Silencing of GhSINAT5 Reduces Drought Resistance and Salt Tolerance in Cotton
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Extraction of Total Cotton RNA and Synthesis of the First Strand of cDNA
2.3. Quantitative Real-Time Polymerase Chain Reaction
2.4. Gene Cloning and Bioinformatics Analysis
2.5. VIGS and Gene Expression Level Detection
2.6. Cotton Stress Treatment
2.7. Determination of Physiological and Biochemical Indicators
2.8. Statistical Data
3. Results
3.1. Cloning and Sequence Analysis of GhSINAT5
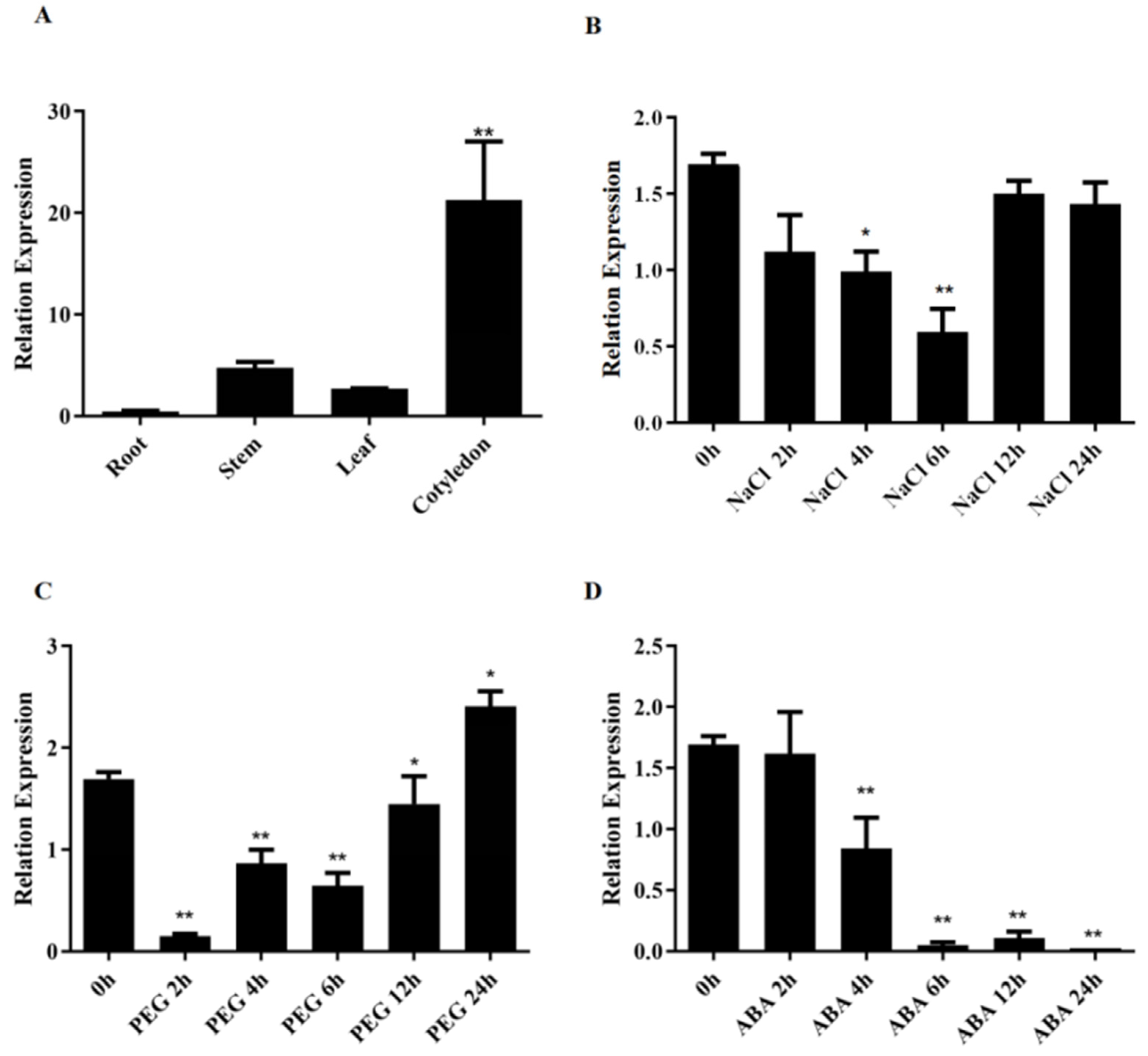
3.2. GhSINAT5 Expression Pattern Analysis
3.3. Silencing GhSINAT5 Reduces Drought Resistance in Cotton
3.4. Silencing of GhSINAT5 Increases the Extent of Membrane Injury and Decreases Antioxidant Enzyme Activity in Cotton Cells
3.5. GhSINAT5 Regulates the Expression of Key Genes Involved in Drought Stress
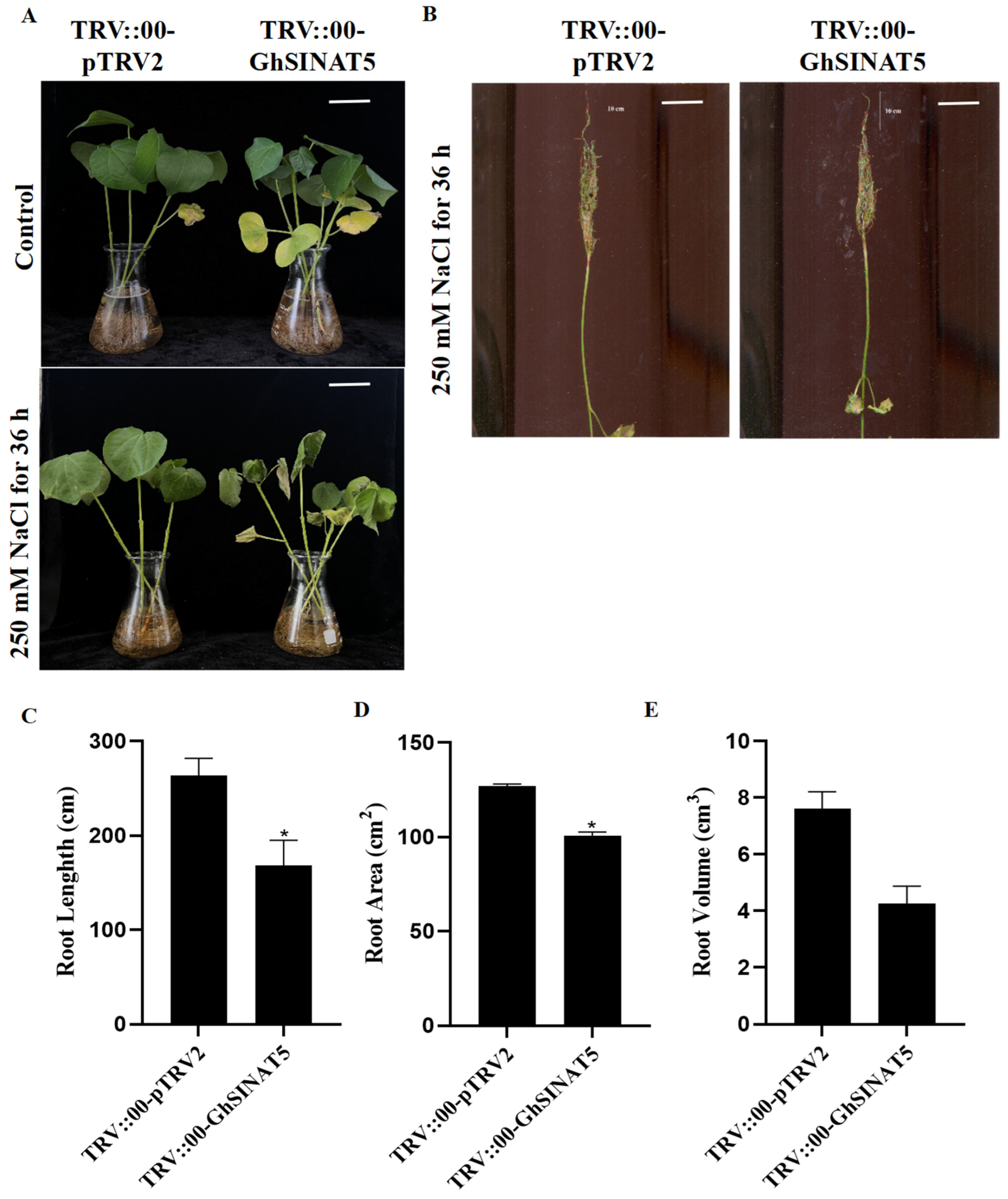
3.6. Silencing of GhSINAT5 Reduces Salt Resistance in Cotton
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vierstra, R.D. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef]
- Harper, J.W.; Schulman, B. Structural complexity in ubiquitin recognition. Cell 2006, 124, 1133–1136. [Google Scholar] [CrossRef]
- Ari, S.; Mark, B.; Richard, E.; John, L.; Stuart, N. The ubiquitin-proteasome system: Central modifier of plant signalling. New Phytol. 2012, 196, 13–28. [Google Scholar]
- Zhang, J.X.; Jiang, X.; Xiao, S.; Jiang, S.; Yao, W.; Zhang, M. Genome-wide identification of SINA gene family in sugarcane and functional analysis of SsSINA1a in drought response. bioRxiv 2022. [Google Scholar] [CrossRef]
- Chen, L.; Hellmann, H. Plant E3 ligases: Flexible enzymes in a sessile world. Mol. Plant 2013, 6, 1388–1404. [Google Scholar] [CrossRef]
- Shu, K.; Yang, W. E3 ubiquitin ligases: Ubiquitous actors in plant development and abiotic stress responses. Plant Cell Physiol. 2017, 58, 1461–1476. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Fearon, E.R. Siah-1 N-terminal RING domain is required for proteolysis function, and C-terminal sequences regulate oligomerization and binding to target proteins. Mol. Cell. Biol. 1999, 19, 724–732. [Google Scholar] [CrossRef] [PubMed]
- Carthew, R.W.; Rubin, G.M. Seven in absentia, a gene required for specification of R7 cell fate in the drosophila eye. Cell 1990, 63, 561–577. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Guo, H.S.; Dallman, G.; Fang, S.; Weissman, A.M.; Chua, N.H. SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 2002, 419, 167–170. [Google Scholar] [CrossRef]
- Wang, M.; Jin, Y.; Fu, J.; Zhu, Y.; Zheng, J.; Hu, J.; Wang, G. Genome-wide analysis of SINA family in plants and their phylogenetic relationships. DNA Seq. 2009, 19, 206–216. [Google Scholar] [CrossRef]
- Wang, W.; Fan, Y.; Niu, X.; Miao, M.; Kud, J.; Zhou, B.; Zeng, L.; Liu, Y.; Xiao, F. Functional analysis of the SEVEN IN ABSENTIA (SINA) ubiquitin ligase family in tomato. Plant Cell Environ. 2018, 4, 689–703. [Google Scholar]
- Meng, Z.H. Cloning and Preliminary Functional Analysis of GhSH1a Gene in Upland Cotton. Master’s Thesis, Shanxi University, Taiyuan, China, 2006. [Google Scholar]
- Ren, Z.; Liu, W.; Wang, X.; Chen, M.; Zhao, J.; Zhang, F.; Feng, H.; Liu, J.; Yang, D.; Ma, X.; et al. SEVEN IN ABSENTIA ubiquitin ligases positively regulate defense against verticillium dahliae in Gossypium hirsutum. Front. Plant Sci. 2021, 12, 760520. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yu, Y.; Wan, H.; Ni, Z. Sea-island cotton (Gossypium barbadense L.) GbTCP5 improves plant adaptation to drought and salt stress by directly activating GbERD7, GbUBC19, and GbGOLS2 expression. Ind. Crops Prod. 2023, 203, 117209. [Google Scholar] [CrossRef]
- Wang, Y. GhMYB4, and GbTCP5 for GbTCP4 Functional Identification of Drought and Salt Stress Response in Cotton. Ph.D. Thesis, Xinjiang Agricultural University, Urumqi, China, 2022. [Google Scholar]
- Zeng, J.C. Screening of GhMYB4 Interacting Proteins and Functional Identification of Interacting Proteins in Response to Drought and Salt Stress. Master’s Thesis, Xinjiang Agricultural University, Urumqi, China, 2023. [Google Scholar]
- Gao, X.Q.; Britt, R.C.J.; Shan, L.B.; He, P. Agrobacterium-mediated virus-induced gene silencing assay in cotton. J. Vis. Exp. 2011, 20, e2938. [Google Scholar]
- Liu, Y.; Li, L.Q.; Zhang, L.; Lv, Q.; Zhao, Y.; Li, X.J. Isolation and identification of wheat gene TaDIS1 encoding a RING finger domain protein, which negatively regulates drought stress tolerance in transgenic Arabidopsis. Plant Sci. 2018, 275, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Ning, Y.; Jantasuriyarat, C.; Zhao, Q.; Zhang, H.; Chen, S.; Liu, J.; Liu, L.; Tang, S.; Park, C.H.; Wang, X. The SINA E3 ligase OsDIS1 negatively regulates drought response in rice. Plant Physiol. 2011, 157, 242. [Google Scholar] [CrossRef] [PubMed]
- Ning, Y.; Xie, Q.; Wang, G.L. OsDIS1 mediated stress response pathway in rice. Plant Signal. Behav. 2011, 6, 1684–1686. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Wang, C.T.; Jiang, C.; Pan, J.; Zhang, G.B.; Liu, H.; Zhang, H.X. The tumor necrosis factor receptor-associated factor (TRAF)-like family protein SEVEN IN ABSENTIA 2 (SINA2) promotes drought tolerance in an ABA-dependent manner in Arabidopsis. New Phytol. 2014, 202, 174–187. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.; Zhou, W.; Liu, Y.F.; Song, Z.; Zheng, S.; Wang, F.; Lu, Z.; Qi, D.; Li, B.; Sun, N. Characterization of RING-type ubiquitin SINA E3 ligases and their responsive expression to salt and osmotic stresses in Brassica napus. Plant Cell Rep. 2023, 42, 859–877. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, Y.H.; Wan, H.N.; Tang, J.; Ni, Z.Y. The sea-island cotton GbTCP4 transcription factor positively regulates drought and salt stress responses. Plant Sci. 2022, 322, 111329. [Google Scholar] [CrossRef]
- Zhang, X.; Dong, J.; Deng, F.; Wang, W.; Shen, F. The long non-coding RNA lncRNA973 is involved in cotton response to salt stress. BMC Plant Biol. 2019, 19, 9998. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.Z.; Li, W.J.; Hu, X.H.; Guo, J.R.; Liu, A.M.; Zhang, B.L. A cotton MYB transcription factor, GbMYB5, is positively involved in plant adaptive response to drought stress. Plant. Cell Physiol. 2015, 56, 917–929. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Zeng, J.; Yu, Y.; Ni, Z. Silencing of GhSINAT5 Reduces Drought Resistance and Salt Tolerance in Cotton. Genes 2024, 15, 1063. https://doi.org/10.3390/genes15081063
Wang Y, Zeng J, Yu Y, Ni Z. Silencing of GhSINAT5 Reduces Drought Resistance and Salt Tolerance in Cotton. Genes. 2024; 15(8):1063. https://doi.org/10.3390/genes15081063
Chicago/Turabian StyleWang, Yi, Jiacong Zeng, Yuehua Yu, and Zhiyong Ni. 2024. "Silencing of GhSINAT5 Reduces Drought Resistance and Salt Tolerance in Cotton" Genes 15, no. 8: 1063. https://doi.org/10.3390/genes15081063
APA StyleWang, Y., Zeng, J., Yu, Y., & Ni, Z. (2024). Silencing of GhSINAT5 Reduces Drought Resistance and Salt Tolerance in Cotton. Genes, 15(8), 1063. https://doi.org/10.3390/genes15081063





