Satellitome Analysis in the Southern Lapwing (Vanellus chilensis) Genome: Implications for SatDNA Evolution in Charadriiform Birds
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples, Chromosome Preparations, and C-Banding
2.2. DNA Extraction and Genome Sequencing
2.3. Characterization of Satellitome and BLAST Search of VchSatDNAs
2.4. Primer Design and VchSatDNAs Amplification by Polymerase Chain Reaction (PCR)
2.5. Fluorescence In Situ Hybridizations (FISH)
2.6. Comparative Genomic Hybridization (CGH): Experimental Design and Probe Preparation
3. Results
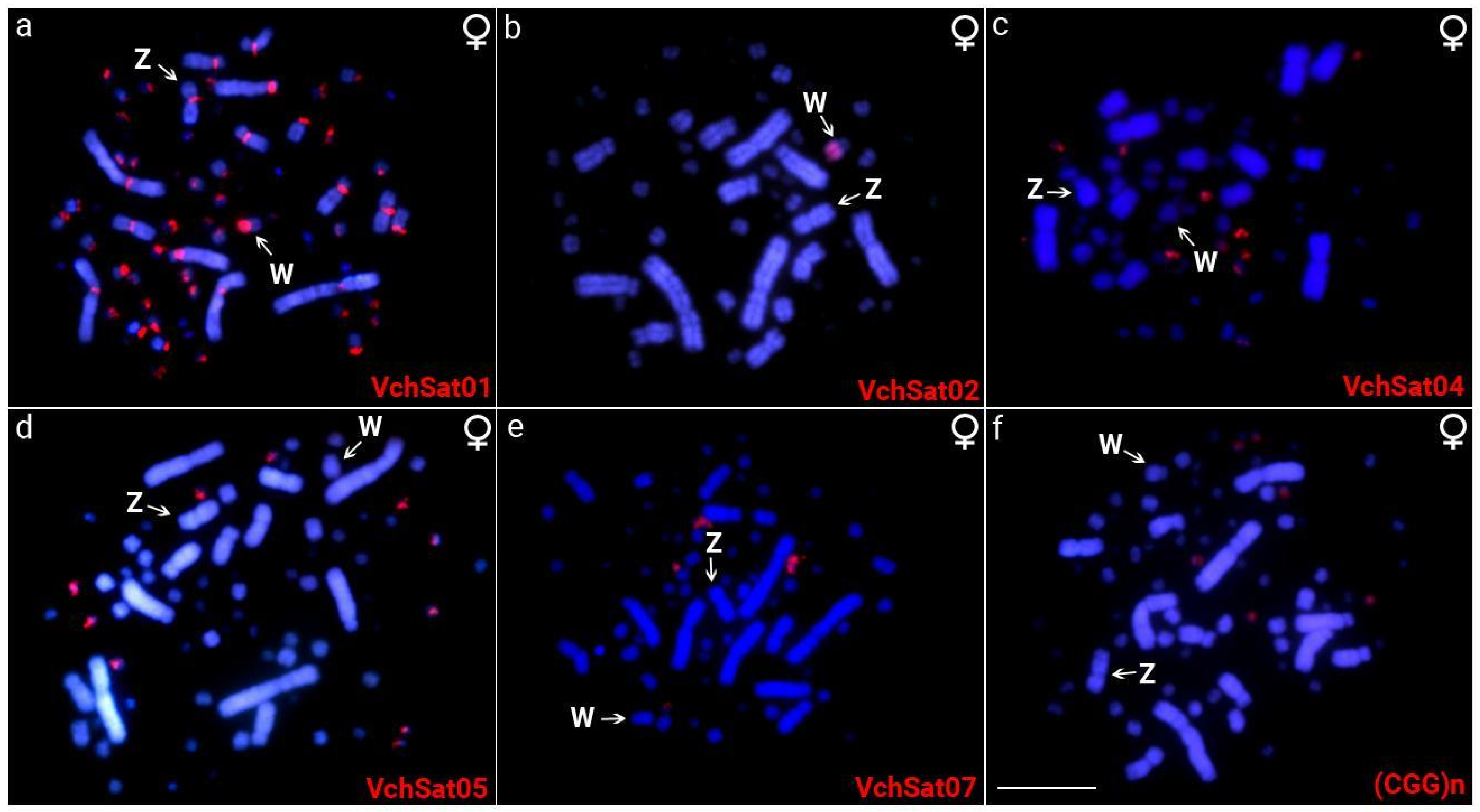
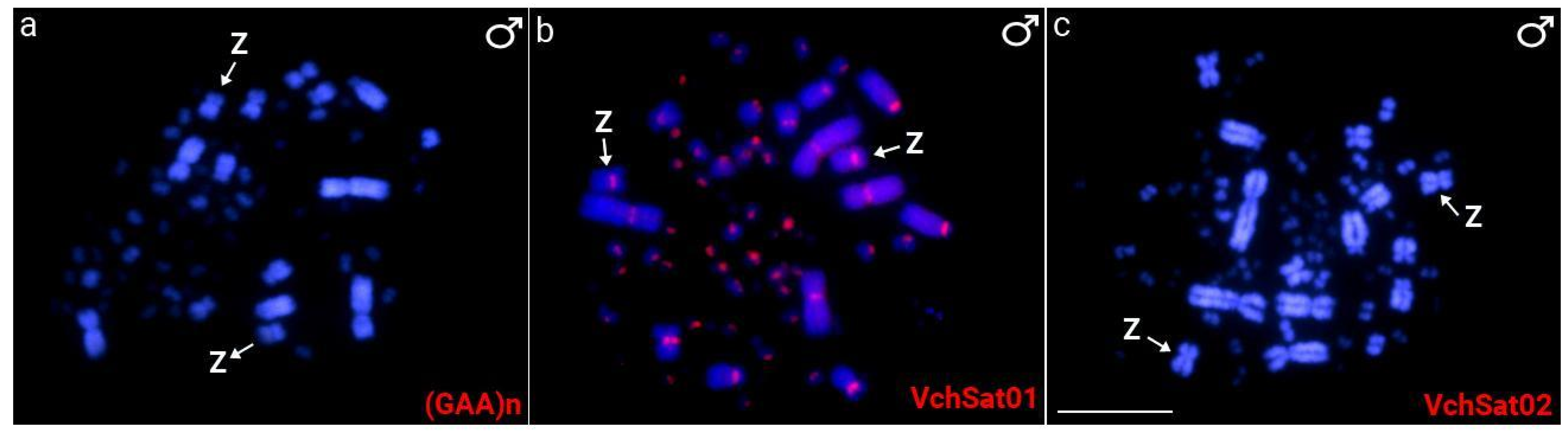
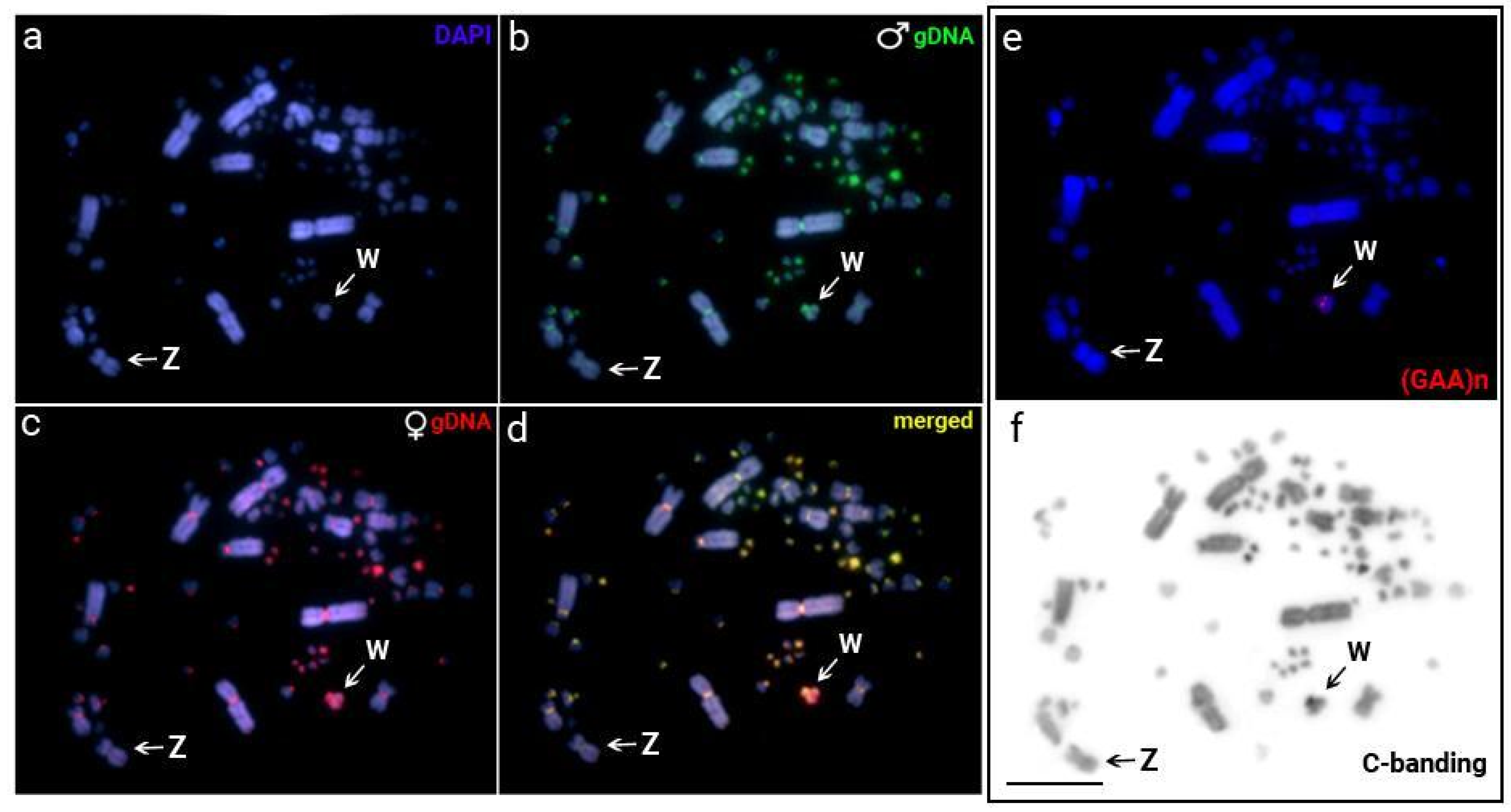
3.1. Cytogenetic Data
3.2. General Characterization of VCH Satellitome
3.3. Chromosomal Distribution of VchSatDNAs and Microsatellites
3.4. Intraspecific Comparative Genomic Hybridizations (CGH)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Griffin, D.K.; Robertson, L.B.W.; Tempest, H.G.; Skinner, B.M. The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenet. Genome Res. 2007, 117, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Kretschmer, R.; Ferguson-Smith, M.A.; De Oliveira, E.H.C. Karyotype Evolution in Birds: From Conventional Staining to Chromosome Painting. Genes 2018, 9, 181. [Google Scholar] [CrossRef] [PubMed]
- Degrandi, T.M.; Barcelos, S.A.; Costa, A.L.; Garnero, A.d.V.; Hass, I.; Gunski, R.J. Introducing the Bird Chromosome Database: An overview of cytogenetic studies on birds. Cytogenet. Genome Res. 2020, 160, 199–205. [Google Scholar] [CrossRef]
- O’Connor, R.E.; Kretschmer, R.; Romanov, M.N.; Griffin, D.K. A bird’s-eye view of chromosomic evolution in the class Aves. Cells 2024, 13, 310. [Google Scholar] [CrossRef]
- Nie, W.; O’Brien, P.C.M.; Ng, B.L.; Fu, B.; Volobouev, V.; Carter, N.P.; Ferguson-Smith, M.A.; Yang, F. Avian comparative genomics: Reciprocal chromosome painting between domestic chicken (Gallus gallus) and the stone curlew (Burhinus oedicnemus, Charadriiformes)—An atypical species with low diploid number. Chromosome Res. 2009, 17, 99–113. [Google Scholar] [CrossRef]
- Hansmann, T.; Nanda, I.; Volobouev, V.; Yang, F.; Schartl, M.; Haaf, T.; Schmid, M. Cross-species chromosome painting corroborates microchromosome fusion during karyotype evolution of Birds. Cytogenet. Genome Res. 2009, 126, 281–304. [Google Scholar] [CrossRef]
- Kretschmer, R.; Gunski, R.J.; Garnero, A.D.V.; O’Brien, P.C.; Ferguson-Smith, M.A.; De Freitas, T.R.O.; de Oliveira, E.H.C. Chromosome painting in Vanellus chilensis: Detection of a fusion common to clade Charadrii (Charadriiformes). Cytogenet. Genome Res. 2015, 146, 58–63. [Google Scholar] [CrossRef]
- Kretschmer, R.; de Souza, M.S.; Barcellos, S.A.; Degrandi, T.M.; Pereira, J.C.; O’Brien, P.C.M.; Ferguson-Smith, M.A.; Gunski, R.J.; Garnero, A.D.V.; de Oliveira, E.H.C.; et al. Novel insights into chromosome evolution of Charadriiformes: Extensive genomic reshuffling in the wattled jacana (Jacana jacana, Charadriiformes, Jacanidae). Genet. Mol. Biol. 2020, 43, e20190236. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, M.L.S.; Nagamachi, C.Y.; Ribas, T.F.A.; Diniz, C.G.; O’Brien, P.C.M.; Ferguson-Smith, M.A.; Yang, F.; Pieczarka, J.C. Chromosomal painting in Charadrius collaris Vieillot, 1818 and Vanellus chilensis Molina, 1782 and an analysis of chromosomal signatures in Charadriiformes. PLoS ONE 2022, 17, e0272836. [Google Scholar] [CrossRef]
- de Souza, M.S.; Barcellos, S.A.; dos Santos, M.d.S.; Gunski, R.J.; Garnero, A.d.V.; de Oliveira, E.H.C.; O’Connor, R.E.; Griffin, D.K.; Kretschmer, R. Microchromosome BAC-FISH Reveals Different Patterns of Genome Organization in Three Charadriiformes Species. Animals 2022, 12, 3052. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ruano, F.J.; López-León, M.D.; Cabrero, J.; Camacho, J.P.M. High-throughput analysis of the satellitome illuminates satellite DNA evolution. Sci. Rep. 2016, 6, 28333. [Google Scholar] [CrossRef]
- Garrido-Ramos, M.A. Satellite DNA: An Evolving Topic. Genes 2017, 8, 230. [Google Scholar] [CrossRef] [PubMed]
- Šatović-Vukšić, E.; Plohl, M. Satellite DNAs—From Localized to Highly Dispersed Genome Components. Genes 2023, 14, 742. [Google Scholar] [CrossRef]
- Ferretti, A.B.S.M.; Milani, D.; Palacios-Gimenez, O.M.; Ruiz-Ruano, F.J.; Cabral-de-Mello, D.C. High dynamism for neo-sex chromosomes: Satellite DNAs reveal complex evolution in a grasshopper. Heredity 2020, 125, 124–137. [Google Scholar] [CrossRef]
- Cabral-de-Mello, D.C.; Zrzavá, M.; Kubíčková, S.; Rendón, P.; Marec, F. The role of satellite DNAs in genome architecture and sex chromosome evolution in Crambidae moths. Front. Genet. 2021, 12, 661417. [Google Scholar] [CrossRef] [PubMed]
- Kretschmer, R.; Goes, C.A.G.; Bertollo, L.A.C.; Ezaz, T.; Porto-Foresti, F.; Toma, G.A.; Utsunomia, R.; Cioffi, M.B. Satellitome analysis illuminates the evolution of ZW sex chromosomes of Triportheidae fishes (Teleostei: Characiformes). Chromosoma 2022, 131, 29–45. [Google Scholar] [CrossRef]
- Flynn, J.M.; Hu, K.B.; Clark, A.G. Three recent sex chromosome-to-autosome fusions in a Drosophila virilis strain with high satellite DNA content. Genetics 2023, 224, iyad062. [Google Scholar] [CrossRef]
- Brown, J.E.; Jones, K.W. Localization of satellite DNA in the microchromosomes of the Japanese quail by in situ hybridization. Chromosoma 1972, 38, 313–318. [Google Scholar] [CrossRef]
- Yamada, K.; Yamada, K.; Nishida-Umehara, C.; Matsuda, Y. Characterization and chromosomal distribution of novel satellite DNA sequences of the lesser rhea (Pterocnemia pennata) and the greater rhea (Rhea americana). Chromosome Res. 2002, 10, 513–523. [Google Scholar] [CrossRef]
- Yamada, K.; Nishida-Umehara, C.; Matsuda, Y. A new family of satellite DNA sequences as a major component of centrometric heterochromatin in owls (Strigiformes). Chromosoma 2004, 112, 277–287. [Google Scholar] [CrossRef]
- Deryusheva, S.; Krasikova, A.; Kulikova, T.; Gaginskaya, E. Tandem 41-bp repeats in chicken and Japanese quail genomes: FISH mapping and transcription analysis on lampbrush chromosomes. Chromosoma 2007, 116, 519–530. [Google Scholar] [CrossRef]
- Liangouzov, I.A.; Derjusheva, S.E.; Saifitdinova, A.F.; Malykh, A.G.; Gaginskaya, E.R. Monomers of a satellite DNA sequence of chaffinch (Fringilla coelebs L., Aves: Passeriformes) contain short clusters of the TTAGGG repeat. Russ. J. Genet. 2002, 38, 1359–1364. [Google Scholar] [CrossRef]
- Weissensteiner, M.H.; Pang, A.W.C.; Bunikis, I.; Höijer, I.; Vinnere-Petterson, O.; Suh, A.; Wolf, J.B.W. Combination of short-read, long-read, and optical mapping assemblies reveals large-scale tandem repeat arrays with population genetic implications. Genome Res. 2017, 27, 697–708. [Google Scholar] [CrossRef]
- Kulak, M.; Komissarov, A.; Fillon, V.; Tsukanova, K.; Saifitdinova, A.; Galkina, S. Genome organization of major tandem repeats and their specificity for heterochromatin of macro- and microchromosomes in Japanese quail. Genome 2022, 65, 391–403. [Google Scholar] [CrossRef]
- Peona, V.; Kutschera, V.E.; Blom, M.P.K.; Irestedt, M.; Suh, A. Satellite DNA evolution in Corvoidea inferred from short and long reads. Mol. Ecol. 2023, 32, 1288–1305. [Google Scholar] [CrossRef]
- Oliveira, A.M.; Souza, G.M.; Toma, G.A.; Santos, N.; Santos, R.Z.; Goes, C.A.G.; Deon, G.A.; Setti, P.G.; Porto-Foresti, F.; Utsunomia, R.; et al. Satellite DNAs, heterochromatin, and sex chromosomes of the wattled jacana (Charadriiformes; Jacanidae): A species with highly rearranged karyotype. Genome, 2024; in press. [Google Scholar] [CrossRef]
- Setti, P.G.; Deon, G.A.; Santos, R.Z.; Goes, C.A.G.; Garnero, A.D.V.; Gunski, R.J.; de Oliveira, E.H.C.; Porto-Foresti, F.; de Freitas, T.R.O.; Silva, F.A.O.; et al. Evolution of bird sex chromosomes: A cytogenomic approach in Palaeognathae species. BMC Ecol. Evol. 2024; in press. [Google Scholar]
- Weissensteiner, M.H.; Suh, A. Repetitive DNA: The Dark Matter of Avian Genomics. In Avian Genomics in Ecology and Evolution; Springer: Berlin/Heidelberg, Germany, 2019; pp. 93–150. [Google Scholar]
- Gill, F.; Donsker, D.; Rasmussen, P. IOC World Bird List (v13.2). 2023. Available online: https://www.worldbirdnames.org/ioc-lists/crossref/ (accessed on 1 December 2023).
- Hammar, B. The karyotypes of thirty-one birds. Hereditas 1970, 65, 29–58. [Google Scholar] [CrossRef]
- Samanta, M.; Prasad, R. Chromosome complement and C-banding patterns in some Indian Chradiform birds. J. Zool. Syst. Evol. 1981, 20, 100–111. [Google Scholar] [CrossRef]
- Bhunya, S.P.; Mohanty, M.K. Chromosome evolution in two families of Charadriiform birds. Caryologia 1990, 43, 79–85. [Google Scholar] [CrossRef]
- Christidis, L. Animal Cytogenetics 4: Chordata 3 B: Aves; Gebrüder Borntraeger: Berlin, Germany, 1990; pp. 20–59. [Google Scholar]
- Bian, X.; Cai, H.; Li, Q.; Shao, T.; Cui, Z.; Wu, C.; Dai, E.; Xu, Z. Studies on the karyotypes of Birds XIV.14 Species of Charadriiform Birds (Aves). Zool. Res. 1993, 14, 86–90. [Google Scholar]
- Ebied, A.M.; Hassan, H.A.; Almaaty, A.H.A.; Yaseen, A.E. Karyotypic characterization of ten species of birds. Cytologia 2005, 70, 181–194. [Google Scholar] [CrossRef]
- Sasaki, M.; Ikeuchi, T.; Makino, S. A feather pulp culture technique for avian chromosomes, with notes on the chromosomes of the peafowl and the ostrich. Experientia 1968, 24, 1292–1293. [Google Scholar] [CrossRef]
- Sumner, A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972, 75, 304–306. [Google Scholar] [CrossRef]
- Lui, R.L.; Blanco, D.R.; Moreira-Filho, O.; Margarido, V.P. Propidium iodide for making heterochromatin more evident in the C-banding technique. Biotech. Histochem. 2012, 87, 433–438. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Novák, P.; Neumann, P.; Macas, J. Global analysis of repetitive DNA from unassembled sequence reads using RepeatExplorer2. Nat. Protoc. 2020, 15, 3745–3776. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Smith, A.F.A.; Hubley, R.; Green, P. RepeatMasker Open-3.0 1996–2010. 2020. Available online: https://repeatmasker.org/ (accessed on 20 December 2023).
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Kretschmer, R.; Santos, M.S.; Furo, I.O.; Oliveira, E.H.C.; Cioffi, M.B. FISH–in birds. In Cytogenetics and Molecular Cytogenetics; Liehr, T., Ed.; CRC Press: Boca Raton, FL, USA, 2023. [Google Scholar]
- Zwick, M.S.; Hanson, R.E.; Islam-Faridi, M.N.; Stelly, D.M.; Wing, R.A.; Price, H.J.; McKnight, T.D. A rapid procedure for the isolation of C 0 t-1 DNA from plants. Genome 1997, 40, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Traut, W.; Sahara, K.; Otto, T.D.; Marec, F. Molecular differentiation of sex chromosomes probed by comparative genomic hybridization. Chromosoma 1999, 108, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Síchová, J.; Nguyen, P.; Dalíková, M.; Marec, F. Chromosomal evolution in tortricid moths: Conserved karyotypes with diverged features. PLoS ONE 2013, 8, e64520. [Google Scholar] [CrossRef] [PubMed]
- Fuková, I.; Nguyen, P.; Marec, F. Codling moth cytogenetics: Karyotype, chromosomal location of rDNA, and molecular differentiation of sex chromosomes. Genome 2005, 48, 1083–1092. [Google Scholar] [CrossRef]
- Burt, D.W. Origin and evolution of avian minichromosomes. Cytogenet. Genome Res. 2002, 96, 97–112. [Google Scholar] [CrossRef]
- Voleníková, A.; Lukšíková, K.; Mora, P.; Pavlica, T.; Altmanová, M.; Štundlová, J.; Pelikánová, S.; Simanovsky, S.A.; Jankásek, M.; Reichard, M.; et al. Fast satellite DNA evolution in Nothobranchius annual killifishes. Chromosome Res. 2023, 31, 33. [Google Scholar] [CrossRef]
- Anjos, A.; Milani, D.; Bardella, V.B.; Paladini, A.; Cabral-de-Mello, D.C. Evolution of satDNAs on holocentric chromosomes: Insights from hemipteran insects of the genus Mahanarva. Chromosome Res. 2023, 31, 5. [Google Scholar] [CrossRef]
- Vozdova, M.; Kubickova, S.; Martínková, N.; Galindo, D.J.; Bernegossi, A.M.; Cernohorska, H.; Kadlcikova, D.; Musilová, P.; Duarte, J.M.; Rubes, J. Satellite DNA in Neotropical Deer Species. Genes 2021, 12, 123. [Google Scholar] [CrossRef]
- Boštjančić, L.L.; Bonassin, L.; Anušić, L.; Lovrenčić, L.; Besendorfer, V.; Maguire, I.; Grandjean, F.; Austin, C.M.; Greve, C.; Hamadou, A.B.; et al. The Pontastacus leptodactylus (Astacidae) Repeatome Provides Insight into Genome Evolution and Reveals Remarkable Diversity of Satellite DNA. Front. Genet. 2021, 11, 611745. [Google Scholar] [CrossRef]
- Garrido-Ramos, M.A. Satellite DNA in plants: More than just rubbish. Cytogenet. Genome Res. 2015, 146, 153–170. [Google Scholar] [CrossRef] [PubMed]
- Talbert, P.B.; Henikoff, S. What makes a centromere? Exp. Cell Res. 2020, 389, 111895. [Google Scholar] [CrossRef]
- Thakur, J.; Packiaraj, J.; Henikoff, S. Sequence, Chromatin and Evolution of Satellite DNA. Int. J. Mol. Sci. 2021, 22, 4309. [Google Scholar] [CrossRef]
- Henikoff, S.; Ahmad, K.; Malik, H.S. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 2001, 293, 1098–1102. [Google Scholar] [CrossRef]
- O’Neill, R.J.; Eldridge, M.D.B.; Metcalfe, C.J. Centromere Dynamics and Chromosome Evolution in Marsupials. J. Hered. 2004, 95, 375–381. [Google Scholar] [CrossRef]
- Plohl, M.; Meštrović, N.; Mravina, B. Centromere identity from the DNA point of view. Chromosoma 2014, 123, 313–325. [Google Scholar] [CrossRef] [PubMed]
- Ferree, P.M.; Barbash, D.A. Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol. 2009, 7, e1000234. [Google Scholar] [CrossRef] [PubMed]
- Plohl, M.; Meštrović, N.; Mravinac, B. Satellite DNA Evolution. Genome Dyn. 2012, 7, 126–152. [Google Scholar] [CrossRef]
- Madsen, C.S.; Brooks, J.E.; de Kloet, E.; de Kloet, S.R. Sequence conservation of an avian centromeric repeated DNA component. Genome 1994, 37, 351–355. [Google Scholar] [CrossRef]
- International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 2004, 432, 695–716. [Google Scholar] [CrossRef] [PubMed]
- Pigozzi, M.I. Diverse stages of sex-chromosome differentiation in tinamid birds: Evidence from crossover analysis in Eudromia elegans and Crypturellus tataupa. Genetica 2011, 139, 771–777. [Google Scholar] [CrossRef] [PubMed]
- Nishida, C.; Ishijima, J.; Ishishita, S.; Yamada, K.; Griffin, D.K.; Yamazaki, T.; Matsuda, Y. Karyotype reorganization with conserved genomic compartmentalization in dot-shaped microchromosomes in the Japanese mountain hawk-eagle (Nisaetus nipalensis orientalis, Accipitridae). Cytogenet. Genome Res. 2013, 141, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, Y.; Nishida-Umehara, C.; Ishijima, J.; Yamada, K.; Matsuda, Y. Comparison of the Z and W sex chromosomal architectures in elegant crested tinamou (Eudromia elegans) and ostrich (Struthio camelus) and the process of sex chromosome differentiation in palaeognathous birds. Chromosoma 2007, 116, 159–173. [Google Scholar] [CrossRef]
- Palacios-Gimenez, O.; Dias, G.B.; de Lima, L.G.; Kuhn, G.C.S.; Ramos, É.; Martins, C.; Cabral-de-Mello, D.C. High-throughput analysis of the satellitome revealed enormous diversity of satellite DNAs in the neo-Y chromosome of the cricket Eneoptera surinamensis. Sci. Rep. 2017, 7, 6422. [Google Scholar] [CrossRef]
- Gatto, K.; Mattos, J.V.; Seger, K.R.; Lourenço, L.B. Sex chromosome differentiation in the frog genus Pseudis involves satellite DNA and chromosome rearrangements. Front. Genet. 2018, 9, 301. [Google Scholar] [CrossRef]
- Crepaldi, C.; Martí, E.; Gonçalves, É.M.; Martí, D.A.; Parise-Maltempi, P.P. Genomic differences between the sexes in a fish species seen through satellite DNAs. Front. Genet. 2021, 12, 728670. [Google Scholar] [CrossRef]
- da Silva, M.J.; Gazoni, T.; Haddad, C.F.B.; Parise-Maltempi, P.P. Analysis in Proceratophrys boiei genome illuminates the satellite DNA content in a frog from the Brazilian Atlantic forest. Front. Genet. 2023, 14, 1101397. [Google Scholar] [CrossRef]
- Li, N.; Li, X.; Zhou, J.; Yu, L.; Li, S.; Zhang, Y.; Qin, R.; Gao, W.; Deng, C. Genome-Wide Analysis of Transposable Elements and Satellite DNAs in Spinacia Species to Shed Light on Their Roles in Sex Chromosome Evolution. Front. Plant Sci. 2021, 11, 575462. [Google Scholar] [CrossRef]
- Cabral-de-Mello, D.C.; Mora, P.; Rico-Porras, J.M.; Ferretti, A.B.S.M.; Palomeque, T.; Lorite, P. The spread of satellite DNAs in euchromatin and insights into the multiple sex chromosome evolution in Hemiptera revealed by repeatome analysis of the bug Oxycarenus hyalinipennis. Insect Mol. Biol. 2023, 32, 725–737. [Google Scholar] [CrossRef]
- Fridolfsson, A.K.; Cheng, H.; Copeland, N.G.; Jenkins, N.A.; Liu, H.C.; Raudsepp, T.; Woodage, T.; Chowdhary, B.; Halverson, J.; Ellegren, H. Evolution of the avian sex chromosomes from an ancestral pair of autosomes. Proc. Natl. Acad. Sci. USA 1998, 95, 8147–8152. [Google Scholar] [CrossRef]
- Stefos, K.; Arrighi, F.E. Heterochromatic nature of W chromosome in birds. Exp. Cell Res. 1971, 68, 228–231. [Google Scholar] [CrossRef] [PubMed]
- Stiglec, R.; Ezaz, T.; Graves, J.A.M. A new look at the evolution of avian sex chromosomes. Cytogenet. Genome Res. 2007, 117, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Nishida, C.; Ishishita, S.; Yamada, K.; Griffin, D.K.; Matsuda, Y. Dynamic chromosome reorganization in the osprey (Pandion haliaetus, Pandionidae, Falconiformes): Relationship between chromosome size and the chromosomal distribution of centromeric repetitive DNA sequences. Cytogenet. Genome Res. 2014, 142, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Schartl, M.; Schmid, M.; Nanda, I. Dynamics of vertebrate sex chromosome evolution: From equal size to giants and dwarfs. Chromosoma 2016, 125, 553–571. [Google Scholar] [CrossRef] [PubMed]
- Raudsepp, T.; Houck, M.L.; O’Brien, P.C.; Ferguson-Smith, M.A.; Ryder, O.A.; Chowdhary, B.P. Cytogenetic analysis of California condor (Gymnogyps californianus) chromosomes: Comparison with chicken (Gallus gallus) macrochromosomes. Cytogenet. Genome Res. 2002, 98, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Bed’Hom, B.; Coullin, P.; Guillier-Gencik, Z.; Moulin, S.; Bernheim, A.; Volobouev, V. Characterization of the atypical karyotype of the black-winged kite Elanus caeruleus (Falconiformes: Accipitridae) by means of classical and molecular cytogenetic techniques. Chromosome Res. 2003, 11, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Handley, L.J.; Ceplitis, H.; Ellegren, H. Evolutionary strata on the chicken Z chromosome: Implications for sex chromosome evolution. Genetics 2004, 167, 367–376. [Google Scholar] [CrossRef]
- Rutkowska, J.; Lagisz, M.; Nakagawa, S. The long and the short of avian W chromosomes: No evidence for gradual W shortening. Biol. Lett. 2012, 8, 636–638. [Google Scholar] [CrossRef] [PubMed]

| Species | Sampling Location | Number and Sex of Individuals |
|---|---|---|
| J. jacana | São Gabriel, Rio Grande do Sul, Brazil (30°18′07.2″ S 54°22′51.9″ W) | 02♀ and 01♂ |
| V. chilensis | São Gabriel, Rio Grande do Sul, Brazil (30°18′07.2″ S 54°22′51.9″ W) | 01♀ and 01♂ |
| C. canutus | Belém, Pará, Brazil (1°17′24.8″ S 48°26′29.9″ W) | 01♀ and 01♂ |
| SatDNA | RUL (bp) | Abundance F (%) | Divergence F (%) | Abundance M (%) | Divergence M (%) | Abundance F/M (%) | A + T (%) |
|---|---|---|---|---|---|---|---|
| VchSat01-179 | 179 | 0.00566813534316 | 3.47 | 0.00734518650644 | 3.20 | 0.77 | 47.4% |
| VchSat02-5351 | 5351 | 0.00153609301945 | 3.92 | 0.000995645031422 | 3.39 | 1.54 | 54.0% |
| VchSat03-2447 | 2447 | 0.000786165893518 | 0.51 | 0.000954216629777 | 0.50 | 0.82 | 56.9% |
| VchSat04-145 | 145 | 0.000559347409739 | 11.61 | 0.000618731320616 | 11.50 | 0.9 | 44.1% |
| VchSat05-958 | 958 | 0.000419615999712 | 6.22 | 0.000496465807068 | 6.25 | 0.84 | 52.4% |
| VchSat06-234 | 234 | 0.000200179487108 | 6.99 | 0.000131052708244 | 10.13 | 1.52 | 37.6% |
| VchSat07-36 | 36 | 0.00000994875019722 | 6.29 | 0.00000619126971609 | 6.02 | 1.6 | 16.7% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kretschmer, R.; Toma, G.A.; Deon, G.A.; dos Santos, N.; dos Santos, R.Z.; Utsunomia, R.; Porto-Foresti, F.; Gunski, R.J.; Garnero, A.D.V.; Liehr, T.; et al. Satellitome Analysis in the Southern Lapwing (Vanellus chilensis) Genome: Implications for SatDNA Evolution in Charadriiform Birds. Genes 2024, 15, 258. https://doi.org/10.3390/genes15020258
Kretschmer R, Toma GA, Deon GA, dos Santos N, dos Santos RZ, Utsunomia R, Porto-Foresti F, Gunski RJ, Garnero ADV, Liehr T, et al. Satellitome Analysis in the Southern Lapwing (Vanellus chilensis) Genome: Implications for SatDNA Evolution in Charadriiform Birds. Genes. 2024; 15(2):258. https://doi.org/10.3390/genes15020258
Chicago/Turabian StyleKretschmer, Rafael, Gustavo A. Toma, Geize Aparecida Deon, Natalia dos Santos, Rodrigo Zeni dos Santos, Ricardo Utsunomia, Fabio Porto-Foresti, Ricardo José Gunski, Analía Del Valle Garnero, Thomas Liehr, and et al. 2024. "Satellitome Analysis in the Southern Lapwing (Vanellus chilensis) Genome: Implications for SatDNA Evolution in Charadriiform Birds" Genes 15, no. 2: 258. https://doi.org/10.3390/genes15020258
APA StyleKretschmer, R., Toma, G. A., Deon, G. A., dos Santos, N., dos Santos, R. Z., Utsunomia, R., Porto-Foresti, F., Gunski, R. J., Garnero, A. D. V., Liehr, T., de Oliveira, E. H. C., de Freitas, T. R. O., & Cioffi, M. d. B. (2024). Satellitome Analysis in the Southern Lapwing (Vanellus chilensis) Genome: Implications for SatDNA Evolution in Charadriiform Birds. Genes, 15(2), 258. https://doi.org/10.3390/genes15020258












