Research on the Effects of the Relationship between Agronomic Traits and Dwarfing Genes on Yield in Colored Wheat
Abstract
1. Introduction
2. Materials and Methods
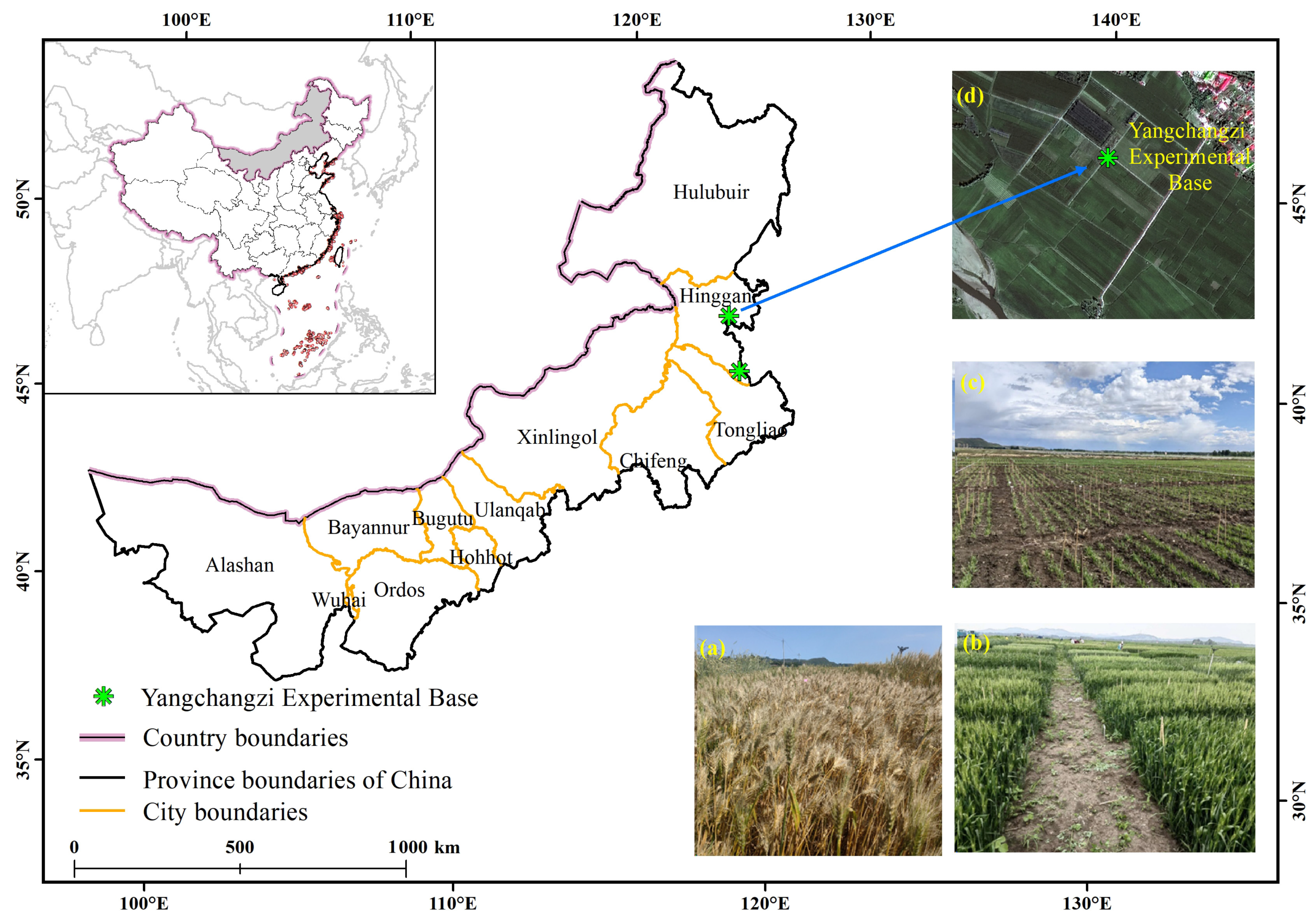
2.1. Experimental Materials
2.2. Data Analysis
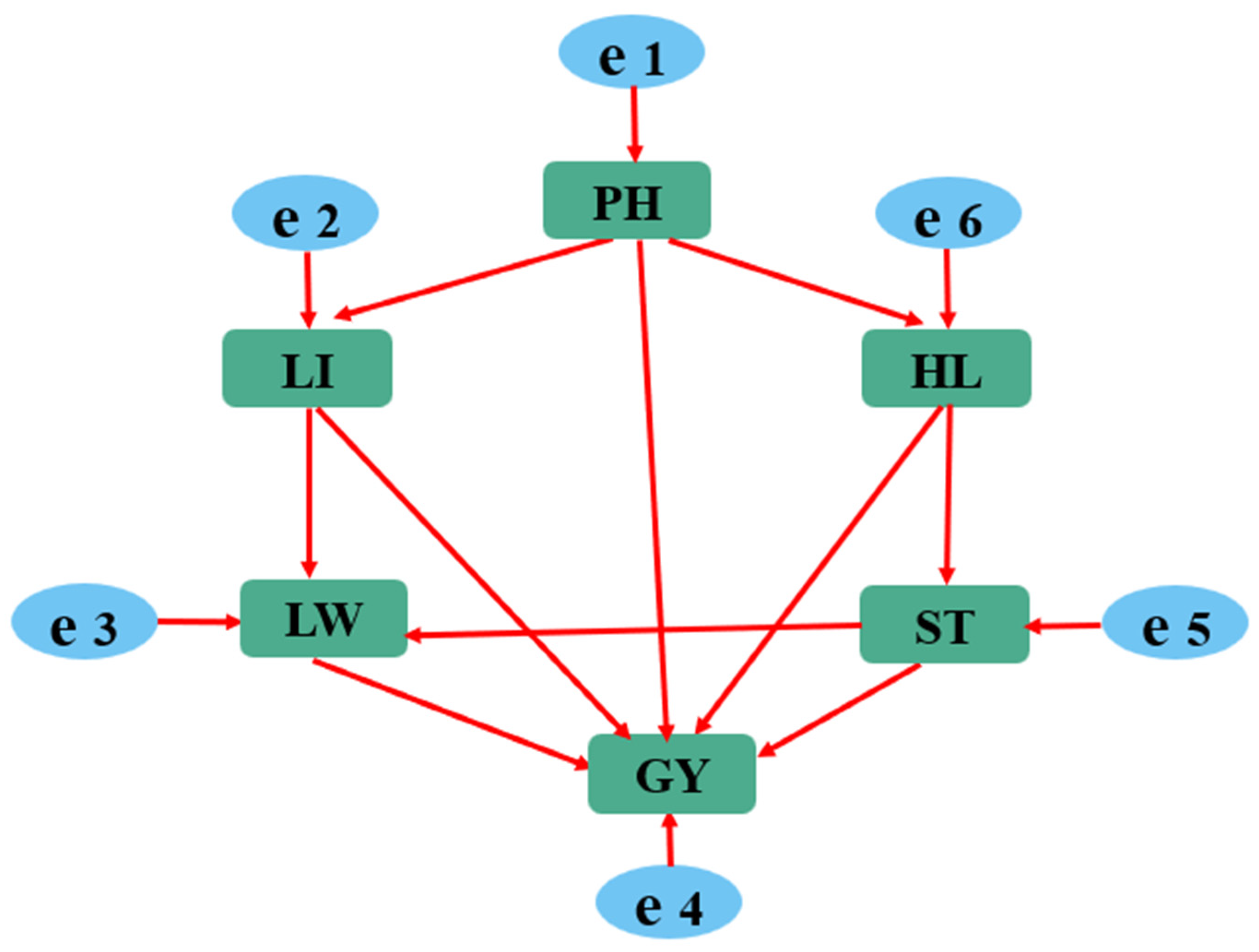
2.2.1. Initial Structural Equation Model
2.2.2. Model Verification
2.2.3. Entropy Method and Indicator Selection
2.3. Transcriptome Sequencing and Real-Time Fluorescence Quantitative Polymerase Chain Reaction (RT-qPCR) Validation of Candidate Genes
2.3.1. RNA Extraction
2.3.2. Reverse Transcription
2.3.3. Fluorescent Quantitative RT-qPCR Amplification
3. Results and Analysis
3.1. Basic Statistical Characteristics of Colored Wheat
3.2. Correlation Analysis of Main Agronomic Traits and Yield in Colored Wheat
3.3. Using Structural Equation Modeling to Analyze the Relationship between the Main Agronomic Traits and Yield in Colored Wheat
3.4. Using the Entropy Method to Screen Colored Wheat Seed Resources
3.5. Screening for Dwarfing Genes in Colored Wheat
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| No. | Resource | Color | Source | No. | Resource | Color | Source |
|---|---|---|---|---|---|---|---|
| 1 | KY2022-1 | Purple | Chinese Inner Mongolia Academy of Agriculture and Animal Husbandry Sciences | 37 | ZNY2022-24 | White | Chinese Academy of Sciences |
| 2 | KY2022-2 | Purple | Chinese Inner Mongolia Academy of Agriculture and Animal Husbandry Sciences | 38 | ZNY2022-25 | White | Chinese Academy of Sciences |
| 3 | LY2022-1 | Purple | Chinese Inner Mongolia Yuanlu Company | 39 | ZNY2022-26 | White | Chinese Academy of Sciences |
| 4 | QY2022-1 | Purple | Northwest A&F University | 40 | ZNY2022-27 | White | Chinese Academy of Sciences |
| 5 | QY2022-2 | Blue | Northwest A&F University | 41 | ZNY2022-28 | White | Chinese Academy of Sciences |
| 6 | QY2022-3 | Purple | Northwest A&F University | 42 | ZNY2022-29 | White | Chinese Academy of Sciences |
| 7 | QY2022-4 | Purple | Northwest A&F University | 43 | ZNY2022-30 | White | Chinese Academy of Sciences |
| 8 | QY2022-5 | Purple | Northwest A&F University | 44 | ZNY2022-31 | White | Chinese Academy of Sciences |
| 9 | QY2022-6 | White | Northwest A&F University | 45 | ZNY2022-32 | White | Chinese Academy of Sciences |
| 10 | NXY2022-1 | Purple | Ningxia University | 46 | ZNY2022-33 | White | Chinese Academy of Sciences |
| 11 | MDY2022-1 | Purple | Inner Mongolia | 47 | ZNY2022-34 | White | Chinese Academy of Sciences |
| 12 | ZKYY2022-1 | Purple | Chinese Academy of Sciences | 48 | ZNY2022-35 | White | Chinese Academy of Sciences |
| 13 | ZKYY2022-2 | Purple | Chinese Academy of Sciences | 49 | ZNY2022-36 | White | Chinese Academy of Sciences |
| 14 | ZNY2022-1 | Purple | Chinese Academy of Sciences | 50 | ZNY2022-37 | White | Chinese Academy of Sciences |
| 15 | ZNY2022-2 | Purple | Chinese Academy of Sciences | 51 | ZNY2022-38 | White | Chinese Academy of Sciences |
| 16 | ZNY2022-3 | Purple | Chinese Academy of Sciences | 52 | ZNY2022-39 | White | Chinese Academy of Sciences |
| 17 | ZNY2022-4 | Purple | Chinese Academy of Sciences | 53 | ZNY2022-40 | White | Chinese Academy of Sciences |
| 18 | ZNY2022-5 | Purple | Chinese Academy of Sciences | 54 | ZNY2022-41 | White | Chinese Academy of Sciences |
| 19 | ZNY2022-6 | Purple | Chinese Academy of Sciences | 55 | ZNY2022-42 | White | Chinese Academy of Sciences |
| 20 | ZNY2022-7 | Purple | Chinese Academy of Sciences | 56 | ZNY2022-43 | White | Chinese Academy of Sciences |
| 21 | ZNY2022-8 | Purple | Chinese Academy of Sciences | 57 | ZNY2022-44 | White | Chinese Academy of Sciences |
| 22 | ZNY2022-9 | Purple | Chinese Academy of Sciences | 58 | ZNY2022-45 | White | Chinese Academy of Sciences |
| 23 | ZNY2022-10 | Purple | Chinese Academy of Sciences | 59 | ZNY2022-46 | White | Inner Mongolia Huhe hot |
| 24 | ZNY2022-11 | Purple | Chinese Academy of Sciences | 60 | ZNY2022-47 | Purple | Inner Mongolia Academy of Agricultural Sciences |
| 25 | ZNY2022-12 | Purple | Chinese Academy of Sciences | 61 | ZNY2022-48 | Purple | Inner Mongolia Academy of Agricultural Sciences |
| 26 | ZNY2022-13 | Purple | Chinese Academy of Sciences | 62 | ZNY2022-49 | Purple | Inner Mongolia Academy of Agricultural Sciences |
| 27 | ZNY2022-14 | Purple | Chinese Academy of Sciences | 63 | ZNY2022-50 | Purple | Inner Mongolia Academy of Agricultural Sciences |
| 28 | ZNY2022-15 | Purple | Chinese Academy of Sciences | 64 | ZNY2022-51 | Purple | Inner Mongolia Academy of Agricultural Sciences |
| 29 | ZNY2022-16 | Purple | Chinese Academy of Sciences | 65 | ZNY2022-52 | Purple | Inner Mongolia Academy of Agricultural Sciences |
| 30 | ZNY2022-17 | Purple | Chinese Academy of Sciences | 66 | ZNY2022-53 | Purple | Inner Mongolia Academy of Agricultural Sciences |
| 31 | ZNY2022-18 | White | Chinese Academy of Sciences | 67 | ZNY2022-54 | Purple | Inner Mongolia Academy of Agricultural Sciences |
| 32 | ZNY2022-19 | White | Chinese Academy of Sciences | 68 | ZNY2022-55 | Purple | Inner Mongolia Academy of Agricultural Sciences |
| 33 | ZNY2022-20 | White | Chinese Academy of Sciences | 69 | ZNY2022-56 | Purple | Inner Mongolia Academy of Agricultural Sciences |
| 34 | ZNY2022-21 | White | Chinese Academy of Sciences | 70 | ZNY2022-57 | Purple | Inner Mongolia Ulanhot |
| 35 | ZNY2022-22 | White | Chinese Academy of Sciences | 71 | ZNY2022-58 | Purple | Inner Mongolia Shangkuli Farm Hulunbuir |
| 36 | ZNY2022-23 | White | Chinese Academy of Sciences | 72 | ZNY2022-59 | Purple | Inner Mongolia Shangkuli Farm Hulunbuir |
| NM | Amplification Length (bp) | Primer Name | Sequence (5′–3′) |
|---|---|---|---|
| FN649763.1 | 114 bp | Triticum RHT1 F-primer | cgccatgttcgattctctgg |
| Triticum RHT1 R-primer | tacacctcggacatgacctg | ||
| OL958546.1 | 116 bp | Triticum Rht8 F-primer | gactctgcgtgatctcgaga |
| Triticum Rht8 R-primer | cgttgttgaagagtgggagc | ||
| KC614643.1 | 60 bp | Triticum Rht-D1 F-primer | tccgaggacaagatgatggt |
| Triticum Rht-D1 R-primer | cagctcgtccacctcctc |
| Name | Reagents | Instrumentation and Equipment | ||||
|---|---|---|---|---|---|---|
| The Names of the Reagents | Brand | Catalog Number | Instrument Name | Brand | Model | |
| Introduction to the Experimental Procedure for Plant Tissue RNA Extraction | TRIzol Reagent | Invitrogen (A biotechnology company in the United States.) | 15596026 | Fully automated sample rapid grinder | Shanghai Jingxin Technology | JXFSTPRP-48 |
| Trichloromethane | Xilong Chemicals (A chemical company in China.) | AR500ML | Centrifuge | Eppendorf | 5417R | |
| Isopentanol | Xilong Chemicals (A chemical company in China.) | AR500ML | Pipette | Eppendorf | ||
| Isopropanol (analytical grade) | Xilong Chemicals (A chemical company in China.) | AR500ML | Vortex mixer | Kylin-Bell | QL-901 | |
| Anhydrous ethanol (analytical grade) | Xilong Chemicals | 72188-01 | ||||
| DEPC-treated water | AMBION | AM9922 | ||||
| Introduction to the Procedure for Chain-Specific mRNA Library Preparation (DNBSEQ) | DNF-471 STANDARD SENSITIVITY RNA ANALYSIS KIT | AATI | DNF-471-0500 | Fragment Analyzer | Agilent | 5300 |
| BGI Optimal series dual-module mRNA library construction kit | BGI | LR00R96 | Microcentrifuge | Baygene | BG Qspin7000 | |
| BGI Plug-In Adapter Kit | BGI | LA00R04 | Vortex mixer | Kylin-Bell | QL-901 | |
| DNA separation magnetic beads | BGI | LB00V60 | Automated pipetting workstation | MGI | MGISP-960 | |
| Qubit® ssDNA Assay Kit | Invitrogen | Q10212 | PCR instrument | ABI APPIED BIOSYSTEMS | 9700 | |
| MGISEQ-2000RS High-throughput sequencing reagent kit (FCL PE100) | MGI | 1000012554 | Pipette | Eppendorf | ||
| MGISEQ-2000RS High-throughput sequencing reagent kit(FCL PE150) | MGI | 1000012555 | Qubit | Thermo Fisher | Q33216 | |
| DNBSEQ-T7RS High-throughput sequencing reagent kit (FCL PE100) | MGI | 1000028455 | Gene sequencer | MGI | MGISEQ-2000 | |
| Reagents and Reagent Kits | Source |
|---|---|
| TRIzol™ Reagent Invitrogen™ | Invitrogen 15596018 (A biotechnology company in the United States.) |
| RNase-Free Water | QIAGEN 129112 (A biotechnology company in the United States.) |
| First Strand cDNA synthesis Kit | Invitrogen K1622 (A biotechnology company in the United States.) |
| QuantityNova SYBR Green PCR Kit | QIAGEN 208052 (A biotechnology company in the United States.) |
| TBE | City of Shanghai Weiao Biological Technology Company Limited in China. |
| 6*Loading buffer | City of Shanghai Weiao Biological Technology Company Limited in China. |
| Instrument Name | Instrument Model | Manufacturer |
|---|---|---|
| Biological Safety Cabinet | BHC-1300 | City of Suzhou Purification Equipment Company Limited in China. |
| Tissue Homogenizer | TL-2010S | City of Beijing Dinghaoyuan Technology Company Limited in China. |
| Tabletop High-Speed Centrifuge | Fresco21 | Thermo Fisher Scientific, headquartered in Wilmington, MA, USA |
| Full Wavelength Microplate Reader | MutiskanTM GO | Thermo Fisher Scientific, headquartered in Wilmington, MA, USA |
| Bioanalyzer | BIO RAD POWER PAC 3000 | BIO RAD, based in Hercules, CA, USA |
| Wide Mini-Sub Cell Horizontal Electrophoresis System | BIO RAD DNA SUB CELL | BIO RAD, based in Hercules, CA, USA |
| Gel Imaging System | GIS-1600 | City of Shanghai Tianneng Technology Company Limited in China |
| Gradient PCR Instrument | Veriti DX | Thermo Fisher Scientific, headquar-tered in Wilmington, MA, USA |
| Fluorescent Quantitative PCR Instrument | Roche480II Real Time PCR System | Roche, headquartered in Basel, Switzerland |
References
- Abdel-Aal, E.-S.M.; Young, J.C.; Rabalski, I. Anthocyanin composition in black, blue, pink, purple, and red cereal grains. J. Agric. Food. Chem. 2006, 54, 4696–4704. [Google Scholar] [CrossRef]
- Kim, M.-J.; Hyun, J.-N.; Kim, J.-A.; Park, J.-C.; Kim, M.-Y.; Kim, J.-G.; Lee, S.-J.; Chun, S.-C.; Chung, I.-M. Relationship between phenolic compounds, anthocyanins content and antioxidant activity in colored barley germplasm. J. Agric. Food. Chem. 2007, 55, 4802–4809. [Google Scholar] [CrossRef]
- Liu, D.; Li, S.; Chen, W.; Zhang, B.; Liu, D.; Liu, B.; Zhang, H. Transcriptome analysis of purple pericarps in common wheat (Triticum aestivum L.). PLoS ONE 2016, 11, e0155428. [Google Scholar] [CrossRef]
- Paznocht, L.; Kotíková, Z.; Šulc, M.; Lachman, J.; Orsák, M.; Eliášová, M.; Martinek, P. Free and esterified carotenoids in pigmented wheat, tritordeum and barley grains. Food Chem. 2018, 240, 670–678. [Google Scholar] [CrossRef]
- Wang, F.; Xu, Z.; Fan, X.; Zhou, Q.; Cao, J.; Ji, G.; Jing, S.; Feng, B.; Wang, T. Transcriptome analysis reveals complex molecular mechanisms underlying UV tolerance of wheat (Triticum aestivum L.). J. Agric. Food. Chem. 2018, 67, 563–577. [Google Scholar] [CrossRef]
- Sharma, S.; Chunduri, V.; Kumar, A.; Kumar, R.; Khare, P.; Kondepudi, K.K.; Bishnoi, M.; Garg, M. Anthocyanin bio-fortified colored wheat: Nutritional and functional characterization. PLoS ONE 2018, 13, e0194367. [Google Scholar] [CrossRef]
- de Oliveira Gondim, T.C.; Rocha, V.S.; Sediyama, C.S.; Miranda, G.V. Path analysis for yield components and agronomic traits of wheat under defoliation. Pesqui. Agropecu. Bras. 2008, 43, 487–493. [Google Scholar] [CrossRef]
- Siney, S.; Saba, J. Analysis of yield and yield components traits in twenty bread wheat genotypes under dryland conditions. Philipp. J. Crop Sci. 2015, 40, 78–84. [Google Scholar]
- Yagdi, K. Path coefficient analysis of some yield components in durum wheat (Triticum durum Desf.). Pak. J. Bot. 2009, 41, 745–751. [Google Scholar]
- Mizuno, N.; Ishikawa, G.; Kojima, H.; Tougou, M.; Kiribuchi-Otobe, C.; Fujita, M.; Nakamura, K. Genetic mechanisms determining grain number distribution along the spike and their effect on yield components in wheat. Mol. Breed. 2021, 41, 62. [Google Scholar] [CrossRef]
- Bian, Y.; Li, L.; Tian, X.; Xu, D.; Sun, M.; Li, F.; Xie, L.; Liu, S.; Liu, B.; Xia, X.; et al. Rht12b, a widely used ancient allele of TaGA2oxA13, reduces plant height and enhances yield potential in wheat. Theor. Appl. Genet. 2023, 136, 253. [Google Scholar] [CrossRef]
- Casebow, R.; Hadley, C.; Uppal, R.; Addisu, M.; Gooding, M. Reduced Height (Rht) Alleles Affect Wheat Grain Quality. PLoS ONE 2016, 11, e0156056. [Google Scholar] [CrossRef]
- Qamar, Z.U.; Bansal, U.K.; Dong, C.M.; Alfred, R.L.; Bhave, M.; Bariana, H.S. Detection of puroindoline (Pina-D1 and Pinb-D1) allelic variation in wheat landraces. J. Cereal Sci. 2014, 60, 610–616. [Google Scholar] [CrossRef]
- Kline, R.B. Principles and Practice of Structural Equation Modeling, 3rd ed.; The Guilford Press: New York, NY, USA, 2011. [Google Scholar]
- Grace, J.B. Structural Equation Modeling and Natural Systems; Cambridge University Press: Cambridge, UK, 2006. [Google Scholar]
- Lamb, E.; Shirtliffe, S.; May, W. Structural equation modeling in the plant sciences: An example using yield components in oat. Can. J. Plant. Sci. 2011, 91, 603–619. [Google Scholar] [CrossRef]
- Schermelleh-Engel, K.; Moosbrugger, H.; Müller, H. Evaluating the fit of structural equation models: Test of significance and descriptive goodness of fit measures. Methods Psychol. Res. 2003, 8, 23–74. [Google Scholar] [CrossRef]
- Fereydouni, F.; Hajian-Tilaki, K.; Meftah, N.; Chehrazi, M. A path causal model in the association between self-efficacy and self-care with quality of life in patients with type 2 diabetes: An application of the structural equation model. Health Sci. Rep. 2022, 5, e534. [Google Scholar] [CrossRef]
- Zhao, W.; Jin, G. Landscape aging in ethnic minority regions based on entropy method-fuzzy comprehensive evaluation—Take Yanji City, China as an example. IOP Conf. Ser. Earth Environ. Sci. 2021, 787, 012067. [Google Scholar] [CrossRef]
- Rebetzke, G.J.; Rattey, A.R.; Bovill, W.D.; Richards, R.A.; Brooks, B.J.; Ellis, M. Agronomic assessment of the durum Rht18 dwarfing gene in bread wheat. Crop Pasture Sci. 2022, 73, 325–336. [Google Scholar] [CrossRef]
- Mathews, K.L.; Chapman, S.C.; Trethowan, R.; Singh, R.P.; Crossa, J.; Pfeiffer, W.; van Ginkel, M.; DeLacy, I. Global adaptation of spring bread and durum wheat lines near-isogenic for major reduced height genes. Crop Sci. 2006, 46, 603–613. [Google Scholar] [CrossRef]
- Hedden, P. The genes of the Green Revolution. Trends Genet. 2003, 19, 5–9. [Google Scholar] [CrossRef]
- Philippa, B.; Rohit, M.; Tianyuan, X. An autoactive NB-LRR gene causes Rht13 dwarfism in wheat. Proc. Natl. Acad. Sci. USA 2022, 119, e2209875119. [Google Scholar] [CrossRef]
- Flintham, J.E.; Börner, A.; Rner, A.J.; Gale, M.D. Optimizing wheat grain yield: Effects of Rht (gibberellin-insensitive) dwarfing genes. J. Agric. Sci. 1997, 128, 11–25. [Google Scholar] [CrossRef]
- Schierenbeck, M.; Alqudah, M.A.; Lantos, E. Green Revolution dwarfing Rht genes negatively affected wheat floral traits related to cross-pollination efficiency. Plant J. 2024, 118, 1071–1085. [Google Scholar] [CrossRef]
- Figueroa, J.D.C.; Maucher, T.; Reule, W.; Peña, R.J. Influence of high molecular weight glutenins on viscoelastic properties of intact wheat kernel and relation to functional properties of wheat dough. Cereal Chem. 2009, 86, 139–144. [Google Scholar] [CrossRef]
- Hernández-Estrada, Z.J.; Rayas-Duarte, P.; Cárdenas, J.d.D.F. Creep recovery of wet gluten and high-molecular-weight glutenin subunit composition: Relationship with viscoelasticity of dough and breadmaking quality of hard red winter wheat. Cereal Chem. 2017, 94, 223–229. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, Y.; Zhang, B.; Florides, C.G.; Gao, Z.; Wang, Z.; Zhang, X.; Wei, Y. Comparison of quality properties between high-molecular-weight glutenin subunits 5+10 and 2+12 near-isogenic lines under three common wheat genetic backgrounds. Cereal Chem. 2018, 95, 575–583. [Google Scholar] [CrossRef]
- Sherman, J.D.; Nash, D.; Lanning, S.P.; Martin, J.M.; Blake, N.K.; Morris, C.F.; Talbert, L.E. Genetics of End-Use Quality Differences between a Modern and Historical Spring Wheat. Crop Sci. 2014, 54, 1972. [Google Scholar] [CrossRef]
- Tang, T.; Botwright-Acuña, T.; Spielmeyer, W.; Richards, R.A. Effect of gibberellin-sensitive Rht18 and gibberellin-insensitive Rht-D1b dwarfing genes on vegetative and reproductive growth in bread wheat. J. Exp. Bot. 2021, 72, 445–458. [Google Scholar] [CrossRef]
- Ravel, C.; Dardevet, M.; Leenhardt, F.; Bordes, J.; Joseph, J.L.; Perretant, M.R.; Exbrayat, F.; Poncet, C.; Balfourier, F.; Chanliaud, E.; et al. Improving the yellow pigment content of bread wheat flour by selecting the three homoeologous copies of Psy1. Mol. Breed. 2013, 31, 87–99. [Google Scholar] [CrossRef]
- Harjes, C.E.; Rocheford, T.R.; Bai, L.; Brutnell, T.P.; Kandianis, C.B.; Sowinski, S.G.; Stapleton, A.E.; Vallabhaneni, R.; Williams, M.; Wurtzel, E.T.; et al. Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science 2008, 319, 330–333. [Google Scholar] [CrossRef]
- Leenhardt, F.; Lyan, B.; Rock, E.; Boussard, A.; Potus, J.; Chanliaud, E.; Remesy, C. Genetic variability of carotenoid concentration, and lipoxygenase and peroxidase activities among cultivated wheat species and bread wheat varieties. Eur. J. Agron. 2006, 25, 170–176. [Google Scholar] [CrossRef]
- He, X.Y.; He, Z.H.; Zhang, L.P.; Sun, D.J.; Morris, C.F.; Fuerst, E.P.; Xia, X.C. Allelic variation of polyphenol oxidase (PPO) genes located on chromosomes 2A and 2D and development of functional markers for the PPO genes in common wheat. Theor. Appl. Genet. 2007, 115, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, A.; Wen, W.; Gao, F.; Zhai, S.; Jin, H.; Liu, J.; Guo, Q.; Zhang, Y.; Dreisigacker, S.; Xia, X.; et al. Development and validation of KASP assays for genes underpinning key economic traits in bread wheat. Theor. Appl. Genet. 2016, 129, 1843–1860. [Google Scholar] [CrossRef]



| Traits | Minimum | Maximum | Mean | SD | CV (%) |
|---|---|---|---|---|---|
| Plant height (cm) | 42 | 110.33 | 71.12 | 16.49 | 23.19 |
| Leaf length (cm) | 13.67 | 33.00 | 24.83 | 3.77 | 15.20 |
| Leaf width (cm) | 0.97 | 2.30 | 1.56 | 0.28 | 17.84 |
| Head length (cm) | 5.33 | 13.83 | 9.01 | 1.91 | 21.18 |
| Stem thickness (mm) | 30.30 | 35.14 | 32.82 | 0.94 | 2.86 |
| Yield (kg hm−2) | 382.05 | 5744.1 | 2709.15 | 101.40 | 56.14 |
| No. | Plant Height (cm) | Leaf Length (cm) | Leaf Width (cm) | Head Length (cm) | Stem Thickness (mm) | Yield (kg hm−2) |
|---|---|---|---|---|---|---|
| Plant height (cm) | 1 | 0.306 ** | −0.038 | 0.404 ** | 0.002 | 0.257 * |
| Leaf length (cm) | 1 | 0.123 | 0.334 ** | −0.084 | 0.184 | |
| Leaf width (cm) | 1 | −0.100 | −0.261 * | −0.097 | ||
| Head length (cm) | 1 | −0.209 | 0.45 * | |||
| Stem thickness (mm) | 1 | 0.100 | ||||
| Yield (kg hm−2) | 1 |
| Model | χ2 (df) | χ2/df | p | RMR | RMSEA | AIC | GFI | NFI | CFI |
|---|---|---|---|---|---|---|---|---|---|
| Initial model | 5.626 (4) | 1.4065 | 0.000 | 0.051 | 0.082 | 819.022 | 0.968 | 0.913 | 0.967 |
| Final model | 12.802 (5) | 2.5604 | 0.025 | 0.021 | 0.041 | 824.197 | 0.933 | 0.803 | 0.844 |
| No. | Variety Serial Number | Color | Plant Height (cm) | Leaf Length (cm) | Leaf Width (cm) | Head Length (cm) | Stem Thickness (mm) | Yield (kg hm−2) | Composite Index |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 70 | Purple–Black | 106.67 ± 1.65 | 26.83 ± 8.36 | 1.70 ± 3.25 | 11.67 ± 5.33 | 33.50 ± 1.11 | 5571.90 ± 0.77 | 87.15 |
| 2 | 6 | Purple | 42.67 ± 0.29 | 24.00 ± 0.84 | 1.80 ± 0.11 | 9.67 ± 1.20 | 32.93 ± 4.87 | 5744.10 ± 0.20 | 81.32 |
| 3 | 11 | Purple–Black | 96.00 ± 0.09 | 28.83 ± 3.0 | 1.73 ± 0.26 | 13.83 ± 1.2 | 33.57 ± 0.87 | 5088 ± 2.29 | 79.91 |
| 4 | 3 | Purple | 72.33 ± 0.16 | 28.00 ± 0.98 | 1.57 ± 3.06 | 10.50 ± 0.42 | 33.60 ± 3.22 | 5170.35 ± 0.20 | 76.64 |
| 5 | 1 | Black | 68.67 ± 0.05 | 27.50 ± 0.44 | 2.03 ± 0.55 | 11.33 ± 0.22 | 33.87 ± 5.21 | 4839.45 ± 3.22 | 72.42 |
| 6 | 5 | Black | 58.33 ± 0.55 | 19.67 ± 0.05 | 1.80 ± 0.11 | 6.67 ± 2.0 | 32.83 ± 5.72 | 5001.30 ± 0.98 | 71.89 |
| 7 | 2 | Purple–Black | 60.33 ± 0.00 | 31.33 ± 0.55 | 1.60 ± 0.05 | 11.00 ± 3.22 | 33.89 ± 1.11 | 4472.55 ± 0.20 | 71.66 |
| 8 | 8 | Purple–Black | 62.67 ± 0.95 | 24.17 ± 1.06 | 1.97 ± 2.01 | 10.83 ± 3.21 | 33.67 ± 0.22 | 4746.60 ± 4.21 | 70.15 |
| 9 | 10 | Purple | 63.33 ± 0.01 | 25.00 ± 1.06 | 1.80 ± 0.06 | 10.67 ± 3.25 | 33.36 ± 2.28 | 4664.85 ± 0.44 | 69.21 |
| 10 | 4 | Purple | 91.00 ± 0.01 | 13.67 ± 0.20 | 1.60 ± 0.06 | 6.83 ± 0.06 | 33.12 ± 6.24 | 4768.65 ± 0.87 | 66.28 |
| Weight | Weight | ||
|---|---|---|---|
| Plant height | 0.16532 | ||
| Leaf area index | 0.55325 | Leaf length | 0.05961 |
| Leaf width | 0.94039 | ||
| Head | 0.10456 | Head length | 0.63697 |
| Stem thickness | 0.36303 | ||
| Yield | 0.17686 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Gao, X.; Qi, G.; Wurilige; Guo, L.; Zhang, M.; Fu, Y.; Wang, Y.; Wang, J.; Wang, Y.; et al. Research on the Effects of the Relationship between Agronomic Traits and Dwarfing Genes on Yield in Colored Wheat. Genes 2024, 15, 649. https://doi.org/10.3390/genes15060649
Li W, Gao X, Qi G, Wurilige, Guo L, Zhang M, Fu Y, Wang Y, Wang J, Wang Y, et al. Research on the Effects of the Relationship between Agronomic Traits and Dwarfing Genes on Yield in Colored Wheat. Genes. 2024; 15(6):649. https://doi.org/10.3390/genes15060649
Chicago/Turabian StyleLi, Wurijimusi, Xinmei Gao, Geqi Qi, Wurilige, Longyu Guo, Mingwei Zhang, Ying Fu, Yingjie Wang, Jingyu Wang, Ying Wang, and et al. 2024. "Research on the Effects of the Relationship between Agronomic Traits and Dwarfing Genes on Yield in Colored Wheat" Genes 15, no. 6: 649. https://doi.org/10.3390/genes15060649
APA StyleLi, W., Gao, X., Qi, G., Wurilige, Guo, L., Zhang, M., Fu, Y., Wang, Y., Wang, J., Wang, Y., Yang, F., Gao, Q., Fan, Y., Wen, L., Li, F., Bai, X., Zhao, Y., Gun-Aajav, B., & Xu, X. (2024). Research on the Effects of the Relationship between Agronomic Traits and Dwarfing Genes on Yield in Colored Wheat. Genes, 15(6), 649. https://doi.org/10.3390/genes15060649





