Highlights
- RNA-seq analysis revealed different aspects of the radiotherapy (RT) approach to canine oral melanoma (COM) according to tissue type. In biopsies, it was possible to appreciate the effect on DNA damage and the cell cycle. Conversely, glycosylation and cell adhesion regulation were observed in blood samples.
- Canonical and non-canonical WNT pathways seemed to be associated with overall survival in COM.
- Tumor microenvironment (TME) composition appeared to have a pivotal role in COM progression and response to RT.
Abstract
Oral melanoma (OM) is the most common malignant oral tumour among dogs and shares similarities with human mucosal melanoma (HMM), validating the role of canine species as an immunocompetent model for cancer research. In both humans and dogs, the prognosis is poor and radiotherapy (RT) represents a cornerstone in the management of this tumour, either as an adjuvant or a palliative treatment. In this study, by means of RNA-seq, the effect of RT weekly fractionated in 9 Gray (Gy), up to a total dose of 36 Gy (4 weeks), was evaluated in eight dogs affected by OM. Furthermore, possible transcriptomic differences in blood and biopsies that might be associated with a longer overall survival (OS) were investigated. The immune response, glycosylation, cell adhesion, and cell cycle were the most affected pathways by RT, while tumour microenvironment (TME) composition and canonical and non-canonical WNT pathways appeared to be modulated in association with OS. Taking these results as a whole, this study improved our understanding of the local and systemic effect of RT, reinforcing the pivotal role of anti-tumour immunity in the control of canine oral melanoma (COM).
1. Introduction
Malignant melanoma is a relatively common cancer of canines, with a high local invasiveness and metastatic propensity, accounting for about 100,000 diagnoses/year in the USA. It most frequently occurs in the oral cavity (COM), hairy skin (cutaneous melanoma), nailbed epithelium, and footpad (subungual and acral melanoma), as well as in ocular tissue (uveal melanoma). The mean age of canines at the diagnosis of malignant melanoma is 11.6 years. Certain canine breeds are more likely to develop melanoma, e.g., Poodles, Beauce Shepherds, Rottweilers, Schnauzers, Scottish Terriers, and Labrador Retrievers; furthermore, black-coated breeds are usually over-represented when compared to white-coated ones. The presence of a protective hair coat suggests that UV does not play a significant role in melanocytic melanoma [1,2,3,4].
As said, COM is the most common oral malignancy in dogs; the most frequent localization is the gingiva, but it can be found in the lip, tongue, and palate. It can be heavily pigmented or amelanotic, ulcerated, and necrotic. Overall, it is an aggressive malignant tumour, locally invasive and highly metastatic, with regional lymph nodes and lungs as preferential metastatic sites [5]. The prognosis is poor, with a median survival time of roughly 65 days in the absence of therapy [6]. As to therapy, the primary conventional treatment is a wide-margin surgery resection, combined (adjuvant) or not (palliative) with radiation therapy (RT), to locally control the tumour. However, such a therapeutic approach is much less successful for metastatic disease [5,6,7]. The use of conventional anticancer drugs is still controversial. For example, platinum compounds and melphalan, administered alone or as adjuvant chemotherapeutics following surgery or RT, show low response rates and did not improve survival times [6,8]. Similarly, unclear and underwhelming results are reported in humans, too [6]. Temozolomide (TEM) deserves a separate discussion. This orally available DNA alkylating prodrug is believed to be a radiosensitizer, and it represents the gold standard for the treatment of human glioblastoma and metastatic melanoma [9,10,11,12]. In canines, TEM shows some anti-tumour activity against melanoma and glioma [13,14]. As classical clinical management approaches are challenging, various immunotherapy strategies such as vaccination (Oncept™, a bacterial plasmid DNA vaccine against human tyrosinase) and monoclonal antibodies (e.g., targeting programmed cell death-1, PD-1) have been used in the therapy of COM. However, controversial or not yet definitive results have been observed. Hence, additional research on the feasibility of different immunotherapy approaches in COM is needed [6,15,16].
Rodent (murine) models classically represent the gold standard for preclinical research in cancer biology and therapeutics; however, they possess some relevant shortcomings and limitations [2,3,8,17,18,19]. Therefore, the need for additional animal models has become of increasing interest within the scientific community. To address this issue, the term ‘comparative oncology’, i.e., the study of naturally occurring cancers in animals as models for human disease, was coined and such a concept represents one of the clearest examples of the “One Health” approach to diseases [8,20,21]. After thirty years, it is a well-established concept that pets, and particularly canines, represent a unique spontaneous animal model for human cancer research [8,17,18,19,22]. COM is among the most common canine malignancies, and today it is considered a clinically faithful animal model for HMM [1,2,3,4,8,18,19,23,24]. At present, there is still a lack of understanding of HMM at the molecular level [25], and understanding the molecular landscapes of cancer is of paramount importance to facilitate early diagnoses and prognoses, develop and optimize effective therapeutic strategies, and improve the patient outcome [19,25,26,27]. As a consequence, obtaining additional information about COM molecular biology and therapy is currently needed and encouraged [8,19,22,28].
In comparative oncology, genomics underwent rapid growth in the past twenty years, particularly with the completion of the first drafts of human and canine genomes in 2003 and 2005, respectively [21,29,30]. Despite the size difference (the canine genome is ~20% smaller than the human one), human and canine genomes are highly homologous (~85%); furthermore (and interestingly), a higher homology exists among the human top 100 genes most frequently mutated in cancer [31]. At present, more complete assemblies of human and canine genomes have been released by the human Genome Reference Consortium and the International Consortium of Canine Genome Sequencing (Dog10K) [32,33].
Radiation therapy is one of the most common approaches to cancer treatment. Either alone or in combination with other therapeutic options (e.g., surgery, anticancer drugs), RT is used in at least two-thirds of cancer treatment regimens, and approximately half of all cancer patients worldwide have undergone RT [34,35,36]. Furthermore, RT is an important curative option to control uncomplicated loco-regional or unresectable/locally advanced tumours, including HMM and COM, too [13,37,38,39]. The RT dogma “One-Size-Fits-All” assumes that every patient has the same opportunity to benefit from RT [35]. However, with such an approach, RT effectiveness is limited by the maximum dose tolerated by adjacent normal tissues, which may result in radiotoxicity [34,36]. Nowadays, the major goal of RT is to maximize cancer cell killing by using dosage regimens that adjacent healthy tissues can tolerate, with minor radiation injury [36]. To achieve this goal, radiation sensitizers (i.e., drugs responsive to ionizing radiation and enhancing RT effectiveness such as TEM and gemcitabine) have been used in the RT–drug combined treatment of human and canine glioblastoma and metastatic melanoma [9,10,11,13,14,40]. All this allowed RT to evolve from the aforementioned dogma to a more dynamic and patient-tailored therapeutic approach [34,36,41,42].
The sequencing of human and canine genomes paved the way for the era of precision or personalized medicine (PM), whose motto is “the right treatment for the right patient at the right time” [43]. In such a context, next-generation sequencing (NGS) methodologies (i.e., genomics, transcriptomics, epigenomics) are increasingly used in oncology to speed up cancer early diagnoses, discover therapeutic targets, and predict the patient outcome, ultimately improving the PM approach to cancer [44,45,46,47]. Conversely, and surprisingly enough, NGS has not yet similarly affected RT [35]. Thus, the concept of personalized radiation therapy, to guide exclusive RT and/or combination therapy, has emerged and has been encouraged in recent years [44,48,49]. By using NGS datasets, radiation oncologists could identify biomarkers (e.g., gene expression profiles) useful to classify radio-sensitive/resistant tumours and/or tumour-surrounding normal tissues before the treatment [36,44,50]. Gene signatures, useful to identify radiosensitive patients and predict the diagnosis, prognosis, or response to RT, have been developed and validated in some human tumours, e.g., glioblastoma and breast and colorectal cancer cells. To achieve this goal, most studies used candidate gene biomarkers or functional assays of DNA damage repair to predict radiosensitivity. However, few genetic biomarkers are being used to tailor human cancer RT [48,49,50].
To the best of our knowledge, there is still no effective radiosensitive gene signature for COM; only one paper matching whole exome and transcriptome sequencing to identify the possible causes of RT and adjuvant chemotherapy failure has been published so far [51]. Therefore, in this study, we utilized an NGS approach (i.e., RNA-seq) to evaluate the transcriptomic landscape of COM in buccal biopsies before and after RT, and its association with the disease outcome.
In addition, there is an increasing demand for molecular tools to provide clinically valuable information about patient outcomes and successful delivery of PM [52]. In such a context, peripheral blood testing is an alternative approach to invasive needle-based biopsy sampling, especially for tumours that are anatomically difficult to be sampled (such as HMM and COM). Blood sampling is also routinely performed in the clinic and is affordable [52,53,54]. As to COM, to our knowledge, there are no biomarkers in routine clinical use to monitor the canine response to RT; hence, we also investigated the usefulness of a peripheral blood transcriptome as a suitable surrogate tissue for COM.
2. Materials and Methods
2.1. Animal Recruitment, Clinico-Pathological Features, Therapy, and Sample Acquisition
This study included client-owned dogs affected by COM and presented at the Centro Oncologico Veterinario (Sasso Marconi, Bologna, Italy). For each dog, data collected included sex, age, breed, tumour localization, and size as well as tumour-node metastasis (TNM) staging, according to WHO classification [23].
For all cases, all owners signed written informed consent for the use of the biological samples for research purposes. Approval by an ethics committee was not required, since the research did not influence any diagnostic or therapeutic decision.
During surgery, dogs underwent a surgical biopsy (TruCut or Punch), and the specimen (T0) was divided into two aliquots: The first one (~100 mg; T0) was placed in Eppendorf tubes filled with 1.8 mL RNAlater™ Stabilizing Solution (Life Technologies, Foster City, CA, USA) and conserved at −20 °C until the analysis. The second aliquot was formalin-fixed (10% buffered formalin) and paraffin-embedded for routine histological and immunohistochemical investigations. A second biopsy (T1) was collected at the end of the hypofractionated RT; once placed in an RNA stabilizing solution, it was kept at −20 °C until RNA extraction. Simultaneously, blood samples (in duplicate) were withdrawn in PAXgene tubes (Qiagen, Milan, Italy) as per the manufacturer’s instructions. These were at first stored overnight at room temperature and then conserved at −20 °C until RNA isolation. Tissue and blood sampling procedures were kept consistent for all the samples and were obtained by the same hospital personnel.
The diagnosis of COM was made on hematoxylin and eosin-stained slides by adopting previously proposed criteria, which include the mitotic index (MI), the melanin abundance, and the nuclear atypia grading [55,56,57]. Confirmatory immunohistochemical investigations were made by using antibodies raised against Melan-A (MEL-A) and PNL-2 melanocytic markers as well as Ki-67 (the latter for prognostic purposes) [23,55,56,57]. Only cases diagnosed as COM were taken into consideration.
As to the chosen therapeutic approach, only dogs receiving RT and adjuvant chemotherapy with TEM were included in this study. Dogs were irradiated weekly with fractions of 9 Gray (Gy), up to a total dose of 36 Gy (4 weeks). Two weeks after the completion of hypofractionated RT, dogs received adjuvant chemotherapy with TEM, i.e., 100 mg/m2 orally administered once a day and for 5 consecutive days.
To assess the possible relationships between RNA-seq outputs, the treatment protocol, and the patient outcome, the OS, which is a gold standard endpoint in oncology [58], was considered. Specifically, according to the oncologist, a mortality threshold value of 6 months (before/after) was applied to divide the clinical cases into two subgroups. Therefore, all dogs were scrupulously followed up with and the disease outcome was recorded.
2.2. RNA Isolation from Biopsies and Blood Samples
RNA from biopsies was isolated using the TRIzol reagent (Life Technologies, Milan, Italy) and subsequently purified using the RNeasy Mini kit (Qiagen, Milan, Italy) as per the manufacturer’s instructions. RNA from blood samples was isolated following the protocol of the PAXgene blood RNA kit (PreAnalytics/Qiagen, Milan, Italy). A DNase treatment was performed on the column for quality assurance before RNA was eluted from the filter. RNA concentration was determined using the NanoDrop ND-1000 UV-Vis spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA), and its quality was measured by the 2100 Bioanalyzer and the RNA 6000 Nano kit (Agilent Technologies, Santa Clara, CA, USA). Only RNA samples with an RNA integrity number (RIN) ≥ 7 were selected for the RNA-seq library preparation. The isolated RNA was stored at −80 °C until further use.
2.3. RNA-Seq Library Preparation and Sequencing
Using 1 (blood samples) or 0.5 µg (biopsies) of total RNA as input, mRNA was enriched using the NEB magnetic mRNA isolation kit (New England BioLabs, Ipswich, MA, USA); hence, RNA-seq libraries were prepared using the NEBNext Ultra RNA Library Prep Kit from Illumina (New England BioLabs, Ipswich, MA, USA) as per the manufacturer’s specifications. The prepared libraries were purified with Agencourt Ampure XP beads (Beckman Coulter, Brea, CA, USA). Individual libraries were multiplexed together in a 6-library pool. The pooled libraries were sequenced on 50-cycle runs using an Illumina Hiseq2500 platform (Genomix4Life, Caserta, Italy).
2.4. Differential Expression Analysis and Functional Analysis
Raw reads underwent quality control with FastQC software (v.0.11.9; [59]) and low-quality reads and adapters were removed using Trimmomatic (v.0.36; [60]).
Trimmed reads were pseudoaligned to the reference canine transcriptome (ROS_Cfam_1.0, Ensembl release 111) using Kallisto (v.0.48.0; [61]). Transcripts were imported in Rstudio (R version 4.3.0) and collapsed to genes using the tximport package (v.1.30.0; [62]) retrieving the annotations from Ensembl through the R interface biomaRt (v.2.58.0; [63]).
The following steps of the differential gene expression (DGE) analysis were carried out using the edgeR package (v.4.0.3; [64]). The data were organized in four different datasets, the first two comparing T1 vs. T0 for each tissue, using a paired design, and the other two taking into consideration the OS of each subject and setting the contrast between the group with long and short survival time (LONG: >6 months, comprising 3 dogs; SHORT: <6 months, comprising 5 dogs). For these last two comparisons, one tissue at a time was considered and only samples belonging to T0 were analyzed. All the datasets were processed with the same approach as reported below. First, genes with very low expression levels were removed (filterByExpr), and the remaining ones were normalized using the calcNormFactors function according to the trimmed mean of M-values (TMM).
After common and tagwise dispersion estimation (estimateDisp), negative binomial generalized linear models were fitted (glmFit), and differentially expressed genes (DEGs) were identified using the likelihood ratio-test (glmLRT) setting these contrasts: blood T1 vs. T0 (B_T1vsT0), tumour T1 vs. T0 (T_T1vsT0), blood LONG vs. SHORT (B_OS), and tumour LONG vs. SHORT (T_OS). Differentially expressed genes were defined as those with a Benjamini–Hochberg adjusted p-value (BHp) < 0.05 and log2 fold change (lfc) > 0.58 or <−0.58 from each dataset.
Moreover, the gseGO function from the clusterProfiler package (v.4.10.0; [65]) was applied to perform the Gene Set Enrichment Analysis (GSEA). A list of genes produced by the glmLRT function was used to create the input file for this computational analysis, pre-ranking all genes according to their BHp using “1-BHp” and “-(1-BHp)” to include the direction of their expression in the analysis (up- or downregulation, respectively).
2.5. Spearman’s Correlation
All DEGs found to be modulated in LONG vs. SHORT comparison in biopsies were submitted to a Spearman nonparametric correlation analysis to check for possible correlation between transcriptional expression (log2 counts per million, logCPMs, after filtering and TMM normalization) and OS (days).
2.6. Statistical Analysis
Except for RNA-seq data, the statistical analysis was made using the GraphPad Prism software (version 9.5.1, San Diego, CA, USA).
3. Results
3.1. Clinico-Pathological Features
In a period of time between 1 September 2014 and 31 March 2016, a total of nineteen presumed COM samples were collected. Following a routine TNM staging, the assessment of MI, the presence of melanin pigment, the grading of nuclear atypia, and the immunohistochemical positivity for Melan-A, PNL-2, and Ki-67, a cohort of twelve confirmed COM-affected dogs were initially considered for this study. All the recruited dogs received RT and adjuvant chemotherapy. Following the adoption of this more stringent criteria to standardize the caseload, including the use of TEM as adjuvant chemotherapy, the number of eligible COM was reduced to eight, specifically, two mixed breeds, two Golden Retrievers, one miniature Schnauzer, one miniature Spitz, one Labrador, and one Italian Hound. Five were males, three were females, and two spayed subjects were considered, one for each sex; the age ranged between 8 and 15 years, and the weight was between 6 and 38 kg. A detailed description of clinical and morphological/immunophenotypical features is reported in Tables S1 and S2, while examples of a positive/negative presence of melanin pigment as well as of MEL-A-, PNL-2-, and ki-67-positive immunohistochemical reactions are reported in Figures S1 and S2. Furthermore, an illustrative picture showing the clinical effects of RT + TEM is shown in Figure S3.
3.2. Differential Expression Analysis and Functional Analysis
For each sample, considering both biopsies and blood, a mean of 35,090,610 raw reads were obtained and more than 24 million reads (24,754,070 reads on average) were pseudoaligned to the reference transcriptome (Tables S3 and S4).
Concerning the comparison between T1 and T0 to assess the transcriptional effect of RT, 8 and 109 DEGs were identified in tumour biopsies and blood samples, respectively (Table S5). DEGs for which a reference gene name was available are reported in Table 1 and Table 2. Forty-eight GO terms were found to be enriched for T_T1vsT0, and 14 for B_T1vsT0 (Table S6). As it is possible to appreciate from Figure 1, in tumour samples, the GO terms related to the cell cycle (GO:0007049; GO:0045786), DNA replication (GO:0006260; GO:0006275), and DNA damage response (GO:0006974) were mostly affected by RT. Such modulation is supported by genes found to be differentially expressed, like Cyclin-Dependent Kinase Inhibitor 1A (CDKN1A; lfcT_T1vsT0 = 1.03), DEAD-Box Helicase 43 (DDX43; lfcT_T1vsT0 = −6.15), Centromere Protein K (CENPK; lfcT_T1vsT0 = −1.25), and Metallothionein 1E (MT1E; lfcT_T1vsT0 = 1.05).

Table 1.
Genes modulated by radiotherapy in biopsies (T) (T1 vs. T0). Only the DEGs for which a reference gene name was available are listed. Ensembl gene ID, gene name, gene description, log2 fold change (lfc), log2 counts per million (logCPMs), and Benjamini–Hochberg adjusted p-value (BHp) are reported for all the DEGs.

Table 2.
Genes modulated by radiotherapy in blood samples (B) (T1 vs. T0). Only the DEGs for which a reference gene name was available are listed. Ensembl gene ID, gene name, gene description, log2 fold change (lfc), log2 counts per million (logCPMs), and Benjamini–Hochberg adjusted p-value (BHp) are reported.
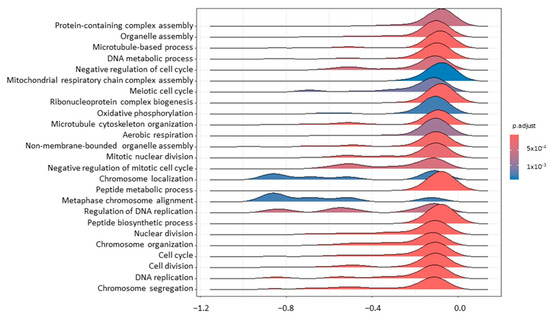
Figure 1.
A ridgeplot of the 25 most significant terms as results of GSEA comparing tumour T1 vs. T0. The colour gradient is related to the level of significance, adjusted with the Benjamini–Hochberg method.
Some of the most interesting GO terms modulated after RT in blood samples (Figure 2) were related to glycosylation (GO:0006487, GO:0070085), translation (GO:0006412), cell adhesion (GO:0007155), and wound healing (GO:0042060). Looking more in detail into the list of DEGs, several transcripts possibly linked to the immune microenvironment and radiosensitivity were noticed, e.g., C-C Motif Chemokine Receptor 7 (CCR7; lfcB_T1vsT0 = −0.89), the Interleukin 21 Receptor (IL21R; lfcB_T1vsT0 = −0.92), the Thymocyte Selection-Associated High-Mobility Group Box (TOX; lfcB_T1vsT0 = −0.81), Layilin (LAYN; lfcB_T1vsT0 = 1.01), Lymphoid Enhancer Binding Factor 1 (LEF1; lfcB_T1vsT0 = −0.9), Purinergic Receptor P2Y2 (P2RY2; lfcB_T1vsT0 = 0.78), MLX Interacting Protein-Like (MLXIPL; lfcB_T1vsT0 = 0.99), Suppressor of Cytokine Signaling 3 (SOCS3; lfcB_T1vsT0 = −0.72), and KLF Transcription Factor 10 (KLF10; lfcB_T1vsT0 = 0.79). In support of GSEA results, it is important to also mention some genes involved in cell adhesion, e.g., Integrin Subunit β 5 (ITGB5; lfcB_T1vsT0 = −0.59) and Junctional Adhesion Molecule 3 (JAM3; lfcB_T1vsT0 = −0.81).
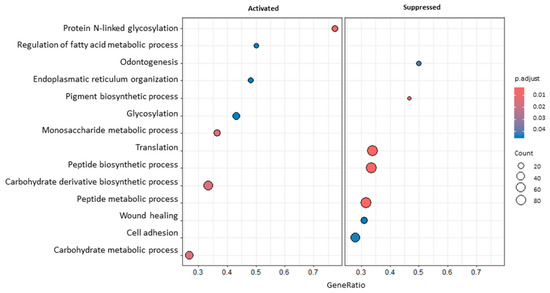
Figure 2.
A dotplot of GSEA results comparing blood T1 vs. T0. The dot size represents the number of genes belonging to each pathway. The colour gradient is related to the level of significance, adjusted with the Benjamini–Hochberg method. The box on the left collects activated pathways, while the box on the right collects suppressed ones.
In order to discern potential transcriptional features of COM specimens that might be suggestive of a prognosis at baseline, LONG vs. SHORT comparisons were considered. In this respect, T0 samples were only analyzed to avoid any biases due to the therapeutic intervention. No terms were enriched either in blood or biopsies. Nevertheless, it was possible to identify 18 and 4 DEGs according to the OS in tumour and blood tissue, respectively (Table S5). The list of DEGs for which a reference gene name was available is reported in Table 3 and Table 4. As to tumour samples, interesting upregulated genes were Keratin 76 (KRT76; lfcT_OS = 8.59); Integrin Subunit α 8 (ITGA8; lfcT_OS = 5.64); Microfibril-Associated Protein 5 (MFAP5; lfcT_OS = 4.52); Sushi, von Willebrand Factor Type A, EGF, and Pentraxin Domain Containing 1 (SVEP1; lfcT_OS = 3.71); and Sclerostin Domain Containing 1 (SOSTDC1; lfcT_OS = 4.66). Among the downregulated ones, worthy of mention is the modulation of Matrix Metallopeptidase 13 (MMP13; lfcT_OS = −4.29) and Wnt Family Member 5B (WNT5B; lfcT_OS = −2.74). Concerning blood samples, we highlight the upregulation of Transketolase-Like 1 (TKTL1; lfcB_OS = 3.04).

Table 3.
Genes modulated in biopsies (T) considering the overall survival (LONG vs. SHORT). Only the DEGs for which a reference gene name was available are listed. Ensembl gene ID, gene name, gene description, log2 fold change (lfc), log2 counts per million (logCPMs), and Benjamini–Hochberg adjusted p-value (BHp) are reported.

Table 4.
Genes modulated in blood samples (B) considering the overall survival (LONG vs. SHORT). Only the DEGs for which a reference gene name was available are listed. Ensembl gene ID, gene name, gene description, log2 fold change (lfc), log2 counts per million (logCPMs), and Benjamini–Hochberg adjusted p-value (BHp) are reported.
3.3. Spearman’s Correlation
The Spearman’s test was applied to investigate the possible correlation between the expression of DEGs regulated in T_OS and OS (Table 5). The expression of MMP13, WNT5B, SVEP1, and phospholipase A2 group IVF (PLA2G4F) was significantly correlated with the OS. As shown in Figure S4, the first and the last two genes of the abovementioned list appear to be positively and negatively correlated with the OS, respectively.

Table 5.
Table summarizing the correlation coefficients (r) and the p-values (p) of Spearman’s correlation analysis between transcriptional expression level (logCPM) of DEGs in T_OS comparison and OS (days). The p-values reported in bold are the significant ones.
4. Discussion
4.1. Transcriptomic Effect of RT on Blood and Tumour Samples
RT represents a cornerstone in the management of COM for local tumour control, through a direct action on cancer cells and immunomodulatory effects. Such an anticancer immune response could be within or outside the radiation field. Nevertheless, this abscopal activity is often incomplete and inefficient without the use of an adjuvant therapy. Canine melanoma as well as osteosarcoma are immunogenic tumours, and could represent priceless models to define possible strategies to amplify the effect of specific anticancer treatments. Just these pieces of evidence pushed us to explore more in-depth how RT could modulate the COM microenvironment and, consequently, increase the patients’ sensitivity to any adjuvant chemotherapies like TEM or to immunotherapies [66,67,68].
Starting from comparing T1 and T0 in biopsies, we could appreciate one of the well-known effects associated with RT. Indeed, radiation causes DNA damage and consequently affects the cell cycle [66]. In the present study, such an effect was confirmed by these specific enriched terms. Moreover, CDKN1A, coding for p21 protein, was found to be upregulated. Specifically, this gene is transcriptionally modulated by p53 in response to DNA damage guiding G1 cell cycle arrest [69].
Furthermore, the treatment downregulated DDX43 and CENPK. The former gene is a DEAD-box RNA-helicase family member associated with adverse clinico-pathological characteristics in breast cancer and lung adenocarcinoma [70,71,72]. Likewise, CENPK is a kinetochore protein and its overexpression in hepatocellular carcinoma promotes proliferation and in ovarian cancer is associated with a poor prognosis [73,74].
Interestingly, MT1E was positively modulated by RT. Metallothioneins are proteins playing a major role in metal ion homeostasis and detoxification. In cancer, they can act as antioxidants, protecting cells from free radicals generated by mutagens, antineoplastic drugs, and radiation [75]. Furthermore, its expression is regulated by methylation in melanoma, being a potential tumour suppressor gene [76].
After exploring the effect of RT on the cell cycle in the tumour tissue, in blood samples, it was possible to investigate other aspects of this therapeutic approach. The enrichment of glycosylation-related terms after RT aligns with what is reported in the literature. Indeed, it has been shown that ionizing radiation causes changes in protein glycosylation; in particular, such a post-translational modification could influence cell adhesion [77,78]. In this case, vasculature and more specifically the endothelium–monocyte interaction could play a pivotal role in the response to RT [78].
It is already known how the TME controls the growth and progression of cancerous tissue; therefore, the impact of RT on vascularization and immune cells could influence the prognosis and the effect of adjuvant treatments. One of the main immune subset populations targeted by RT are CD8+ T cells, and these often contribute to the abovementioned abscopal effect [79]. In line with this, in the present study, several genes possibly involved in CD8+ function and differentiation were modulated by RT, e.g., P2RY2, LAYN, and MKXIPL. Specifically, the first one is a receptor activated by ATP released passively from dead cells and acting as a pro-inflammatory signal. We could speculate that RT, causing the death of cancerous cells, leads to an ATP-driven and purigenic receptor-dependent accumulation of CD11c + CD11b + Ly6Chi tumour-infiltrating leukocytes. These cells appear to be particularly efficient at presenting tumour antigens to CD8+ T cells, according to what was previously reported by Ma et al. [80,81]. In human melanoma, LAYN is one of the most enriched genes among the phenotypically exhausted yet clonally expanded tumour-infiltrating lymphocytes. The deletion of this gene causes enhanced tumour growth and the coded protein facilitates the effector capability of cytotoxic T cells, pointing out that the amount and the specific type of CD8+ T cells present in a cancerous tissue contribute to the patient’s immune response to cancer. However, in contrast with our findings, it seems to not be present in peripheral blood cells [82]. In support of these data, and in line with a possible higher infiltration of CD8+ T cells, MLXIPL was also upregulated by RT, despite its low constitutive expression level. This gene has a multifaceted role depending on the type of tumour and in prostate cancer, its expression is induced by T cell infiltration [83]. Furthermore, an additional sign of the modulation of CD8+ population function comes from the downregulation of IL21R, LEF1, TOX, and CCR7 genes [84,85,86].
Other genes modulated by RT in blood samples and worthy to be cited are SOCS3 and KLF10. The first one, found to be inhibited in the present work, is supposed to play a role in radioresistance; indeed, the silencing of this gene affects radioresistance in glioblastoma cell lines [87]. Intriguingly, it is inducible by different interleukins (e.g., IL-21) through the JAK/STAT pathway, on which it acts as a negative feedback regulator [88]. KLF10, upregulated by RT, acts as a tumour suppressor gene in several cancers through the TGF-β signalling pathway [89]. Furthermore, like SOCS3, it is considered a possible marker of radiosensitivity in pancreatic adenocarcinoma, transcriptionally suppressing the UV radiation resistance-associated gene (UVRAG) and modulating apoptosis, DNA repair, and autophagy [90].
Another aspect that emerged from our analysis is the impact on cell adhesion molecules. The JAM3 and ITGB5 genes were downregulated by RT and the adhesion-related pathways were enriched, too. In a former study, ITGB5 was found to be differentially expressed in the NCI-60 cancer cell panel, thus suggesting that adhesion molecules could have a major role in radiosensitivity [91]. The mRNA levels of JAM3 are higher in metastatic malignant melanomas and its expression is correlated with invasive properties and metastatic potential either in fibrosarcoma or melanoma and also in bladder cancer cell lines [92,93,94,95]. Additionally, it has been demonstrated that the occurrence of lung metastasis of melanoma could be partially mediated by the interaction between jam-b (JAM2) and jam-c (JAM3) [96]. Finally, JAM3 is negatively associated with CD8+ cell infiltration in bladder cancer samples [95], while integrins significantly correlate with immune cell infiltration in skin cutaneous melanoma [97], bringing our discussion back to antineoplastic immunity, the leitmotif of these results.
4.2. Transcriptomic Differences Associated with Overall Survival Time
To identify potential biomarkers of aggressiveness, which could be related to OS, the transcriptome of subjects with long vs. short OS was compared. Some hints suggest that TME is probably one of the main factors affecting the OS. As a matter of fact, it comprises immune and stromal cells, the extracellular matrix, and blood and lymphatic vessels, and plays a critical role in survival and therapeutic response [98]. Two possible markers of cancer-associated fibroblasts (CAFs), MFAP5 and SVEP1, were upregulated in biopsies, and the expression of the last one was positively correlated with a longer OS. Cancer-associated fibroblasts belong to an abundant and heterogeneous cell population of tumour stroma possessing several pro-tumour and tumour-suppressing functions, according to a specific cell subpopulation. In skin cancers, CAFs express different markers and in advanced melanoma, they could be involved in resistance to immunotherapy and BRAF inhibitors [98,99,100]. In a former study, MFAP5 and SVEP1 have been related to a subset of CAFs with a strong extracellular matrix signature in a mouse model of MMTV-PyMT (mouse mammary tumour virus–polyoma middle tumour antigen) [101]. The first gene (MFAP5) is a microfibril-associated glycoprotein involved in the deposition of elastic microfibrils. Its dysregulation seems to assume different meanings according to the compartment of the tumour involved (epithelial or stromal part) and the tumour type taken into consideration. Overexpression in the epithelial counterpart of this gene in head and neck squamous cell carcinoma, ovarian, and breast cancer is related to a worse prognosis [102,103,104]. Moreover, in ovarian cancer, there is a stromal upregulation of its coded protein (microfibrillar-associated protein 5), being associated with poor survival [105]. However, contradictory results have been published, too: a significant reduction in MFAP5/mfap5 was noticed in the stroma of prostate cancer, invasive colonic cancer, and gallbladder adenocarcinoma [106,107,108]. On the other hand, SVEP1 is an adhesion molecule. In hepatocellular carcinoma, its downregulation leads to a poor prognosis and early recurrence, correlating negatively with cancer stem cell markers (i.e., CD44, CD133) [109,110]. In line with this evidence, in intrahepatic cholangiocarcinoma the level of SVEP1 dramatically decreases in tumour tissue if compared with the adjacent one, thus correlating with abnormal neovascularization [111]. In the present study, the expression of this gene was positively correlated with OS (p < 0.0004), thus confirming the studies cited above. On the other hand, an upregulation of SVEP1 in CAFs compared to normal fibroblasts, and its involvement in chemoresistance to gemcitabine and cisplatin, has been recently described in urothelial bladder cancer [112]. Nevertheless, it should be emphasized that the present study considered entire tumour samples including both parenchyma and stroma; thus, it is not possible to attribute these transcriptomic changes specifically to one of the two compartments. To propose a definite conclusion, a more in-depth analysis on a homogeneous cell population should be performed.
Also, other genes associated with metastasis and cancer progression were differentially regulated in tumour tissue, i.e., MMP13, WNT5B, and SOSTDC1. Long OS was negatively correlated with MMP13 and WNT5B, an interstitial collagenase and a WNT ligand, respectively. In line with this, in several human cancers including melanoma, MMP13 is associated with metastasis and poor survival. Intriguingly, in melanoma cell lines this gene enhances invasiveness and proliferation, surprisingly limiting vasculogenic mimicry through the degradation of vascular endothelial cadherin (VE-cadherin or cadherin-5), probably leading to the release of β-catenin in the cytoplasm and nucleus [113,114,115]. Also, WNT5B may decrease VE-cadherin expression, having a pro-metastatic role and causing functional and transcriptional changes in lymphatic endothelial cells in oral squamous cell carcinoma (OSCC) and melanoma [116,117]. In addition, its expression correlates with OS in osteosarcoma, hepatocellular carcinoma, and breast cancer [118,119,120]. An antagonist of the WNT/b-catenin pathway, i.e., SOSTDC1, was upregulated. SOSTDC1 restrains the proliferative ability by promoting the apoptotic rate in acute myeloid leukemia [121]. Its downregulation is related to a poor prognosis in several cancer types, e.g., breast and gastric cancer [122]. Overall, this could let us speculate that the canonical and non-canonical WNT pathways could be involved in determining the OS in COM.
In the biopsies belonging to the group with longer OS, it is interesting to highlight the upregulation of genes like KRT76 and ITGA8. The first one, encoding for a structural protein and in agreement with other authors’ opinion, is the most significantly downregulated gene in human OSCC, strongly correlating with a poor prognosis [123]; moreover, it possesses an immunomodulatory role, too [124]. On the other hand, according to the published literature, the level of expression of ITGA8 is considered to be closely related to metastasis in skin cutaneous melanoma and also correlated with CD8+ infiltration [97]. This gene belongs to the integrin family of transmembrane cell surface receptors, and it is retained to be involved in the process of carcinogenesis in different and sometimes opposite ways, according also to the type of cancer. In ER-positive breast and bladder cancers, it seems to be regulated by methylation, and its consequent lower expression is associated with adverse OS [125].
Finally, taking into consideration the results related to OS in blood samples, TKTL1 was induced in subjects with longer OS. This gene codes for a homodimeric transketolase, generally overexpressed in cancer cells for the acquisition of a glycolytic phenotype (the Warburg effect); in melanoma, it was proved to be regulated by methylation in vitro [126]. However, apparently in contrast with what emerged in our study, in which a higher expression of TKTL1 seemed to be associated with longer OS, this gene could be considered a marker of a poor prognosis in different types of cancers [127].
5. Conclusions
Drawing some of the main lines of the discussion, it was possible to speculate a possible role of TME in COM progression and response to RT, starting from the modulation of the immune response after the ionizing treatment and up to the role of different TME populations in OS. Noteworthily, the glycosylation, cell adhesion, and cell cycle also appeared to be involved in the response to RT, while the canonical and non-canonical WNT pathways could be essential factors in determining OS. In any case, it is fundamental to highlight that the hypotheses of this study are based on a limited number of subjects, even if characterized by a homogeneous therapeutic approach. Evaluations on a wider dataset of samples should be performed to confirm these results and other types of analyses could be considered to corroborate them. For example, the involvement of different immune populations and the expression peculiarities of normal/cancerous melanocytes and the stroma could be examined, taking into consideration the mutational burden of each tumour.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes15081065/s1, Table S1: Clinical data of dogs included in this study; Table S2: Histological and immunohistochemical parameters evaluated for each COM enrolled in this study; Table S3: Sequencing and mapping results of blood samples; Table S4: Sequencing and mapping results of tumour samples; Table S5: Differential gene expression analysis results; Table S6: GSEA results; Figure S1: Histopathology of a melanotic and amelanotic melanoma; Figure S2: Immunohistochemistry of Melan-A and Ki-67; Figure S3: Pictures of a patient with oral melanoma before and after radiation and temozolomide therapy; Figure S4: Dotplots representing Spearman’s correlation between SVEP1, MMP13, WNT5B, PLA2GA7, and OS.
Author Contributions
Conceptualization, G.M., M.G., M.D. and A.G.; methodology, G.M., R.E., M.G., P.L. and V.F.L.; software, G.M., L.M., R.E. and M.P.; validation, G.M., R.E. and M.P.; formal analysis, G.M., L.M., R.E. and M.P.; investigation, G.M., R.E., P.L. and V.F.L.; resources, P.L., V.F.L., M.D. and A.G.; data curation, G.M. and R.E.; writing—original draft preparation, G.M., L.M., R.E. and M.D.; writing—review and editing, G.M., L.M., M.G., P.L., M.P., F.M., M.V., V.F.L., M.D. and A.G.; visualization, G.M., R.E., M.P. and M.D.; supervision, M.G. and M.D.; project administration, M.G. and A.G.; funding acquisition, M.G. and A.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by (i) a grant from the Italian Ministry of Health, grant number IZSVe 11/13 RC “Genetic characterization of canine oral melanoma”, to A.G.; (ii) a grant from the University of Padua, grant number DOR1877477 “Molecular characterization (RNA-seq) of canine oral melanoma”, to M.G.; (iii) PhD scholarships from Fatima Al Fihri Erasmus Mundus Scholarship Program, Action 2, EMA2, Lot 1, and the Italian Association for the Research on Cancer (AIRC, IG 2021) to R.E. and G.M., respectively.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Raw Illumina sequencing data have been deposited in GenBank (SRA) under the BioProject accession PRJNA421895.
Acknowledgments
Authors are very grateful to Luca Aresu and Laura Marconato for their helpful contribution and technical support to clinico-pathological assessment of the case load.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Gillard, M.; Cadieu, E.; De Brito, C.; Abadie, J.; Vergier, B.; Devauchelle, P.; Degorce, F.; Dréano, S.; Primot, A.; Dorso, L.; et al. Naturally Occurring Melanomas in Dogs as Models for Non-UV Pathways of Human Melanomas. Pigment. Cell Melanoma Res. 2014, 27, 90–102. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, B.; Adissu, H.A.; Wei, B.R.; Michael, H.T.; Merlino, G.; Simpson, R.M. Naturally Occurring Canine Melanoma as a Predictive Comparative Oncology Model for Human Mucosal and Other Triple Wild-Type Melanomas. Int. J. Mol. Sci. 2018, 19, 394. [Google Scholar] [CrossRef] [PubMed]
- van der Weyden, L.; Brenn, T.; Patton, E.E.; Wood, G.A.; Adams, D.J. Spontaneously Occurring Melanoma in Animals and Their Relevance to Human Melanoma. J. Pathol. 2020, 252, 4–21. [Google Scholar] [CrossRef] [PubMed]
- van der Weyden, L.; Patton, E.E.; Wood, G.A.; Foote, A.K.; Brenn, T.; Arends, M.J.; Adams, D.J. Cross-species Models of Human Melanoma. J. Pathol. 2016, 238, 152–165. [Google Scholar] [CrossRef] [PubMed]
- Tuohy, J.L.; Selmic, L.E.; Worley, D.R.; Ehrhart, N.P.; Withrow, S.J. Outcome Following Curative-Intent Surgery for Oral Melanoma in Dogs: 70 Cases (1998–2011). J. Am. Vet. Med. Assoc. 2014, 245, 1266–1273. [Google Scholar] [CrossRef]
- Kim, W.S.; Vinayak, A.; Powers, B. Comparative Review of Malignant Melanoma and Histologically Well-Differentiated Melanocytic Neoplasm in the Oral Cavity of Dogs. Vet. Sci. 2021, 8, 261. [Google Scholar] [CrossRef]
- Fonseca-Alves, C.E.; Ferreira, Ê.; de Oliveira Massoco, C.; Strauss, B.E.; Fávaro, W.J.; Durán, N.; Oyafuso da Cruz, N.; dos Santos Cunha, S.C.; Castro, J.L.C.; Rangel, M.M.M.; et al. Current Status of Canine Melanoma Diagnosis and Therapy: Report from a Colloquium on Canine Melanoma Organized by ABROVET (Brazilian Association of Veterinary Oncology). Front. Vet. Sci. 2021, 8, 707025. [Google Scholar] [CrossRef] [PubMed]
- Di Palma, S.; McConnell, A.; Verganti, S.; Starkey, M. Review on Canine Oral Melanoma: An Undervalued Authentic Genetic Model of Human Oral Melanoma? Vet. Pathol. 2021, 58, 881–889. [Google Scholar] [CrossRef] [PubMed]
- Kaina, B.; Beltzig, L.; Strik, H. Temozolomide—Just a Radiosensitizer? Front. Oncol. 2022, 12, 912821. [Google Scholar] [CrossRef]
- Marconato, L.; Rohrer Bley, C.; Leone, V.F.; Finotello, R. An Open-Label Dose Escalation Study Evaluating Tolerability and Safety of a Single 5-Days Course of Temozolomide in Dogs with Advanced Cancer. Vet. Comp. Oncol. 2020, 18, 838–842. [Google Scholar] [CrossRef]
- McAleavey, P.G.; Walls, G.M.; Chalmers, A.J. Radiotherapy-Drug Combinations in the Treatment of Glioblastoma: A Brief Review. CNS Oncol. 2022, 11, CNS86. [Google Scholar] [CrossRef] [PubMed]
- Tas, F.; Erturk, K. Single-Agent Temozolomide May Be an Effective Option for Late Adjuvant Therapy in Patients with Melanoma. J. Oncol. Pharm. Pract. 2021, 27, 40–45. [Google Scholar] [CrossRef]
- Cancedda, S.; Rohrer Bley, C.; Aresu, L.; Dacasto, M.; Leone, V.F.; Pizzoni, S.; Gracis, M.; Marconato, L. Efficacy and Side Effects of Radiation Therapy in Comparison with Radiation Therapy and Temozolomide in the Treatment of Measurable Canine Malignant Melanoma. Vet. Comp. Oncol. 2016, 14, e146–e157. [Google Scholar] [CrossRef]
- Dolera, M.; Malfassi, L.; Bianchi, C.; Carrara, N.; Finesso, S.; Marcarini, S.; Mazza, G.; Pavesi, S.; Sala, M.; Urso, G. Frameless Stereotactic Radiotherapy Alone and Combined with Temozolomide for Presumed Canine Gliomas. Vet. Comp. Oncol. 2018, 16, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Almela, R.M.; Ansón, A. A Review of Immunotherapeutic Strategies in Canine Malignant Melanoma. Vet. Sci. 2019, 6, 15. [Google Scholar] [CrossRef]
- Pazzi, P.; Steenkamp, G.; Rixon, A.J. Treatment of Canine Oral Melanomas: A Critical Review of the Literature. Vet. Sci. 2022, 9, 196. [Google Scholar] [CrossRef] [PubMed]
- Garden, O.A.; Volk, S.W.; Mason, N.J.; Perry, J.A. Companion Animals in Comparative Oncology: One Medicine in Action. Vet. J. 2018, 240, 6–13. [Google Scholar] [CrossRef]
- Giuliano, A. Companion Animal Model in Translational Oncology; Feline Oral Squamous Cell Carcinoma and Canine Oral Melanoma. Biology 2021, 11, 54. [Google Scholar] [CrossRef] [PubMed]
- Prouteau, A.; André, C. Canine Melanomas as Models for Human Melanomas: Clinical, Histological, and Genetic Comparison. Genes 2019, 10, 501. [Google Scholar] [CrossRef]
- Boss, M.K. Canine Comparative Oncology for Translational Radiation Research. Int. J. Radiat. Biol. 2022, 98, 496–505. [Google Scholar] [CrossRef]
- Harrison, B.M.; Loukopoulos, P. Genomics and Transcriptomics in Veterinary Oncology (Review). Oncol. Lett. 2021, 21, 336. [Google Scholar] [CrossRef] [PubMed]
- Gardner, H.L.; Fenger, J.M.; London, C.A. Dogs as a Model for Cancer. Annu. Rev. Anim. Biosci. 2016, 4, 199–222. [Google Scholar] [CrossRef] [PubMed]
- Bergman, P.J. Canine Oral Melanoma. Clin. Tech. Small Anim. Pract. 2007, 22, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Simpson, R.M.; Bastian, B.C.; Michael, H.T.; Webster, J.D.; Prasad, M.L.; Conway, C.M.; Prieto, V.M.; Gary, J.M.; Goldschmidt, M.H.; Esplin, D.G.; et al. Sporadic Naturally Occurring Melanoma in Dogs as a Preclinical Model for Human Melanoma. Pigment. Cell Melanoma Res. 2014, 27, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhang, J.; Guo, J.; Si, L.; Bai, X. Evolving Treatment Approaches to Mucosal Melanoma. Curr. Oncol. Rep. 2022, 24, 1261–1271. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Qi, Z.; Zhang, L.; Guo, J.; Si, L. Immunotherapy in Acral and Mucosal Melanoma: Current Status and Future Directions. Front. Immunol. 2021, 12, 680407. [Google Scholar] [CrossRef] [PubMed]
- Flory, A.; Kruglyak, K.M.; Tynan, J.A.; McLennan, L.M.; Rafalko, J.M.; Fiaux, P.C.; Hernandez, G.E.; Marass, F.; Nakashe, P.; Ruiz-Perez, C.A.; et al. Clinical Validation of a Next-Generation Sequencing-Based Multi-Cancer Early Detection “Liquid Biopsy” Blood Test in over 1000 Dogs Using an Independent Testing Set: The CANcer Detection in Dogs (CANDiD) Study. PLoS ONE 2022, 17, e0266623. [Google Scholar] [CrossRef]
- Prouteau, A.; Mottier, S.; Primot, A.; Cadieu, E.; Bachelot, L.; Botherel, N.; Cabillic, F.; Houel, A.; Cornevin, L.; Kergal, C.; et al. Canine Oral Melanoma Genomic and Transcriptomic Study Defines Two Molecular Subgroups with Different Therapeutical Targets. Cancers 2022, 14, 276. [Google Scholar] [CrossRef] [PubMed]
- International Human Genome Sequencing Consortium. Finishing the Euchromatic Sequence of the Human Genome. Nature 2004, 431, 931–945. [Google Scholar] [CrossRef]
- Lindblad-Toh, K.; Wade, C.M.; Mikkelsen, T.S.; Karlsson, E.K.; Jaffe, D.B.; Kamal, M.; Clamp, M.; Chang, J.L.; Kulbokas, E.J.; Zody, M.C.; et al. Genome Sequence, Comparative Analysis and Haplotype Structure of the Domestic Dog. Nature 2005, 438, 803–819. [Google Scholar] [CrossRef]
- Chibuk, J.; Flory, A.; Kruglyak, K.M.; Leibman, N.; Nahama, A.; Dharajiya, N.; van den Boom, D.; Jensen, T.J.; Friedman, J.S.; Shen, M.R.; et al. Horizons in Veterinary Precision Oncology: Fundamentals of Cancer Genomics and Applications of Liquid Biopsy for the Detection, Characterization, and Management of Cancer in Dogs. Front. Vet. Sci. 2021, 8, 664718. [Google Scholar] [CrossRef] [PubMed]
- Nurk, S.; Koren, S.; Rhie, A.; Rautiainen, M.; Bzikadze, A.V.; Mikheenko, A.; Vollger, M.R.; Altemose, N.; Uralsky, L.; Gershman, A.; et al. The Complete Sequence of a Human Genome. Science 2022, 376, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Jagannathan, V.; Hitte, C.; Kidd, J.M.; Masterson, P.; Murphy, T.D.; Emery, S.; Davis, B.; Buckley, R.M.; Liu, Y.H.; Zhang, X.Q.; et al. Dog10K_Boxer_Tasha_1.0: A Long-Read Assembly of the Dog Reference Genome. Genes 2021, 12, 847. [Google Scholar] [CrossRef]
- Pavlopoulou, A.; Bagos, P.G.; Koutsandrea, V.; Georgakilas, A.G. Molecular Determinants of Radiosensitivity in Normal and Tumor Tissue: A Bioinformatic Approach. Cancer Lett. 2017, 403, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Scott, J.G.; Berglund, A.; Schell, M.J.; Mihaylov, I.; Fulp, W.J.; Yue, B.; Welsh, E.; Caudell, J.J.; Ahmed, K.; Strom, T.S.; et al. A Genome-Based Model for Adjusting Radiotherapy Dose (GARD): A Retrospective, Cohort-Based Study. Lancet Oncol. 2017, 18, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.H.W.; Kuo, M.T. Improving Radiotherapy in Cancer Treatment: Promises and Challenges. Oncotarget 2017, 8, 62742–62758. [Google Scholar] [CrossRef] [PubMed]
- Fiorentino, A.; Gregucci, F.; Desideri, I.; Fiore, M.; Marino, L.; Errico, A.; Di Rito, A.; Borghetti, P.; Franco, P.; Greto, D.; et al. Radiation Treatment for Adult Rare Cancers: Oldest and Newest Indication. Crit. Rev. Oncol. Hematol. 2021, 159, 103228. [Google Scholar] [CrossRef] [PubMed]
- Kawabe, M.; Mori, T.; Ito, Y.; Murakami, M.; Sakai, H.; Yanai, T.; Maruo, K. Outcomes of Dogs Undergoing Radiotherapy for Treatment of Oral Malignant Melanoma: 111 Cases (2006–2012). J. Am. Vet. Med. Assoc. 2015, 247, 1146–1153. [Google Scholar] [CrossRef]
- Yde, S.S.; Sjoegren, P.; Heje, M.; Stolle, L.B. Mucosal Melanoma: A Literature Review. Curr. Oncol. Rep. 2018, 20, 28. [Google Scholar] [CrossRef]
- Lawrence, T.S.; Blackstock, A.W.; McGinn, C. The Mechanism of Action of Radiosensitization of Conventional Chemotherapeutic Agents. Semin. Radiat. Oncol. 2003, 13, 13–21. [Google Scholar] [CrossRef]
- Azria, D.; Bourgier, C.; Brengues, M. One Size Fits All: Does the Dogma Stand in Radiation Oncology? eBioMedicine 2016, 10, 19–20. [Google Scholar] [CrossRef] [PubMed]
- Dolgin, E. Using DNA, Radiation Therapy Gets Personal. Science 2016, 353, 1348–1349. [Google Scholar] [CrossRef] [PubMed]
- Mills, J.R. Precision Medicine—Right Treatment, Right Patient, Right Time, Wrong Approach? Clin. Chem. 2017, 63, 928–929. [Google Scholar] [CrossRef] [PubMed]
- Bristow, R.G.; Alexander, B.; Baumann, M.; Bratman, S.V.; Brown, J.M.; Camphausen, K.; Choyke, P.; Citrin, D.; Contessa, J.N.; Dicker, A.; et al. Combining Precision Radiotherapy with Molecular Targeting and Immunomodulatory Agents: A Guideline by the American Society for Radiation Oncology. Lancet Oncol. 2018, 19, e240–e251. [Google Scholar] [CrossRef] [PubMed]
- Guhan, S.; Klebanov, N.; Tsao, H. Melanoma Genomics: A State-of-the-art Review of Practical Clinical Applications. Br. J. Dermatol. 2021, 185, 272–281. [Google Scholar] [CrossRef]
- Hussen, B.M.; Abdullah, S.T.; Salihi, A.; Sabir, D.K.; Sidiq, K.R.; Rasul, M.F.; Hidayat, H.J.; Ghafouri-Fard, S.; Taheri, M.; Jamali, E. The Emerging Roles of NGS in Clinical Oncology and Personalized Medicine. Pathol. Res. Pract. 2022, 230, 153760. [Google Scholar] [CrossRef] [PubMed]
- Mery, B.; Vallard, A.; Rowinski, E.; Magne, N. High-Throughput Sequencing in Clinical Oncology: From Past to Present. Swiss Med. Wkly. 2019, 149, w20057. [Google Scholar] [CrossRef] [PubMed]
- Bibault, J.E.; Tinhofer, I. The Role of Next-Generation Sequencing in Tumoral Radiosensitivity Prediction. Clin. Transl. Radiat. Oncol. 2017, 3, 16–20. [Google Scholar] [CrossRef]
- Tang, Z.; Zeng, Q.; Li, Y.; Zhang, X.; Ma, J.; Suto, M.J.; Xu, B.; Yi, N. Development of a Radiosensitivity Gene Signature for Patients with Soft Tissue Sarcoma. Oncotarget 2017, 8, 27428–27439. [Google Scholar] [CrossRef]
- Tinhofer, I.; Niehr, F.; Konschak, R.; Liebs, S.; Munz, M.; Stenzinger, A.; Weichert, W.; Keilholz, U.; Budach, V. Next-Generation Sequencing: Hype and Hope for Development of Personalized Radiation Therapy? Radiat. Oncol. 2015, 10, 183. [Google Scholar] [CrossRef]
- Giannuzzi, D.; Marconato, L.; Elgendy, R.; Ferraresso, S.; Scarselli, E.; Fariselli, P.; Nicosia, A.; Pegolo, S.; Leoni, G.; Laganga, P.; et al. Longitudinal Transcriptomic and Genetic Landscape of Radiotherapy Response in Canine Melanoma. Vet. Comp. Oncol. 2019, 17, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Tivey, A.; Church, M.; Rothwell, D.; Dive, C.; Cook, N. Circulating Tumour DNA—Looking beyond the Blood. Nat. Rev. Clin. Oncol. 2022, 19, 600–612. [Google Scholar] [CrossRef]
- Yuan, D.; Zhu, K.; Dang, C.; Zheng, Y.; Yan, R.; Shi, L.; Li, K. NS5ATP9 mRNA Levels in Peripheral Blood Mononuclear Cells Predict Prognosis in Patients with Gastric Cancer. Med. Oncol. 2014, 31, 106. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Park, Y.S.; Lee, H.W.; Park, T.Y.; Lee, J.K.; Heo, E.Y. Peripheral Lymphocyte Count as a Surrogate Marker of Immune Checkpoint Inhibitor Therapy Outcomes in Patients with Non-Small-Cell Lung Cancer. Sci. Rep. 2022, 12, 626. [Google Scholar] [CrossRef] [PubMed]
- Bergin, I.L.; Smedley, R.C.; Esplin, D.G.; Spangler, W.L.; Kiupel, M. Prognostic Evaluation of Ki67 Threshold Value in Canine Oral Melanoma. Vet. Pathol. 2011, 48, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Smedley, R.C.; Spangler, W.L.; Esplin, D.G.; Kitchell, B.E.; Bergman, P.J.; Ho, H.Y.; Bergin, I.L.; Kiupel, M. Prognostic Markers for Canine Melanocytic Neoplasms: A Comparative Review of the Literature and Goals for Future Investigation. Vet. Pathol. 2011, 48, 54–72. [Google Scholar] [CrossRef]
- Spangler, W.L.; Kass, P.H. The Histologic and Epidemiologic Bases for Prognostic Considerations in Canine Melanocytic Neoplasia. Vet. Pathol. 2006, 43, 136–149. [Google Scholar] [CrossRef] [PubMed]
- Gyawali, B.; Booth, C.M. Defining Clinically Important Overall Survival Thresholds: Lessons from Quality of Life. Nat. Rev. Clin. Oncol. 2022, 19, 613–614. [Google Scholar] [CrossRef]
- Andrews, S.; Krueger, F.; Seconds-Pichon, A.; Biggins, F.; Wingett, S.F. A Quality Control Tool for High Throughput Sequence Data. Babraham Bioinformatics; Babraham Institute: Babraham, UK, 2015. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-Optimal Probabilistic RNA-Seq Quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential Analyses for RNA-Seq: Transcript-Level Estimates Improve Gene-Level Inferences. F1000Research 2015, 4, 1521. [Google Scholar] [CrossRef]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping Identifiers for the Integration of Genomic Datasets with the R/Bioconductor Package BiomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. ClusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Fan, T.M.; Selting, K.A. Exploring the Potential Utility of Pet Dogs with Cancer for Studying Radiation-Induced Immunogenic Cell Death Strategies. Front. Oncol. 2019, 8, 680. [Google Scholar] [CrossRef]
- Polton, G.; Borrego, J.F.; Clemente-Vicario, F.; Clifford, C.A.; Jagielski, D.; Kessler, M.; Kobayashi, T.; Lanore, D.; Queiroga, F.L.; Rowe, A.T.; et al. Melanoma of the Dog and Cat: Consensus and Guidelines. Front. Vet. Sci. 2024, 11, 1359426. [Google Scholar] [CrossRef]
- Hoopes, P.J.; Wagner, R.J.; Duval, K.; Kang, K.; Gladstone, D.J.; Moodie, K.L.; Crary-Burney, M.; Ariaspulido, H.; Veliz, F.A.; Steinmetz, N.F.; et al. Treatment of Canine Oral Melanoma with Nanotechnology-Based Immunotherapy and Radiation. Mol. Pharm. 2018, 15, 3717–3722. [Google Scholar] [CrossRef]
- Engeland, K. Cell Cycle Regulation: P53-P21-RB Signaling. Cell Death Differ. 2022, 29, 946–960. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Xu, H.E.; Luo, Z.; Zhou, J.; Zhou, Y.; Liu, M. Expression and Significance of DDX43 in Lung Adenocarcinoma. Pak. J. Pharm. Sci. 2017, 30, 1491–1496. [Google Scholar]
- Amer, N.N.; Khairat, R.; Hammad, A.M.; Kamel, M.M. DDX43 mRNA Expression and Protein Levels in Relation to Clinicopathological Profile of Breast Cancer. PLoS ONE 2023, 18, e0284455. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Fatah, T.M.A.; McArdle, S.E.B.; Johnson, C.; Moseley, P.M.; Ball, G.R.; Pockley, A.G.; Ellis, I.O.; Rees, R.C.; Chan, S.Y.T. HAGE (DDX43) Is a Biomarker for Poor Prognosis and a Predictor of Chemotherapy Response in Breast Cancer. Br. J. Cancer 2014, 110, 2450–2461. [Google Scholar] [CrossRef]
- Wang, H.; Liu, W.; Liu, L.; Wu, C.; Wu, W.; Zheng, J.; Zhang, M.; Chen, X.; Zhou, B.; Gao, Z.; et al. Overexpression of Centromere Protein K (CENP-K) Gene in Hepatocellular Carcinoma Promote Cell Proliferation by Activating AKT/TP53 Signal Pathway. Oncotarget 2017, 8, 73994–74005. [Google Scholar] [CrossRef]
- Lee, Y.C.; Huang, C.C.; Lin, D.Y.; Chang, W.C.; Lee, K.H. Overexpression of Centromere Protein K (CENPK) in Ovarian Cancer Is Correlated with Poor Patient Survival and Associated with Predictive and Prognostic Relevance. PeerJ 2015, 3, e1386. [Google Scholar] [CrossRef]
- Si, M.; Lang, J. The Roles of Metallothioneins in Carcinogenesis. J. Hematol. Oncol. 2018, 11, 107. [Google Scholar] [CrossRef]
- Faller, W.J.; Rafferty, M.; Hegarty, S.; Gremel, G.; Ryan, D.; Fraga, M.F.; Esteller, M.; Dervan, P.A.; Gallagher, W.M. Metallothionein 1E Is Methylated in Malignant Melanoma and Increases Sensitivity to Cisplatin-Induced Apoptosis. Melanoma Res. 2010, 20, 392–400. [Google Scholar] [CrossRef]
- Tóth, E.; Vékey, K.; Ozohanics, O.; Jeko, A.; Dominczyk, I.; Widlak, P.; Drahos, L. Changes of Protein Glycosylation in the Course of Radiotherapy. J. Pharm. Biomed. Anal. 2016, 118, 380–386. [Google Scholar] [CrossRef][Green Version]
- Jaillet, C.; Morelle, W.; Slomianny, M.C.; Paget, V.; Tarlet, G.; Buard, V.; Selbonne, S.; Caffin, F.; Rannou, E.; Martinez, P.; et al. Radiation-Induced Changes in the Glycome of Endothelial Cells with Functional Consequences. Sci. Rep. 2017, 7, 5290. [Google Scholar] [CrossRef]
- Jeon, S.H.; Song, C.; Eom, K.Y.; Kim, I.A.; Kim, J.S. Modulation of CD8+ T Cell Responses by Radiotherapy—Current Evidence and Rationale for Combination with Immune Checkpoint Inhibitors. Int. J. Mol. Sci. 2023, 24, 16691. [Google Scholar] [CrossRef]
- Ma, Y.; Adjemian, S.; Mattarollo, S.R.; Yamazaki, T.; Aymeric, L.; Yang, H.; Portela Catani, J.P.; Hannani, D.; Duret, H.; Steegh, K.; et al. Anticancer Chemotherapy-Induced Intratumoral Recruitment and Differentiation of Antigen-Presenting Cells. Immunity 2013, 38, 729–741. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Pitt, J.M.; Li, Q.; Yang, H. The Renaissance of Anti-neoplastic Immunity from Tumor Cell Demise. Immunol. Rev. 2017, 280, 194–206. [Google Scholar] [CrossRef] [PubMed]
- Mahuron, K.M.; Moreau, J.M.; Glasgow, J.E.; Boda, D.P.; Pauli, M.L.; Gouirand, V.; Panjabi, L.; Grewal, R.; Luber, J.M.; Mathur, A.N.; et al. Layilin Augments Integrin Activation to Promote Antitumor Immunity. J. Exp. Med. 2020, 217, e20192080. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Ge, Y.; Niu, K.; Li, Y.; Qi, L.W.; Zhu, H.; Ma, G. MLXIPL Associated with Tumor-Infiltrating CD8+ T Cells Is Involved in Poor Prostate Cancer Prognosis. Front. Immunol. 2024, 15, 1364329. [Google Scholar] [CrossRef] [PubMed]
- Le, P.T.; Ha, N.; Tran, N.K.; Newman, A.G.; Esselen, K.M.; Dalrymple, J.L.; Schmelz, E.M.; Bhandoola, A.; Xue, H.H.; Singh, P.B.; et al. Targeting Cbx3/HP1γ Induces LEF-1 and IL-21R to Promote Tumor-Infiltrating CD8 T-Cell Persistence. Front. Immunol. 2021, 12, 738958. [Google Scholar] [CrossRef] [PubMed]
- Meiser, P.; Knolle, M.A.; Hirschberger, A.; de Almeida, G.P.; Bayerl, F.; Lacher, S.; Pedde, A.M.; Flommersfeld, S.; Hönninger, J.; Stark, L.; et al. A Distinct Stimulatory CDC1 Subpopulation Amplifies CD8+ T Cell Responses in Tumors for Protective Anti-Cancer Immunity. Cancer Cell 2023, 41, 1498–1515. [Google Scholar] [CrossRef] [PubMed]
- Roberts, E.W.; Broz, M.L.; Binnewies, M.; Headley, M.B.; Nelson, A.E.; Wolf, D.M.; Kaisho, T.; Bogunovic, D.; Bhardwaj, N.; Krummel, M.F. Critical Role for CD103+/CD141+ Dendritic Cells Bearing CCR7 for Tumor Antigen Trafficking and Priming of T Cell Immunity in Melanoma. Cancer Cell 2016, 30, 324–336. [Google Scholar] [CrossRef] [PubMed]
- Ventero, M.P.; Fuentes-Baile, M.; Quereda, C.; Perez-Valeciano, E.; Alenda, C.; Garcia-Morales, P.; Esposito, D.; Dorado, P.; Barbera, V.M.; Saceda, M. Radiotherapy Resistance Acquisition in Glioblastoma. Role of SOCS1 and SOCS3. PLoS ONE 2019, 14, e0212581. [Google Scholar] [CrossRef]
- de Totero, D.; Capaia, M.; Fabbi, M.; Croce, M.; Meazza, R.; Cutrona, G.; Zupo, S.; Loiacono, F.; Truini, M.; Ferrarini, M.; et al. Heterogeneous Expression and Function of IL-21R and Susceptibility to IL-21-Mediated Apoptosis in Follicular Lymphoma Cells. Exp. Hematol. 2010, 38, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Memon, A.; Lee, W.K. KLF10 as a Tumor Suppressor Gene and Its TGF-β Signaling. Cancers 2018, 10, 161. [Google Scholar] [CrossRef]
- Chang, V.H.S.; Tsai, Y.C.; Tsai, Y.L.; Peng, S.L.; Chen, S.L.; Chang, T.M.; Yu, W.C.Y.; Ch’ang, H.J. Krüpple-like Factor 10 Regulates Radio-Sensitivity of Pancreatic Cancer via UV Radiation Resistance-Associated Gene. Radiother. Oncol. 2017, 122, 476–484. [Google Scholar] [CrossRef]
- Kim, H.S.; Kim, S.C.; Kim, S.J.; Park, C.H.; Jeung, H.C.; Kim, Y.B.; Ahn, J.B.; Chung, H.C.; Rha, S.Y. Identification of a Radiosensitivity Signature Using Integrative Metaanalysis of Published Microarray Data for NCI-60 Cancer Cells. BMC Genom. 2012, 13, 348. [Google Scholar] [CrossRef]
- Fuse, C.; Ishida, Y.; Hikita, T.; Asai, T.; Oku, N. Junctional Adhesion Molecule-C Promotes Metastatic Potential of HT1080 Human Fibrosarcoma. J. Biol. Chem. 2007, 282, 8276–8283. [Google Scholar] [CrossRef] [PubMed]
- Langer, H.F.; Orlova, V.V.; Xie, C.; Kaul, S.; Schneider, D.; Lonsdorf, A.S.; Fahrleitner, M.; Choi, E.Y.; Dutoit, V.; Pellegrini, M.; et al. A Novel Function of Junctional Adhesion Molecule-C in Mediating Melanoma Cell Metastasis. Cancer Res. 2011, 71, 4096–4105. [Google Scholar] [CrossRef] [PubMed]
- Ghislin, S.; Obino, D.; Middendorp, S.; Boggetto, N.; Alcaide-Loridan, C.; Deshayes, F. Junctional Adhesion Molecules Are Required for Melanoma Cell Lines Transendothelial Migration in Vitro. Pigment. Cell Melanoma Res. 2011, 24, 504–511. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Wang, J.; Wang, J.; Wang, Y.; Ji, B.; Xu, Y.; He, J.; Zhang, L.; Zhang, L.; Ding, B.; et al. JAM3: A Prognostic Biomarker for Bladder Cancer via Epithelial–Mesenchymal Transition Regulation. Biomol. Biomed. 2024, 24, 853–867. [Google Scholar] [CrossRef] [PubMed]
- Arcangeli, M.L.; Frontera, V.; Bardin, F.; Thomassin, J.; Chetaille, B.; Adams, S.; Adams, R.H.; Aurrand-Lions, M. The Junctional Adhesion Molecule-B Regulates JAM-C-dependent Melanoma Cell Metastasis. FEBS Lett. 2012, 586, 4046–4051. [Google Scholar] [CrossRef] [PubMed]
- Nurzat, Y.; Su, W.; Min, P.; Li, K.; Xu, H.; Zhang, Y. Identification of Therapeutic Targets and Prognostic Biomarkers Among Integrin Subunits in the Skin Cutaneous Melanoma Microenvironment. Front. Oncol. 2021, 11, 751875. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Shen, M.; Wu, L.; Yang, H.; Yao, Y.; Yang, Q.; Du, J.; Liu, L.; Li, Y.; Bai, Y. Stromal Cells in the Tumor Microenvironment: Accomplices of Tumor Progression? Cell Death Dis. 2023, 14, 587. [Google Scholar] [CrossRef]
- Fujimura, T. Stromal Factors as a Target for Immunotherapy in Melanoma and Non-Melanoma Skin Cancers. Int. J. Mol. Sci. 2022, 23, 4044. [Google Scholar] [CrossRef] [PubMed]
- Kennel, K.B.; Bozlar, M.; De Valk, A.F.; Greten, F.R. Cancer-Associated Fibroblasts in Inflammation and Antitumor Immunity. Clin. Cancer Res. 2023, 29, 1009–1016. [Google Scholar] [CrossRef]
- Bartoschek, M.; Oskolkov, N.; Bocci, M.; Lövrot, J.; Larsson, C.; Sommarin, M.; Madsen, C.D.; Lindgren, D.; Pekar, G.; Karlsson, G.; et al. Spatially and Functionally Distinct Subclasses of Breast Cancer-Associated Fibroblasts Revealed by Single Cell RNA Sequencing. Nat. Commun. 2018, 9, 5150. [Google Scholar] [CrossRef]
- Yang, X.; Wu, K.; Li, S.; Hu, L.; Han, J.; Zhu, D.; Tian, X.; Liu, W.; Tian, Z.; Zhong, L.; et al. MFAP5 and TNNC1: Potential Markers for Predicting Occult Cervical Lymphatic Metastasis and Prognosis in Early Stage Tongue Cancer. Oncotarget 2017, 8, 2525–2535. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wu, P.; Zhang, Q.; Chen, W.; Liu, X.; Zheng, W. MFAP5 Promotes Basal-like Breast Cancer Progression by Activating the EMT Program. Cell Biosci. 2019, 9, 24. [Google Scholar] [CrossRef] [PubMed]
- Kujawa, K.A.; Zembala-Nożynska, E.; Syrkis, J.P.; Cortez, A.J.; Kupryjańczyk, J.; Lisowska, K.M. Microfibril Associated Protein 5 (MFAP5) Is Related to Survival of Ovarian Cancer Patients but Not Useful as a Prognostic Biomarker. Int. J. Mol. Sci. 2022, 23, 15994. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.S.; Yeung, T.L.; Yip, K.P.; Pradeep, S.; Balasubramanian, L.; Liu, J.; Wong, K.K.; Mangala, L.S.; Armaiz-Pena, G.N.; Lopez-Berestein, G.; et al. Calcium-Dependent FAK/CREB/TNNC1 Signalling Mediates the Effect of Stromal MFAP5 on Ovarian Cancer Metastatic Potential. Nat. Commun. 2014, 5, 5092. [Google Scholar] [CrossRef]
- Jia, Z.; Wang, Y.; Sawyers, A.; Yao, H.; Rahmatpanah, F.; Xia, X.Q.; Xu, Q.; Pio, R.; Turan, T.; Koziol, J.A.; et al. Diagnosis of Prostate Cancer Using Differentially Expressed Genes in Stroma. Cancer Res. 2011, 71, 2476–2487. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Westerhoff, M.; Hornick, J.L.; Krausz, T.; Antic, T.; Xiao, S.Y.; Hart, J. Loss of Microfibril-Associated Protein 5 (MFAP5) Expression in Colon Cancer Stroma. Virchows Arch. 2020, 476, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Xu, L.; Hemmerich, A.; Ferguson, N.L.; Guy, C.D.; McCall, S.J.; Cardona, D.M.; Westerhoff, M.; Pai, R.K.; Xiao, S.Y.; et al. Reduced MFAP5 Expression in Stroma of Gallbladder Adenocarcinoma and Its Potential Diagnostic Utility. Virchows Arch. 2021, 478, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.C.; Han, Z.Q.; Guo, M.X.; Zhao, S.; Guo, Y.H.; Meng, B.; Sun, Y.; Chen, L. Decreased Expression of SVEP1 Is Closely Related to a Cancer Stem Cell-like Phenotype and Poor Prognosis in Hepatocellular Carcinoma. Neoplasma 2022, 69, 1209–1216. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, D.; Yi, X.; Qi, L.; Tian, X.; Sun, B.; Dong, Q.; Han, Z.; Li, Q.; Song, T.; et al. The Novel MiR-1269b-Regulated Protein SVEP1 Induces Hepatocellular Carcinoma Proliferation and Metastasis Likely through the PI3K/Akt Pathway. Cell Death Dis. 2020, 11, 320. [Google Scholar] [CrossRef]
- Chen, L.; He, Y.; Han, Z.; Gong, W.; Tian, X.; Guo, L.; Guo, H.; Song, T.; Chen, L. The Impact of Decreased Expression of SVEP1 on Abnormal Neovascularization and Poor Prognosis in Patients with Intrahepatic Cholangiocarcinoma. Front. Genet. 2023, 13, 1127753. [Google Scholar] [CrossRef]
- Zhuang, J.; Shen, L.; Li, M.; Sun, J.; Hao, J.; Li, J.; Zhu, Z.; Ge, S.; Zhang, D.; Guo, H.; et al. Cancer-Associated Fibroblast–Derived MiR-146a-5p Generates a Niche That Promotes Bladder Cancer Stemness and Chemoresistance. Cancer Res. 2023, 83, 1611–1627. [Google Scholar] [CrossRef]
- Li, S.; Pritchard, D.M.; Yu, L.G. Regulation and Function of Matrix Metalloproteinase-13 in Cancer Progression and Metastasis. Cancers 2022, 14, 3263. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Sun, B.; Li, Y.; Liu, Y.; Zhang, D.; Wang, X.; Gu, Q.; Zhao, J.; Dong, X.; Liu, Z.; et al. Dual Effects of Collagenase-3 on Melanoma: Metastasis Promotion and Disruption of Vasculogenic Mimicry. Oncotarget 2015, 6, 8890–8899. [Google Scholar] [CrossRef] [PubMed]
- Meierjohann, S.; Hufnagel, A.; Wende, E.; Kleinschmidt, M.A.; Wolf, K.; Friedl, P.; Gaubatz, S.; Schartl, M. MMP13 Mediates Cell Cycle Progression in Melanocytes and Melanoma Cells: In Vitro Studies of Migration and Proliferation. Mol. Cancer 2010, 9, 201. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.H.; Chang, J.S.; Hsiao, J.R.; Yen, Y.C.; Jiang, S.S.; Liu, S.H.; Chen, Y.L.; Shen, Y.Y.; Chang, J.Y.; Chen, Y.W. Tumour Cell-Derived WNT5B Modulates in Vitro Lymphangiogenesis via Induction of Partial Endothelial-Mesenchymal Transition of Lymphatic Endothelial Cells. Oncogene 2017, 36, 1503–1515. [Google Scholar] [CrossRef] [PubMed]
- Alve, S.; Gramolelli, S.; Jukonen, J.; Juteau, S.; Pink, A.; Manninen, A.A.; Hänninen, S.; Monto, E.; Lackman, M.H.; Carpén, O.; et al. DLL4/Notch3/WNT5B Axis Mediates Bidirectional Prometastatic Crosstalk between Melanoma and Lymphatic Endothelial Cells. JCI Insight 2024, 9, e171821. [Google Scholar] [CrossRef] [PubMed]
- Perkins, R.S.; Murray, G.; Suthon, S.; Davis, L.; Perkins, N.B.; Fletcher, L.; Bozzi, A.; Schreiber, S.L.; Lin, J.; Laxton, S.; et al. WNT5B Drives Osteosarcoma Stemness, Chemoresistance and Metastasis. Clin. Transl. Med. 2024, 14, 14. [Google Scholar] [CrossRef] [PubMed]
- Suthon, S.; Perkins, R.S.; Bryja, V.; Miranda-Carboni, G.A.; Krum, S.A. WNT5B in Physiology and Disease. Front. Cell Dev. Biol. 2021, 9, 667581. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.J.; Ying, L.; Shi, K.Q. Expression of the Wnt Ligands Gene Family and Its Relationship to Prognosis in Hepatocellular Carcinoma. Cancer Cell Int. 2019, 19, 34. [Google Scholar] [CrossRef]
- Li, C.; Wang, M.; Shi, Y.; Xin, H. SOSTDC1 Acts as a Tumor Inhibitor in Acute Myeloid Leukemia by Downregulating the Wnt/Β-catenin Pathway. Environ. Toxicol. 2022, 37, 1934–1943. [Google Scholar] [CrossRef]
- Tong, X.; Zhu, C.; Liu, L.; Huang, M.; Xu, J.; Chen, X.; Zou, J. Role of Sostdc1 in Skeletal Biology and Cancer. Front. Physiol. 2022, 13, 1029646. [Google Scholar] [CrossRef] [PubMed]
- Ambatipudi, S.; Bhosale, P.G.; Heath, E.; Pandey, M.; Kumar, G.; Kane, S.; Patil, A.; Maru, G.B.; Desai, R.S.; Watt, F.M.; et al. Downregulation of Keratin 76 Expression during Oral Carcinogenesis of Human, Hamster and Mouse. PLoS ONE 2013, 8, e70688. [Google Scholar] [CrossRef] [PubMed]
- Sequeira, I.; Neves, J.F.; Carrero, D.; Peng, Q.; Palasz, N.; Liakath-Ali, K.; Lord, G.M.; Morgan, P.R.; Lombardi, G.; Watt, F.M. Immunomodulatory Role of Keratin 76 in Oral and Gastric Cancer. Nat. Commun. 2018, 9, 3437. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, L.; Liu, L.; Ruan, D.; Wang, C. Hypermethylated ITGA8 Facilitate Bladder Cancer Cell Proliferation and Metastasis. Appl. Biochem. Biotechnol. 2024, 196, 245–260. [Google Scholar] [CrossRef] [PubMed]
- Jayachandran, A.; Lo, P.H.; Chueh, A.C.; Prithviraj, P.; Molania, R.; Davalos-Salas, M.; Anaka, M.; Walkiewicz, M.; Cebon, J.; Behren, A. Transketolase-like 1 Ectopic Expression Is Associated with DNA Hypomethylation and Induces the Warburg Effect in Melanoma Cells. BMC Cancer 2016, 16, 134. [Google Scholar] [CrossRef] [PubMed]
- Hao, S.; Meng, Q.; Sun, H.; Li, Y.; Li, Y.; Gu, L.; Liu, B.; Zhang, Y.; Zhou, H.; Xu, Z.; et al. The Role of Transketolase in Human Cancer Progression and Therapy. Biomed. Pharmacother. 2022, 154, 113607. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).