Genome-Wide Association Analysis Identified Quantitative Trait Loci (QTLs) Underlying Drought-Related Traits in Cultivated Peanut (Arachis hypogaea L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Experimental Treatments
2.2. Phenotypic Data Collection
2.3. SNP Genotyping Utilizing SNP Gene Array
2.4. Statistical Data Analysis
2.5. Filtered Markers for LD Decay and Population Structure
2.6. Genome-Wide Association Mapping Analysis
2.7. Candidate Genes Search
3. Results
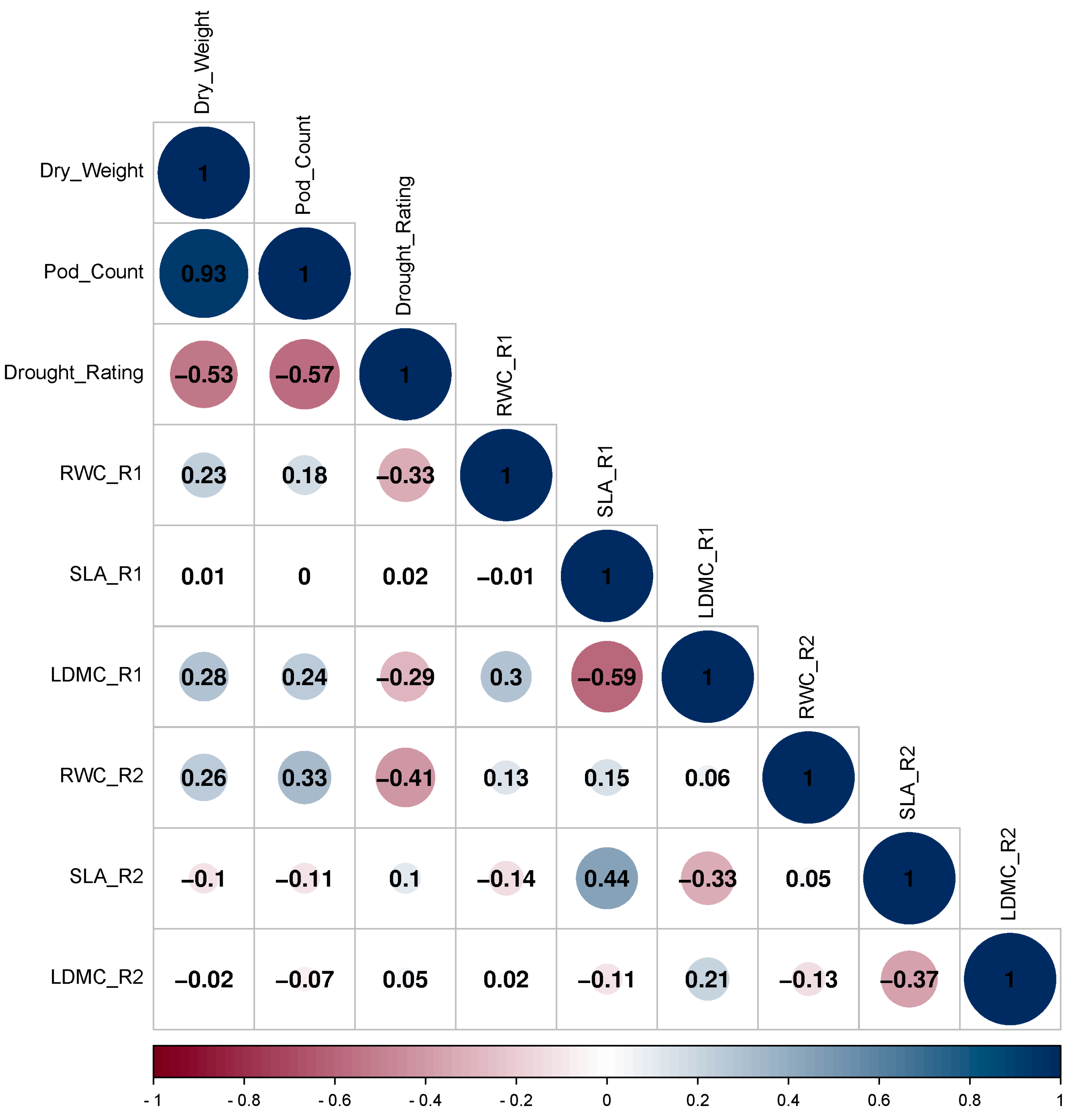
3.1. Correlation of RWC, SLA, LDMC, Drought Rating, Pod Count, and Pod Weight
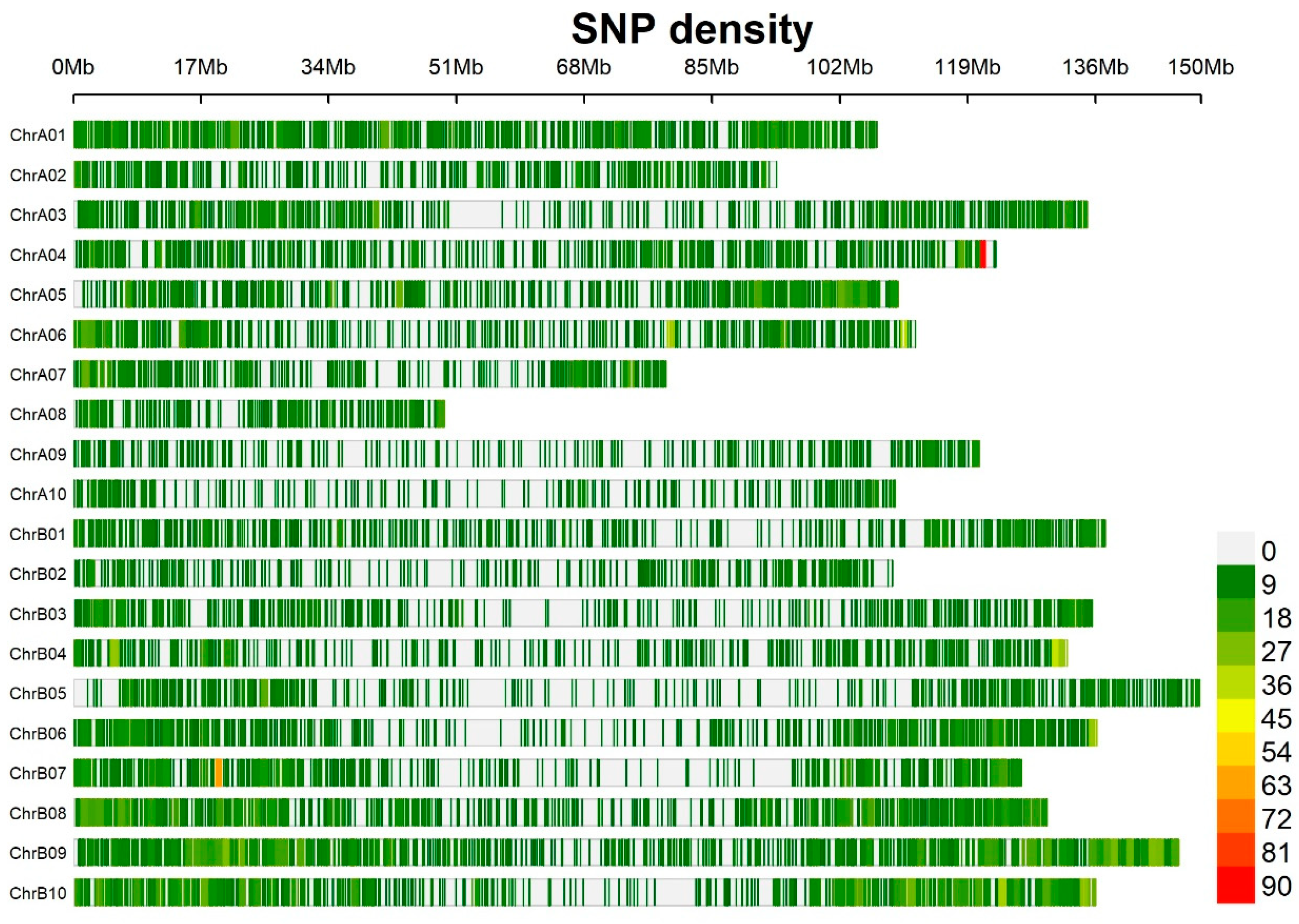
3.2. Distribution of SNP Markers, Linkage Disequilibrium, and Population Structure
3.3. Genomic Regions Associated with Quantitative Traits and Search for Candidate Genes
3.3.1. Relative Water Content Stage 1
3.3.2. Relative Water Content Stage 2
3.3.3. Specific Leaf Area Stage 1
3.3.4. Pod Count Number
3.3.5. Dry Pod Weight Measurements
3.3.6. Determination of Drought Ratings
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moharana, A.; Lenka, B.; Singh, A.; Kumar, N.; Nagaraju, B.; Das, S. Peanut as a food source: A review. J. Pharmacog. Phytochem. 2020, 9, 225–232. [Google Scholar]
- U.S. Department of Agriculture; Foreign Agricultural Service. Available online: https://ipad.fas.usda.gov (accessed on 10 May 2024).
- Puppala, N.; Nayak, S.N.; Sanz-Saez, A.; Chen, C.; Devi, M.J.; Nivedita, N.; Bao, Y.; He, G.; Traore, S.M.; Wright, D.A.; et al. Sustaining yield and nutritional quality of peanuts in harsh environments: Physiological and molecular basis of drought and heat stress tolerance. Front. Genet. 2023, 14, 1121462. [Google Scholar] [CrossRef]
- Vadez, V.; Kholova, J.; Medina, S.; Kakkera, A.; Anderberg, H. Transpiration efficiency: New insights into an old story. J. Exp. Bot. 2014, 65, 6141–6153. [Google Scholar] [CrossRef]
- Vadez, V.; Ratnakumar, P. High transpiration efficiency increases pod yield under intermittent drought in dry and hot atmospheric conditions but less so under wetter and cooler conditions in groundnut (Arachis hypogaea (L.)). Field Crops Res. 2016, 193, 16–23. [Google Scholar] [CrossRef]
- Reddy, T.Y.; Reddy, V.R.; Anbumozhi, V. Physiological responses of groundnut (Arachis hypogaea L.) to drought stress and its amelioration: A critical review. Plant Growth Regul. 2003, 41, 75–88. [Google Scholar] [CrossRef]
- Chavarro, C.; Chu, Y.; Holbrook, C.; Isleib, T.; Bertioli, D.; Hovav, R.; Butts, C.; Lamb, M.; Sorensen, R.; AJackson, S.; et al. Pod and Seed Trait QTL Identification to Assist Breeding for Peanut Market Preferences. G3 2020, 10, 2297–2315. [Google Scholar] [CrossRef]
- Chu, Y.; Chee, P.; Isleib, T.G.; Holbrook, C.C.; Ozias-Akins, P. Major seed size QTL on chromosome A05 of peanut (Arachis hypogaea) is conserved in the US mini core germplasm collection. Mol. Breed. 2019, 40, 6. [Google Scholar] [CrossRef]
- Yang, Y.Q.; Su, Q.; Li, Y.R.; Cheng, Z.S.; Song, Y.H.; Jin, X.X.; Wang, J. Fine mapping of a major QTL qHYF_B06 for peanut yield. Crop J. 2023, 11, 1533–1540. [Google Scholar] [CrossRef]
- Patel, J.D.; Wang, M.L.; Dang, P.; Butts, C.; Lamb, M.; Chen, C.Y. Insights into the Genomic Architecture of Seed and Pod Quality Traits in the U.S. Peanut Mini-Core Diversity Panel. Plants 2022, 11, 837. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li Wang, M.; Dang, P.; Jiang, T.; Zhao, S.; Lamb, M.; Chen, C. Identification of potential QTLs and genes associated with seed composition traits in peanut (Arachis hypogaea L.) using GWAS and RNA-Seq analysis. Gene 2021, 769, 145215. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, M.L.; Schaefer, R.; Dang, P.; Jiang, T.; Chen, C. GWAS and Coexpression Network Reveal Ionomic Variation in Cultivated Peanut. J. Agric. Food Chem. 2019, 67, 12026–12036. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Barkley, N.A.; Wang, M.L.; Holbrook, C.C.; Dang, P.M. Registration of Purified Accessions for the US Peanut Mini-Core Germplasm Collection. J. Plant Regist. 2014, 8, 77–85. [Google Scholar] [CrossRef]
- Blankenship, P.; Mitchell, B.; Layton, R.; Cole, R.; Sanders, T. A low-cost microcomputer system to monitor and control an environmental control plot facility. Comput. Electron. Agric. 1989, 4, 6. [Google Scholar] [CrossRef]
- Barrs, H.D.; Weatherley, P. A re-examination of the relative turgidity technique for estimating water deficit in leaves. Aust. J. Biol. Sci. 1962, 15, 15. [Google Scholar] [CrossRef]
- Clevenger, J.; Chu, Y.; Chavarro, C.; Agarwal, G.; Bertioli, D.J.; Leal-Bertioli, S.C.M.; Pandey, M.K.; Vaughn, J.; Abernathy, B.; Barkley, N.A.; et al. Genome-wide SNP Genotyping Resolves Signatures of Selection and Tetrasomic Recombination in Peanut. Mol. Plant 2017, 10, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Holland, J.B.; Nyquist, W.E.; Cervantes-Martinez, C.T. Estimating and interpreting heritability for plant breeding: An update. Plant Breed. Rev. 2003, 22, 11–112. [Google Scholar]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.S.; Xu, J.Y.; He, W.M.; Yang, T.L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef]
- Earl, D.A.; VonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Z. GAPIT Version 3: Boosting power and accuracy for genomic association and prediction. Genom. Proteom. Bioinform. 2021, 19, 629–640. [Google Scholar] [CrossRef]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.D.; Farmer, A.D.; Cannon, E.K.S.; Liu, X.; Gao, D.Y.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef]
- Tiwari, P.N.; Tiwari, S.; Sapre, S.; Babbar, A.; Tripathi, N.; Tiwari, S.; Tripathi, M.K. Screening and Selection of Drought-Tolerant High-Yielding Chickpea Genotypes Based on Physio-Biochemical Selection Indices and Yield Trials. Life 2023, 13, 1405. [Google Scholar] [CrossRef] [PubMed]
- Kramp, R.E.; Liancourt, P.; Herberich, M.M.; Saul, L.; Weides, S.; Tielbörger, K.; Májeková, M. Functional traits and their plasticity shift from tolerant to avoidant under extreme drought. Ecology 2022, 103, e3826. [Google Scholar] [CrossRef]
- Wahab, A.; Abdi, G.; Saleem, M.H.; Ali, B.; Ullah, S.; Shah, W.D.; Mumtaz, S.; Yasin, G.; Muresan, C.C.; Marc, R.A. Plants’ Physio-Biochemical and Phyto-Hormonal Responses to Alleviate the Adverse Effects of Drought Stress: A Comprehensive Review. Plants 2022, 11, 1620. [Google Scholar] [CrossRef]
- Dang, P.M.; Chen, C.Y.; Holbrook, C.C. Evaluation of five peanut (Arachis hypogaea) genotypes to identify drought responsive mechanisms utilising candidate-gene approach. Funct. Plant Biol. 2013, 40, 1323–1333. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Dang, P.; Chen, C.; Feng, Y.C.; Batchelor, W.; Lamb, M.; Sanz-Saez, A. Tolerance to mid-season drought in peanut can be achieved by high water use efficiency or high efficient use of water. Crop Sci. 2022, 62, 1948–1966. [Google Scholar] [CrossRef]
- Duan, J.Z.; Zhang, M.H.; Zhang, H.L.; Xiong, H.Y.; Liu, P.L.; Ali, J.; Li, J.J.; Li, Z. OsMIOX, a myo-inositol oxygenase gene, improves drought tolerance through scavenging of reactive oxygen species in rice (Oryza sativa L.). Plant Sci. 2012, 196, 143–151. [Google Scholar] [CrossRef]
- Mathan, J.; Singh, A.; Ranjan, A. Sucrose transport in response to drought and salt stress involves ABA-mediated induction of OsSWEET13 and OsSWEET15 in rice. Physiol. Plant 2021, 171, 620–637. [Google Scholar] [CrossRef]
- Ku, Y.S.; Cheng, S.S.; Cheung, M.Y.; Lam, H.M. The Roles of Multidrug and Toxic Compound Extrusion (MATE) Transporters in Regulating Agronomic Traits. Agronomy 2022, 12, 878. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Zhou, W.K.; Chen, Q.; Fang, M.M.; Zheng, S.S.; Scheres, B.; Li, C.Y. Mediator subunit MED31 is required for radial patterning of Arabidopsis roots. Proc. Natl. Acad. Sci. USA 2018, 115, E5624–E5633. [Google Scholar] [CrossRef] [PubMed]
- Atif, R.M.; Shahid, L.; Waqas, M.; Ali, B.; Rashid, M.A.R.; Azeem, F.; Nawaz, M.A.; Wani, S.H.; Chung, G. Insights on Calcium-Dependent Protein Kinases (CPKs) Signaling for Abiotic Stress Tolerance in Plants. Int. J. Mol. Sci. 2019, 20, 5298. [Google Scholar] [CrossRef]
- Han, G.L.; Qiao, Z.Q.; Li, Y.X.; Yang, Z.R.; Wang, C.F.; Zhang, Y.Y.; Liu, L.L.; Wang, B.S. RING Zinc Finger Proteins in Plant Abiotic Stress Tolerance. Front. Plant Sci. 2022, 13, 877011. [Google Scholar] [CrossRef] [PubMed]
- Mutti, G.; Raveane, A.; Pagano, A.; Bertolini, F.; Semino, O.; Balestrazzi, A.; Macovei, A. Plant TDP1 (Tyrosyl-DNA Phosphodiesterase 1): A Phylogenetic Perspective and Gene Expression Data Mining. Genes 2020, 11, 1465. [Google Scholar] [CrossRef]
- Huang, J.; Ghosh, R.; Bankaitis, V.A. Sec14-like phosphatidylinositol transfer proteins and the biological landscape of phosphoinositide signaling in plants. BBA Mol. Cell Biol. Lipids 2016, 1861, 1352–1364. [Google Scholar] [CrossRef]
- Montag, K.; Ivanov, R.; Bauer, P. Role of SEC14-like phosphatidylinositol transfer proteins in membrane identity and dynamics. Front. Plant Sci. 2023, 14, 1181031. [Google Scholar] [CrossRef] [PubMed]
- Matos, A.R.; Gigon, A.; Laffray, D.; Petres, S.; Zuily-Fodil, Y.; Pham-Thi, A.T. Effects of progressive drought stress on the expression of patatin-like lipid acyl hydrolase genes in Arabidopsis leaves. Physiol. Plant 2008, 134, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Zhang, X.; Zhao, K.; Zhao, K.; Cao, D.; Ma, Q.; Zhu, S.; Qu, C.; Ma, Y.; Gong, F.; et al. Genome wide identification and expression analysis of patatin-like protein family members in peanut (Arachis hypogaea L.). Reprod. Breed. 2021, 1, 48–54. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Q.Q.; Liu, M.L.; Bo, C.; Wang, X.; Ma, Q.; Cheng, B.J.; Cai, R.H. Overexpression of a maize MYB48 gene confers drought tolerance in transgenic Arabidopsis plants. J. Plant Biol. 2017, 60, 612–621. [Google Scholar] [CrossRef]
- Lubkowitz, M. The Oligopeptide Transporters: A Small Gene Family with a Diverse Group of Substrates and Functions? Mol. Plant 2011, 4, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.H.; Wang, X.Q.; Li, J.X.; Guan, J.H.; Tan, Z.J.; Zhang, Z.; Shi, G.R. Genome-Wide Identification and Transcript Analysis Reveal Potential Roles of Oligopeptide Transporter Genes in Iron Deficiency Induced Cadmium Accumulation in Peanut. Front. Plant Sci. 2022, 13, 894848. [Google Scholar] [CrossRef] [PubMed]
- Bu, Q.; Lv, T.; Shen, H.; Luong, P.; Wang, J.; Wang, Z.; Huang, Z.; Xiao, L.; Engineer, C.; Kim, T.H.; et al. Regulation of drought tolerance by the F-box protein MAX2 in Arabidopsis. Plant Physiol. 2014, 164, 424–439. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.L.; Qiao, L.; Su, X.H.; Ji, B.Y.; Dong, C.M. Drought Stress Stimulates the Terpenoid Backbone and Triterpenoid Biosynthesis Pathway to Promote the Synthesis of Saikosaponin in Bupleurum chinense DC. Roots. Molecules 2022, 27, 5470. [Google Scholar] [CrossRef] [PubMed]
- Songsri, P.; Jogloy, S.; Holbrook, C.C.; Vorasoot, N.; Kesmala, T.; Akkasaeng, C.; Patanothai, A. Evaluation of yield and reproductive efficiency in peanut (Arachis hypogaea L.) under different available soil water. Asian J. Plant Sci. 2009, 8, 465–473. [Google Scholar] [CrossRef][Green Version]
- Önemli, F. The correlation analyses of some climate values with flowering and earliness index in peanut (Arachis hypogaea L.). J. Tekirdag. Agric. Faculty 2005, 2, 273–281. [Google Scholar]
- Pallas, J.E., Jr.; Stansell, J.R.; Bruce, R.R. Peanut seed germination as related to soil water regime during pod development. Agron. J. 1977, 69, 381–383. [Google Scholar] [CrossRef]
- Foltyn, V.N.; Bendikov, I.; De Miranda, J.; Panizzutti, R.; Dumin, E.; Shleper, M.; Li, P.; Toney, M.D.; Kartvelishvily, E.; Wolosker, H. Serine racemase modulates intracellular D-serine levels through an α,β-elimination activity. J. Biol. Chem. 2005, 280, 1754–1763. [Google Scholar] [CrossRef] [PubMed]
- Michard, E.; Lima, P.T.; Borges, F.; Silva, A.C.; Portes, M.T.; Carvalho, J.E.; Gilliham, M.; Liu, L.H.; Obermeyer, G.; Feijó, J.A. Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil D-serine. Science 2011, 332, 434–437. [Google Scholar] [CrossRef]
- Cai, Y.C.; Li, S.F.; Jiao, G.; Sheng, Z.H.; Wu, Y.W.; Shao, G.N.; Xie, L.H.; Peng, C.; Xu, J.F.; Tang, S.Q.; et al. OsPK2 encodes a plastidic pyruvate kinase involved in rice endosperm starch synthesis, compound granule formation and grain filling. Plant Biotechnol. J. 2018, 16, 1878–1891. [Google Scholar] [CrossRef]
- Riemer, E.; Qiu, D.Y.; Laha, D.; Harmel, R.K.; Gaugler, P.; Gaugler, V.; Frei, M.; Hajirezaei, M.R.; Laha, N.P.; Krusenbaum, L.; et al. ITPK1 is an InsP(6)/ADP phosphotransferase that controls phosphate signaling. Mol. Plant 2021, 14, 1864–1880. [Google Scholar] [CrossRef] [PubMed]
- Stuhrwohldt, N.; Buhler, E.; Sauter, M.; Schaller, A. Phytosulfokine (PSK) precursor processing by subtilase SBT3.8 and PSK signaling improve drought stress tolerance in Arabidopsis. J. Exp. Bot. 2021, 72, 3427–3440. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Xu, S.; Qu, D.; Yang, L.; Wang, J.; Liu, H.; Xin, W.; Zou, D.; Zheng, H. Identification and Functional Analysis of the Caffeic Acid O-Methyltransferase (COMT) Gene Family in Rice (Oryza sativa L.). Int. J. Mol. Sci. 2022, 23, 8491. [Google Scholar] [CrossRef]
- Blum, A. Plant Breeding for Water-Limited Environments; Springer: New York, NY, USA, 2011. [Google Scholar]


| Traits 1 | Dry Wt | Pod # | Rate | RWC1 | SLA1 | LDMC1 | RWC2 | SLA2 | LDMC2 |
|---|---|---|---|---|---|---|---|---|---|
| σ2e | 2446.48 | 2101.65 | 0.07 | 3.99 | 84.14 | 0 | 0.67 | 174.55 | 0.00621 |
| σ2g | 2831.4 | 2059.81 | 0.15 | 0.46 | 68.56 | 0.00009 | 2.54 | 37.06 | 0 |
| σ2g×e | 172.19 | 84.02 | 0.16 | 3.66 | 108.75 | 0.00016 | 4.94 | 33.79 | 0.0008 |
| σ2error | 4566.38 | 4018.68 | 1.21 | 11.41 | 573.05 | 0.00057 | 20.79 | 616.79 | 0.00056 |
| Heritability | 69.75 | 66.31 | 28.89 | 14.11 | 40.17 | 41.18 | 37.71 | 35.18 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dang, P.; Patel, J.; Sorensen, R.; Lamb, M.; Chen, C.Y. Genome-Wide Association Analysis Identified Quantitative Trait Loci (QTLs) Underlying Drought-Related Traits in Cultivated Peanut (Arachis hypogaea L.). Genes 2024, 15, 868. https://doi.org/10.3390/genes15070868
Dang P, Patel J, Sorensen R, Lamb M, Chen CY. Genome-Wide Association Analysis Identified Quantitative Trait Loci (QTLs) Underlying Drought-Related Traits in Cultivated Peanut (Arachis hypogaea L.). Genes. 2024; 15(7):868. https://doi.org/10.3390/genes15070868
Chicago/Turabian StyleDang, Phat, Jinesh Patel, Ron Sorensen, Marshall Lamb, and Charles Y. Chen. 2024. "Genome-Wide Association Analysis Identified Quantitative Trait Loci (QTLs) Underlying Drought-Related Traits in Cultivated Peanut (Arachis hypogaea L.)" Genes 15, no. 7: 868. https://doi.org/10.3390/genes15070868
APA StyleDang, P., Patel, J., Sorensen, R., Lamb, M., & Chen, C. Y. (2024). Genome-Wide Association Analysis Identified Quantitative Trait Loci (QTLs) Underlying Drought-Related Traits in Cultivated Peanut (Arachis hypogaea L.). Genes, 15(7), 868. https://doi.org/10.3390/genes15070868








