; Ma, F.; Miao, X.; Shi, Z. OsmiR396/Growth Regulating Factor modulate rice grain size through direct regulation of embryo-specific miR408. Plant Physiol. 2021, 186, 519–533. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, G.; Wang, Y.; Anwar, A.; He, B.; Zhang, J.; Chen, C.; Hao, Y.; Chen, R.; Song, S. Genome-wide identification of BcGRF genes in flowering Chinese cabbage and preliminary functional analysis of BcGRF8 in nitrogen metabolism. Front. Plant Sci. 2023, 14, 1144748. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.K.; He, Y.N.; Yang, X.Y.; Tang, X.; Wang, M.; Dai, W.S. Genome-wide identification of the GRF family in sweet orange (Citrus sinensis) and functional analysis of the CsGRF04 in response to multiple abiotic stresses. BMC Genom. 2024, 25, 37. [Google Scholar] [CrossRef]
- Zhang, B.; Tong, Y.; Luo, K.; Zhai, Z.; Liu, X.; Shi, Z.; Zhang, D.; Li, D. Identification of GROWTH-REGULATING FACTOR transcription factors in lettuce (Lactuca sativa) genome and functional analysis of LsaGRF5 in leaf size regulation. BMC Plant Biol. 2021, 21, 485. [Google Scholar] [CrossRef]
- Zhu, Y.; Kuang, W.; Leng, J.; Wang, X.; Qiu, L.; Kong, X.; Wang, Y.; Zhao, Q. The apple 14-3-3 gene MdGRF6 negatively regulates salt tolerance. Front. Plant Sci. 2023, 14, 1161539. [Google Scholar] [CrossRef]
- Wang, S.; Nie, S.; Zhu, F. Chemical constituents and health effects of sweet potato. Food Res. Int. 2016, 89, 90–116. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Moeinzadeh, M.H.; Kuhl, H.; Helmuth, J.; Xiao, P.; Haas, S.; Liu, G.; Zheng, J.; Sun, Z.; Fan, W.; et al. Haplotype-resolved sweet potato genome traces back its hexaploidization history. Nat. Plants 2017, 3, 696–703. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q. Improvement for agronomically important traits by gene engineering in sweetpotato. Breed. Sci. 2017, 67, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Wan, F.X.; Gao, J.; Wang, G.L.; Niu, Y.; Wang, L.Z.; Zhang, X.G.; Wang, Y.Q.; Pan, Y. Genome-wide identification of NAC transcription factor family and expression analysis of ATAF subfamily members under abiotic stress in eggplant. Sci. Hortic. 2021, 289, 110424. [Google Scholar] [CrossRef]
- Liu, Z.; Fu, M.; Li, H.; Chen, Y.; Wang, L.; Liu, R. Systematic analysis of NAC transcription factors in Gossypium barbadense uncovers their roles in response to Verticillium wilt. PeerJ 2019, 7, e7995. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Zhang, Q.; Liu, S.; Liu, Z.; Bian, X.; Yu, T. De novo transcriptome sequencing and gene expression profiling of sweetpotato leaves during low temperature stress. Plant Biotechnol. Rep. 2023, 17, 875–888. [Google Scholar] [CrossRef]
- Wu, S.; Lau, K.H.; Cao, Q.; Hamilton, J.P.; Sun, H.; Zhou, C.; Eserman, L.; Gemenet, D.C.; Olukolu, B.A.; Wang, H.; et al. Genome sequences of two diploid wild relatives of cultivated sweetpotato reveal targets for genetic improvement. Nat. Commun. 2018, 9, 4580. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Tsukaya, H. Regulation of plant growth and development by the GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR duo. J. Exp. Bot. 2015, 66, 6093–6107. [Google Scholar] [CrossRef]
- Dai, Z.; Yan, P.; He, S.; Jia, L.; Wang, Y.; Liu, Q.; Zhai, H.; Zhao, N.; Gao, S.; Zhang, H. Genome-Wide Identification and Expression Analysis of SWEET Family Genes in Sweet Potato and Its Two Diploid Relatives. Int. J. Mol. Sci. 2022, 23, 15848. [Google Scholar] [CrossRef]
- Jiang, Z.; Zhang, H.; Gao, S.; Zhai, H.; He, S.; Zhao, N.; Liu, Q. Genome-Wide Identification and Expression Analysis of the Sucrose Synthase Gene Family in Sweet Potato and Its Two Diploid Relatives. Int. J. Mol. Sci. 2023, 24, 12493. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Shi, L.; Lin, X.; Tang, B.; Xing, M.; Zhu, H. Genome-Wide Identification and Expression Analysis of Malate Dehydrogenase Gene Family in Sweet Potato and Its Two Diploid Relatives. Int. J. Mol. Sci. 2023, 24, 16549. [Google Scholar] [CrossRef]
- Houjun, Z.; Kaili, W.; Cheng, J.; Yanqiu, Z.; Xueqin, S.; Mengzhu, L. Negative Regulation of GRF1/2d on the Formation and Development of Adventitious Roots in Populus alba × P. glandulosa ‘84K’. Sci. Silvae Sin. 2017, 53, 33–39. [Google Scholar]
- Khisti, M.; Avuthu, T.; Yogendra, K.; Valluri, V.K.; Kudapa, H.; Reddy, P.S.; Tyagi, W. Genome-wide identification and expression profiling of growthregulating factor (GRF) and GRFinteracting factor (GIF) gene families in chickpea and pigeonpea. Sci. Rep. 2024, 14, 17178. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Yang, Y.; Luo, X.; Zhou, W.; Shu, K. Genome-wide identification of GRF transcription factors in soybean and expression analysis of GmGRF family under shade stress. BMC Plant Biol. 2019, 19, 269. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H. Biological roles and an evolutionary sketch of the GRF-GIF transcriptional complex in plants. BMB Rep. 2019, 52, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Bolduc, N.; Hake, S. The Maize Transcription Factor KNOTTED1 Directly Regulates the Gibberellin Catabolism Gene ga2ox1. Plant Cell 2009, 21, 1647–1658. [Google Scholar] [CrossRef]
- Daviere, J.M.; Achard, P. Gibberellin signaling in plants. Development 2013, 140, 1147–1151. [Google Scholar] [CrossRef]
- Kuijt, S.J.H.; Greco, R.; Agalou, A.; Shao, J.; Hoen, C.C.J.T.; Overnas, E.; Osnato, M.; Curiale, S.; Meynard, D.; Gulik, R.V. Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX Families of Transcription Factors. Plant Physiol. 2014, 164, 1952–1966. [Google Scholar] [CrossRef]
- Modulating plant growth-metabolism coordination for sustainable agriculture. Sci. Found. China 2018.
- Adam, K.; Hupp, T.R.; Matej, L.; Borivoj, V.; Petr, M. Hammock: A hidden Markov model-based peptide clustering algorithm to identify protein-interaction consensus motifs in large datasets. Bioinformatics 2016, 32, 9–16. [Google Scholar]
- Altschul, S.; Gish, W.; Miller, W.; Myers, E.; Lipman, D. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Jrg, S.; Copley, R.R.; Tobias, D.; Ponting, C.P.; Peer, B. SMART: A web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000, 28, 231–234. [Google Scholar]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Paul, H.; Keun-Joon, P.; Takeshi, O.; Naoya, F.; Hajime, H.; Adams-Collier, C.J.; Kenta, N. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, 585–587. [Google Scholar]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Ma, G.; Silva, J.A.T.D.; Qiu, L.; Xu, J.; Zhou, H.; Wei, M.; Xiong, J.; Li, M.; Zhou, S.; et al. Genome-Wide Identification and Analysis of NAC Transcription Factor Family in Two Diploid Wild Relatives of Cultivated Sweet Potato Uncovers Potential NAC Genes Related to Drought Tolerance. Front. Genet. 2021, 12, 744220. [Google Scholar] [CrossRef]
- Sudhir, K.; Glen, S.; Koichiro, T. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nuclc Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Mering, C.V.; Jensen, L.J.; Snel, B.; Hooper, S.D.; Krupp, M.; Foglierini, M.; Jouffre, N.; Huynen, M.A.; Bork, P. STRING: Known and predicted protein–protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005, 33, D433–D437. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Rauluseviciute, I.; Riudavets-Puig, R.; Blanc-Mathieu, R.; Castro-Mondragon, J.A.; Ferenc, K.; Kumar, V.; Lemma, R.B.; Lucas, J.; Chèneby, J.; Baranasic, D.; et al. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2024, 52, D174–D182. [Google Scholar] [CrossRef] [PubMed]
- Florea, L.; Song, L.; Salzberg, S.L. Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Research 2013, 2, 188. [Google Scholar] [CrossRef] [PubMed]
- Simms, D.; Chomczynski, P. TRIzolTM: A new reagent for optimal single-step isolation of RNA. Focus 1992, 15, 532–535. [Google Scholar]
- Shao, Z.; He, M.; Zeng, Z.; Chen, Y.; Hanna, A.D.; Zhu, H. Genome-Wide Identification and Expression Analysis of the MADS-Box Gene Family in Sweet Potato [Ipomoea batatas (L.) Lam]. Front. Genet. 2021, 12, 750137. [Google Scholar] [CrossRef] [PubMed]
- Ding, N.; Wang, A.; Zhang, X.; Wu, Y.; Wang, R.; Cui, H.; Huang, R.; Luo, Y. Identification and analysis of glutathione S-transferase gene family in sweet potato reveal divergent GST-mediated networks in aboveground and underground tissues in response to abiotic stresses. BMC Plant Biol. 2017, 17, 225. [Google Scholar] [CrossRef]










| Sequence ID | Genomic Length (bp) | CDS Length (bp) | Protein Size (aa) | Molecular Weight (kDa) | Isoelectric Point (pI) | Instability Index | Aliphatic Index | Hydropathicity | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|
| IbGRF1 | 6470 | 1158 | 385 | 42.99 | 8.2 | 59.18 | 50.99 | −0.731 | nucl |
| IbGRF2 | 3970 | 1752 | 583 | 62.20 | 7.01 | 46.43 | 63.09 | −0.555 | nucl |
| IbGRF3 | 2902 | 1542 | 513 | 54.62 | 5.89 | 60.57 | 55.59 | −0.567 | nucl |
| IbGRF4 | 3050 | 1584 | 527 | 56.49 | 9.11 | 58.57 | 56.51 | −0.559 | nucl |
| IbGRF5 | 2444 | 1296 | 431 | 48.02 | 9.22 | 53.28 | 54.94 | −0.762 | nucl |
| IbGRF6 | 4991 | 1383 | 460 | 49.72 | 8.77 | 54.66 | 65.78 | −0.491 | nucl |
| IbGRF7 | 4380 | 3672 | 611 | 64.87 | 8.47 | 49.88 | 62.52 | −0.473 | nucl |
| IbGRF8 | 7869 | 1605 | 534 | 57.54 | 8.57 | 45.99 | 61.63 | −0.556 | nucl |
| IbGRF9 | 2473 | 1104 | 367 | 39.79 | 8.4 | 55.87 | 59.13 | −0.633 | nucl |
| IbGRF10 | 5407 | 1071 | 356 | 39.16 | 8.43 | 57.69 | 50.67 | −0.697 | nucl |
| ItfGRF1 | 3184 | 1605 | 534 | 57.51 | 8.72 | 45.53 | 60.54 | −0.578 | nucl |
| ItfGRF2 | 3321 | 1098 | 365 | 40.78 | 8.52 | 58.11 | 49.51 | −0.767 | nucl |
| ItfGRF3 | 2756 | 1560 | 519 | 55.58 | 8.78 | 58.89 | 56.99 | −0.551 | nucl |
| ItfGRF4 | 2266 | 1095 | 364 | 40.57 | 7.29 | 54.87 | 51.46 | −0.887 | nucl |
| ItfGRF5 | 4507 | 1065 | 354 | 38.95 | 8.43 | 57.24 | 50.96 | −0.678 | nucl |
| ItfGRF6 | 3116 | 957 | 318 | 35.42 | 8.39 | 57.17 | 48.46 | −0.772 | nucl |
| ItfGRF7 | 3339 | 1815 | 604 | 64.02 | 8.47 | 49.99 | 62.12 | −0.466 | nucl |
| ItfGRF8 | 2279 | 1098 | 365 | 39.53 | 7.85 | 54.56 | 57.56 | −0.653 | nucl |
| ItfGRF9 | 2009 | 1254 | 417 | 45.05 | 8.87 | 54.31 | 55.73 | −0.653 | nucl |
| ItfGRF10 | 1979 | 1302 | 433 | 48.28 | 9.1 | 55.21 | 54.92 | −0.748 | nucl |
| ItfGRF11 | 3318 | 1722 | 573 | 61.40 | 6.99 | 43.62 | 64.35 | −0.541 | nucl |
| ItfGRF12 | 2138 | 1527 | 508 | 54.42 | 5.89 | 62.52 | 55.55 | −0.6 | nucl |
| ItbGRF1 | 3355 | 1605 | 534 | 57.61 | 8.72 | 45.88 | 61.44 | −0.556 | nucl |
| ItbGRF2 | 2741 | 1578 | 525 | 56.50 | 8.98 | 58.43 | 57.09 | −0.565 | nucl |
| ItbGRF3 | 2707 | 1158 | 385 | 42.89 | 8.46 | 57.02 | 52.23 | −0.722 | nucl |
| ItbGRF4 | 2440 | 1083 | 360 | 40.26 | 7.29 | 53.61 | 52.03 | −0.891 | nucl |
| ItbGRF5 | 4623 | 960 | 319 | 35.24 | 8.65 | 59.88 | 51.66 | −0.706 | nucl |
| ItbGRF6 | 3487 | 957 | 318 | 35.36 | 8.4 | 55.98 | 49.69 | −0.749 | nucl |
| ItbGRF7 | 3626 | 1830 | 609 | 64.63 | 8.47 | 49.76 | 62.09 | −0.469 | nucl |
| ItbGRF8 | 3701 | 1278 | 425 | 46.31 | 8.24 | 58.5 | 59.55 | −0.682 | nucl |
| ItbGRF9 | 2213 | 1263 | 420 | 45.38 | 8.83 | 54.27 | 55.31 | −0.663 | nucl |
| ItbGRF10 | 2640 | 1317 | 438 | 48.68 | 9.22 | 49.34 | 59.38 | −0.658 | nucl |
| ItbGRF11 | 3394 | 1749 | 582 | 62.17 | 6.79 | 45.9 | 62.85 | −0.551 | nucl |
| ItbGRF12 | 2075 | 1170 | 389 | 42.70 | 6.26 | 67.38 | 59.2 | −0.656 | nucl |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, W.; Lin, X.; Li, Z.; Mai, J.; Hu, M.; Zhu, H. Genome-Wide Identification and Expression Analysis of Growth-Regulating Factor Family in Sweet Potato and Its Two Relatives. Genes 2024, 15, 1064. https://doi.org/10.3390/genes15081064
Huang W, Lin X, Li Z, Mai J, Hu M, Zhu H. Genome-Wide Identification and Expression Analysis of Growth-Regulating Factor Family in Sweet Potato and Its Two Relatives. Genes. 2024; 15(8):1064. https://doi.org/10.3390/genes15081064
Chicago/Turabian StyleHuang, Wenhui, Xiongjian Lin, Zhenqin Li, Jinglin Mai, Mengqin Hu, and Hongbo Zhu. 2024. "Genome-Wide Identification and Expression Analysis of Growth-Regulating Factor Family in Sweet Potato and Its Two Relatives" Genes 15, no. 8: 1064. https://doi.org/10.3390/genes15081064
APA StyleHuang, W., Lin, X., Li, Z., Mai, J., Hu, M., & Zhu, H. (2024). Genome-Wide Identification and Expression Analysis of Growth-Regulating Factor Family in Sweet Potato and Its Two Relatives. Genes, 15(8), 1064. https://doi.org/10.3390/genes15081064





