Evaluation of Suitable Reference Genes for Quantitative Real-Time PCR in Various Tissues of Apocynum venetum
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and PEG6000 Treatment
2.2. Total RNA Extraction and cDNA Synthesis
2.3. qRT-PCR Primer Design and Analysis
2.4. Stability Analysis
2.5. Statistical Analysis
3. Results
3.1. Selection of Candidate Reference Genes of A. venetum
3.2. Expression Levels of Candidate Reference Genes
3.3. Stability Analysis of Candidate Reference Genes
3.4. Determination of the Optimal Number of Reference Genes for Normalization
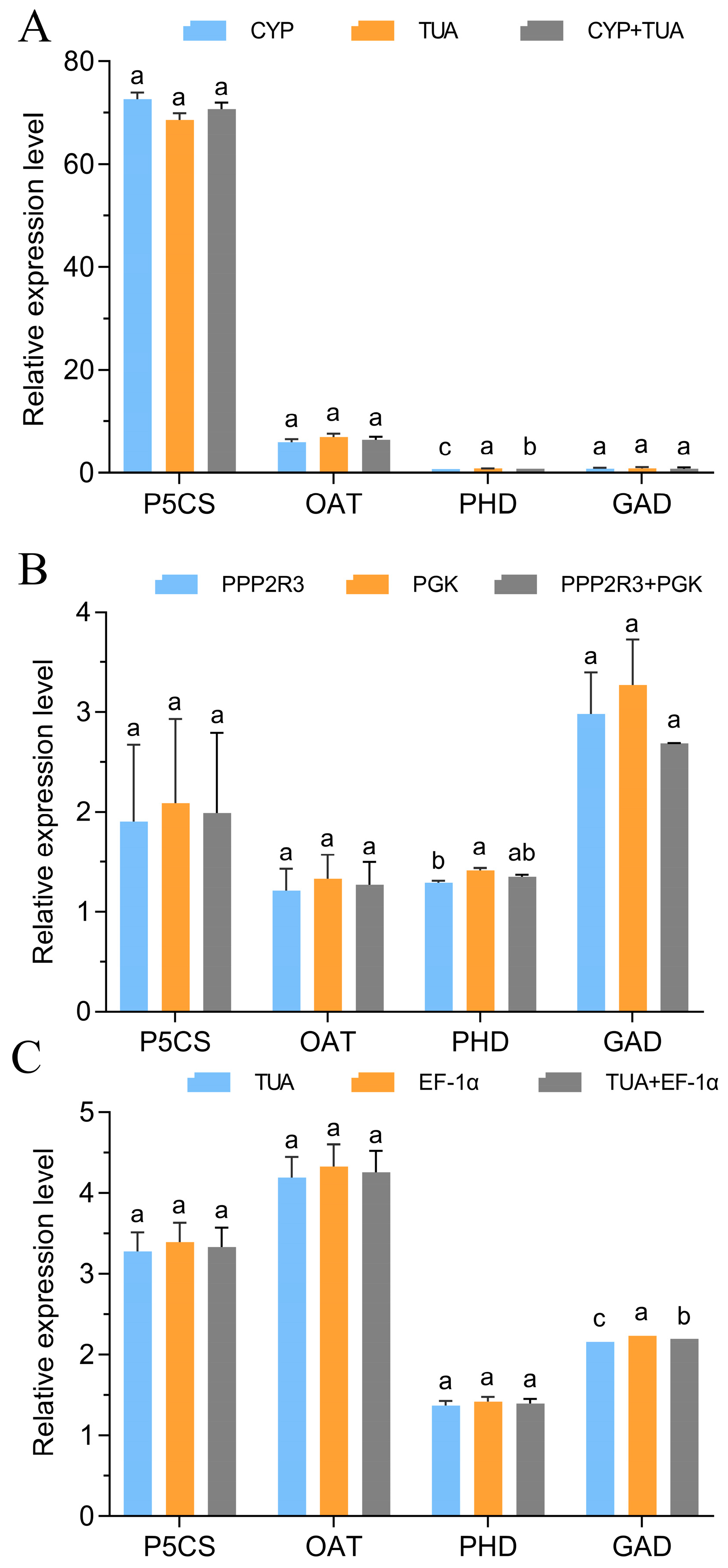
3.5. Validation of the Stability of the Chosen Reference Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| qRT-PCR | Real-time quantitative polymerase chain reaction |
| TUA | α-tubulin actin |
| TUB | β-tubulin |
| ACT | Actin |
| CYP | Cyclophilin |
| EF-1α | Elongation factor-1α |
| PPP2R | B family of regulatory subunits of the protein phosphatase |
| PGK | Phosphoglycerate kinase |
| P5CS | Δ1-pyrroline-5-carboxylate synthase |
| OAT | Ornithine δ-aminotransferase |
| PHD | Proline hydroxylase |
| GAD | Glutamic acid decarboxylase |
References
- Yan, X.; Qian, C.J.; Yin, X.Y.; Fan, X.K.; Zhao, X.Y.; Gu, M.H.; Wang, T.; Ma, X.F. A whole-transcriptome approach to evaluate reference genes for quantitative diurnal gene expression studies under natural field conditions in Tamarix ramosissima leaves. Electron. J. Biotechnol. 2018, 35, 48–56. [Google Scholar] [CrossRef]
- Gan, Y.B.; Zhou, Z.J.; An, L.J.; Bao, S.J.; Forde, B.G. A comparison between northern blotting and quantitative real-time PCR as a means of detecting the nutritional regulation of genes expressed in roots of Arabidopsis thaliana. Agr. Sci. China 2011, 3, 335–342. [Google Scholar] [CrossRef]
- Nashima, K.; Shimizu, T.; Nishitani, C.; Yamamoto, T.; Takahashi, H.; Nakazono, M.; Itai, A.; Isuzugawa, K.; Hanada, T.; Takashina, T.; et al. Microarray analysis of gene expression patterns during fruit development in European pear (Pyrus communis). Sci. Hortic. 2013, 164, 466–473. [Google Scholar] [CrossRef]
- Liu, X.; Gao, Y.B.; Zhao, X.Y.; Zhang, X.X.; Ben, L.L.; Li, Z.L.; Dong, G.C.; Zhou, J.; Huang, J.Y.; Yao, Y.L. Validation of Novel Reference Genes in Different Rice Plant Tissues through Mining RNA-Seq Datasets. Plants 2023, 12, 3946. [Google Scholar] [CrossRef]
- Hindson, C.M.; Chevillet, J.R.; Briggs, H.A.; Gallichotte, E.N.; Ruf, I.K.; Hindson, B.J.; Vessella, R.L.; Tewari, M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 2013, 10, 1003–1005. [Google Scholar] [CrossRef]
- Zeng, A.; Xu, Y.Y.; Song, L.X.; Li, J.Q.; Yan, J.Y. Validation of suitable reference genes for qRT-PCR in cabbage (Brassica oleracea L.) under different abiotic stress experimental conditions. J. Plant Biochem. Biotechnol. 2021, 30, 184–195. [Google Scholar] [CrossRef]
- Jiang, Q.; Wang, F.; Li, M.Y.; Ma, J.; Tan, G.F.; Xiong, A.S. Selection of suitable reference genes for qPCR normalization under abiotic stresses in Oenanthe javanica (BI.) DC. PLoS ONE 2014, 9, 92262. [Google Scholar] [CrossRef] [PubMed]
- Petriccione, M.; Mastrobuoni, F.; Zampella, L.; Scortichini, M. Reference gene selection for normalization of qRT-PCR gene expression data from Actinidia deliciosa leaves infected with Pseudomonas syringae pv. actinidiae. Sci. Rep. 2015, 5, 16961. [Google Scholar] [CrossRef]
- Aminfar, Z.; Rabiei, B.; Tohidfar, M.; Mirjalili, M.H. Selection and validation of reference genes for quantitative real-time PCR in Rosmarinus officinalis L. in various tissues and under elicitation. Biocatal. Agric. Biotechnol. 2019, 20, 101246. [Google Scholar] [CrossRef]
- de Almeida, M.R.; Ruedell, C.M.; Ricachenevsky, F.K.; Sperotto, R.A.; Pasquali, G.; Fett-Neto, A.G. Reference gene selection for quantitative reverse transcription-polymerase chain reaction normalization during in vitro adventitious rooting in Eucalyptus globulus Labill. BMC Mol. Biol. 2010, 11, 73. [Google Scholar] [CrossRef]
- Jia, D.H.; Wang, B.; Li, X.L.; Tan, W.; Gan, B.C.; Peng, W.H. Validation of reference genes for quantitative gene expression analysis in Auricularia cornea. J. Microbiol. Methods 2019, 163, 105658. [Google Scholar] [CrossRef]
- Xie, W.Y.; Zhang, X.Y.; Wang, T.; Hu, J.J. Botany, traditional uses, phytochemistry and pharmacology of Apocynum venetum L. (Luobuma): A review. J. Ethnopharmacol. 2012, 141, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.C.; Zhou, J.H.; Zhang, C.S.; Li, Y.Q. Review of Current Research and Utilization Status of Apocynum venetum Germplasm in China. Chin. Bull. Bot. 2018, 53, 382–390. [Google Scholar]
- Yang, J.L.; Zhang, L.X.; Jiang, L.; Zhan, Y.G.; Fan, G.Z. Quercetin alleviates seed germination and growth inhibition in Apocynum venetum and Apocynum pictum under mannitol-induced osmotic stress. Plant Physiol. Biotechnol. 2021, 159, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Kong, N.N.; Fang, S.T.; Liu, Y.; Wang, J.H.; Yang, C.Y.; Xia, C.H. Flavonoids from the halophyte Apocynum venetum and their antifouling activities against marine biofilm-derived bacteria. Nat. Prod. Res. 2014, 28, 928–931. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Dong, Z.; Chang, X.J.; Zhang, C.H.; Rong Gh Gao, X.D.; Zeng, Z.; Wang, C.P.; Chen, Y.; Rong, Y.H.; Qu, J.H.; et al. Protective effect of the total flavonoids from Apocynum venetum L. on carbon tetrachloride-induced hepatotoxicity in vitro and in vivo. J. Physiol. Biochem. 2018, 74, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Wang, L.; Tanveer, M.; Tian, C.Y. Lithium biofortification of medicinal tea Apocynum venetum. Sci. Rep. 2019, 9, 8182. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.Z.; Nie, T.T.; Huang, Y.T.; Zhan, Y.G. GSNOR deficiency enhances betulin production in Betula platyphylla. Trees 2018, 32, 847–853. [Google Scholar] [CrossRef]
- Wang, Z.L.; Xu, J.Y.; Liu, Y.H.; Chen, J.Y.; Lin, H.F.; Huang, Y.L.; Bian, X.H.; Zhao, Y.C. Selection and validation of appropriate reference genes for real-time quantitative PCR analysis in Momordica charantia. Phytochemistry 2019, 164, 1–11. [Google Scholar] [CrossRef]
- Xie, F.L.; Wang, J.Y.; Zhang, B.H. RefFinder: A web-based tool for comprehensively analyzing and identifying reference genes. Funct. Integr. Genom. 2023, 23, 125. [Google Scholar] [CrossRef]
- Per, T.S.; Khan, N.A.; Reddy, P.S.; Masood, A.; Hasanuzzaman, M.; Khan, M.I.R.; Anjum, N.A. Approaches in modulating proline metabolism in plants for salt and drought stress tolerance: Phytohormones, mineral nutrients and transgenics. Plant Physiol. Biochem. 2017, 115, 126–140. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time uantitative PCR and the 2-ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Gong, H.; Sun, L.; Chen, B.D.; Han, Y.W.; Pang, J.; Wu, W.; Qi, R.M.; Zhang, T.M. Evaluation of candidate reference genes for qRT-PCR studies in three metabolism related tissues of mice after caloric restriction. Sci. Rep. 2016, 6, 38513. [Google Scholar] [CrossRef]
- Zhang, T.T.; Li, M.; Zhan, Y.G.; Fan, G.Z. Dataset of full-length transcriptome assembly and annotation of apocynum venetum using pacbio sequel II. Data Brief 2020, 33, 106494. [Google Scholar] [CrossRef]
- Xiang, T.; Wu, L.J.; Isah, M.B.; Chen, C.; Zhang, X.Y. Apocynum venetum, a medicinal, economical and ecological plant: A review update. PeerJ 2023, 11, e14966. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, F.J.; Guo, H.Y.; Ma, H.; Chen, H.; Song, Y.Y.; Chen, P.; Xu, Q.L. Selection of reference genes for quantitative real-time PCR analysis in Lathyrus sativus L. under different development stages and drought stress. Genet. Resour. Crop Evol. 2022, 69, 2319–2330. [Google Scholar] [CrossRef]
- Clark, A.R.; Ohlmeyer, M. Protein phosphatase 2A as a therapeutic target in inflammation and neurodegeneration. Pharmacol. Therapeut 2019, 201, 181–201. [Google Scholar] [CrossRef]
- Luo, X.Y.; Shi, T.; Sun, H.L.; Song, J.; Ni, Z.J.; Gao, Z.H. Selection of suitable inner reference genes for normalisation of microRNA expression response to abiotic stresses by qRT-PCR in leaves, flowers and young stems of peach. Sci. Hortic. 2014, 165, 281–287. [Google Scholar] [CrossRef]
- Liu, M.; Wu, Z.; Jiang, F.L. Selection and validation of garlic reference genes for quantitative real-time PCR normalization. Plant Cell Tissue Organ Cult. (PCTOC) 2015, 122, 435–444. [Google Scholar] [CrossRef]
- Zhou, X.H.; Liu, J.; Zhuang, Y. Selection of Appropriate Reference Genes in Solanum aculeatissimum for Quantitative Gene Expression Studies Under Different Experimental Conditions. Acta Hortic. Sin. 2014, 41, 1731–1738. [Google Scholar]
- Jia, Y.; Liu, S.C.; Zhao, J.; Chen, X.X.; Sun, L.X.; Yu, X.F.; Li, X. Reference gene selection and validation by qRT-PCR during flower development and in different organs of Primula forbesii. J. Hortic. Sci. Biotech. 2020, 95, 383–394. [Google Scholar] [CrossRef]
- da Silva, F.L.; Lucho, S.R.; Klumb, E.K.; Bianchi, V.J. Selection of reference genes and expression of key genes involved in response to short-term soil flooding stress in Prunus persica rootstock ‘Tsukuba 1’. Gene Rep. 2023, 33, 101843. [Google Scholar] [CrossRef]
- Yuan, X.Y.; Jiang, S.H.; Wang, M.F.; Ma, J.; Zhang, X.Y.; Cui, B. Evaluation of Internal Control for Gene Expression in Phalaenopsis by Quantitative Real-Time PCR. Appl. Biochem. Biotechnol. 2014, 173, 1431–1445. [Google Scholar] [CrossRef]
- Cao, L.Y.; Li, X.X.; Wang, D.; Sun, H.F.; Gao, J.P. Validation of reliable reference genes for accutrate for accurate normalization in qRT-PCR analysis of Codonopsis pilosula. Chin. Herbal. Med. 2017, 9, 226–235. [Google Scholar] [CrossRef]
- Lin, Y.; Liu, G.F.; Rao, Y.; Wang, B.; Tian, R.F.; Tan, Y.Y.; Peng, T. Identification and validation of reference genes for qRT-PCR analyses under different experimental conditions in Allium wallichii. J. Plant Physiol. 2023, 281, 153925. [Google Scholar] [CrossRef]
- Chen, Y.L.; Yuan, Y.T.; Qiu, Z.H.; Hong, W.J.; Mao, K.; Yao, C.C.; Liu, Z.D.; Wang, N.N. Identification of reference genes for RT-qPCR analysis across kiwifruit species under iron deficiency conditions. Sci. Hortic. 2023, 312, 111865. [Google Scholar] [CrossRef]
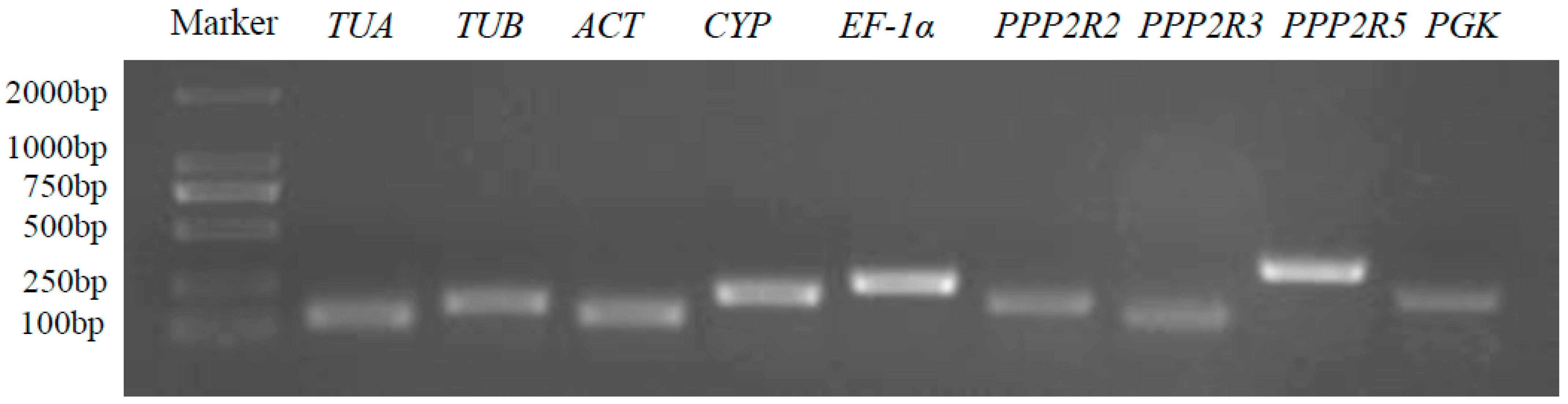
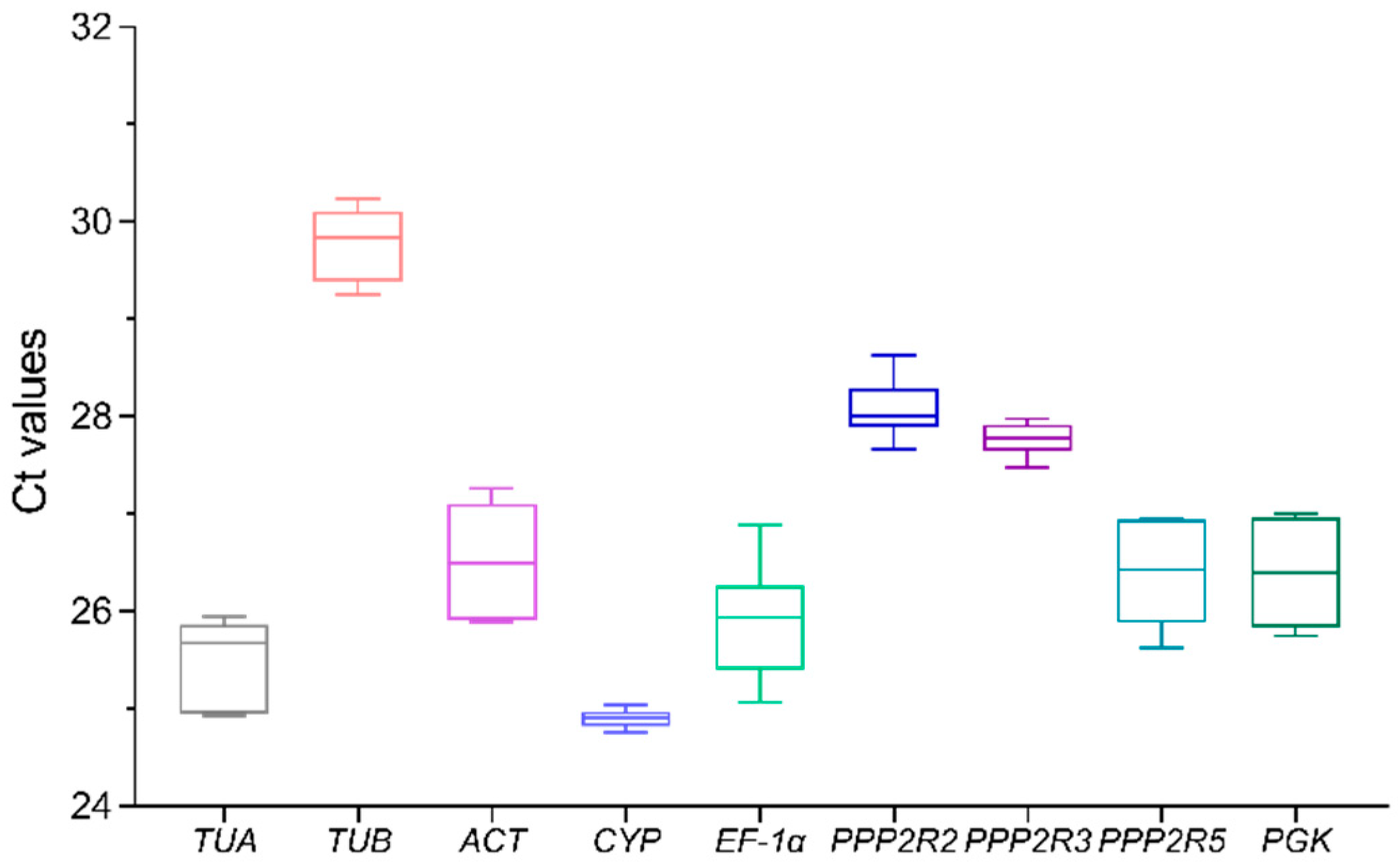
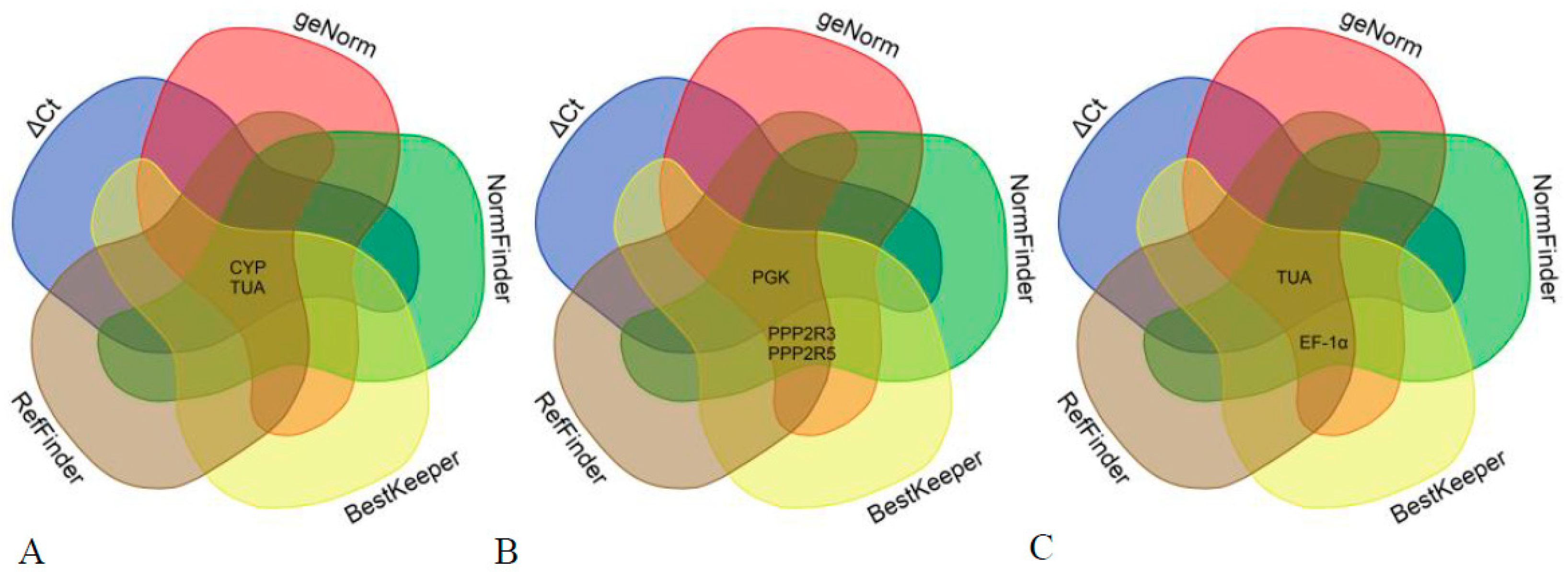
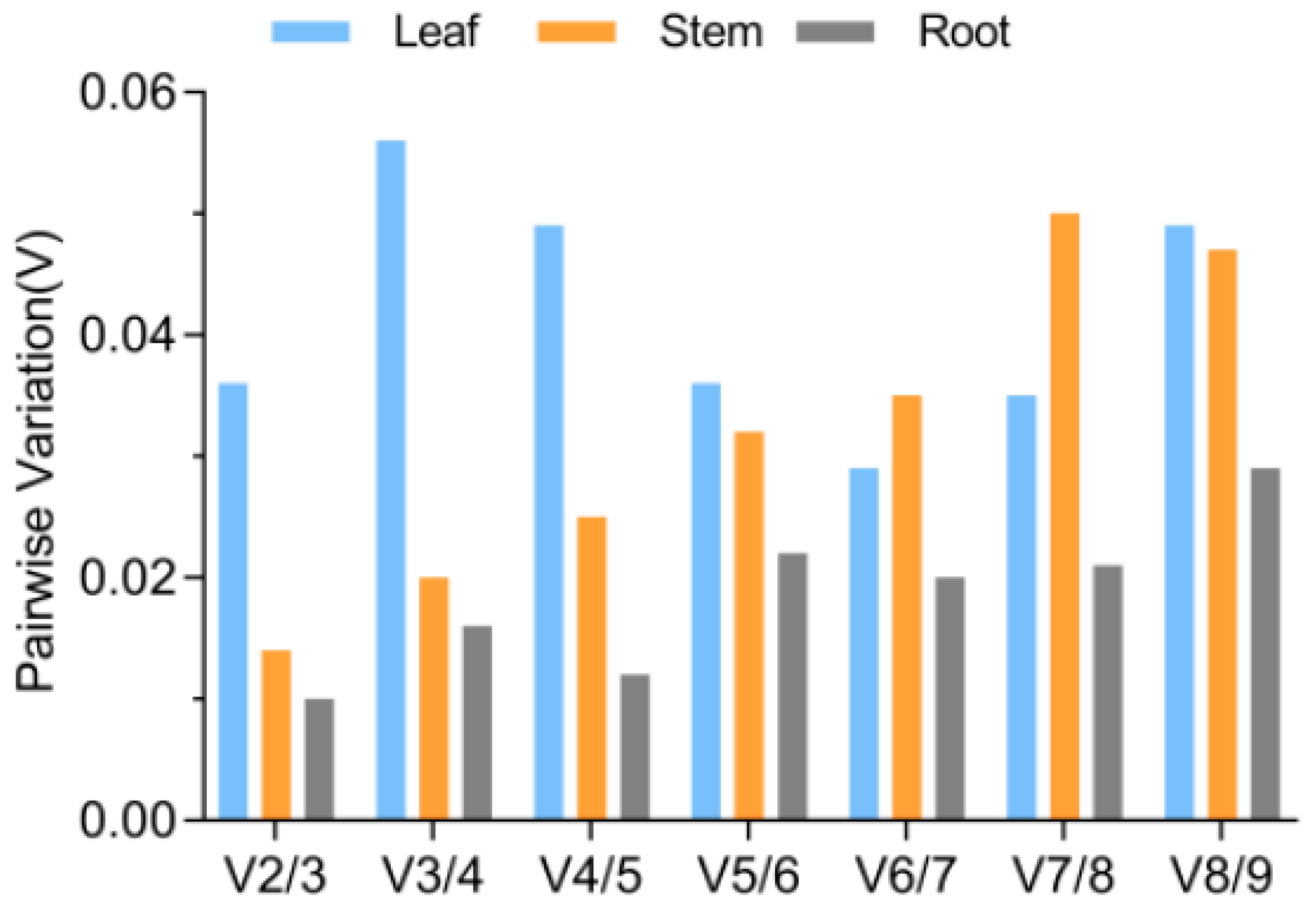
| Gene | Gene Name | Primer Sequences (5′-3′) | Amplicon Product Length (bp) |
|---|---|---|---|
| Abbreviation | (Forward/Reverse) | ||
| TUA | α-tubulin | F:CTTCTTCAGCGAAACTGGGTC; | 152 |
| R:TGTTAGCGGCGTCTTCCTTT | |||
| TUB | β-tubulin | F:GTGGTCGTTATGTCCCTCGTG; | 176 |
| R:ATCAATCAACTCGGCACCCT | |||
| ACT | Actin | F:TTATCATTACCTACACCACCTCC; | 135 |
| R:TGTAAGACTGAAGCCATCTCG | |||
| CYP | Cyclophilin | F:AGGGTGAAAAGGAAGATGAAG; | 206 |
| R:GTACAACAAGTTCTGGGGTGATA | |||
| EF-1α | Elongation factor-1α | F:TTGGAGGTATTGACAAGCGAGTA; | 252 |
| R:TCAACACAGCACAATCAGCCTG | |||
| PPP2R2 | Encoding B/R2/PR55 | F:TTACATGACCCTTAAGTTATGGG; | 173 |
| R:AGGAACCAGTTGCTACACGCA | |||
| PPP2R3 | Encoding B″/R3/PR72 | F:TTTCTTATTCGTCAGGAGCGT; | 129 |
| R:CCAAACATCTGCACTTCCGT | |||
| PPP2R5 | Encoding B′/R5/PR61/B56 | F:CTTTCTTCCGTGATGTTCCAGTC; | 279 |
| R:GGTCCACATTCTCAGACCCTGTA | |||
| PGK | 3-phosphoglycerat kinase | F:TGGCAGACAAGTTTGCTCCT; | 181 |
| R:AAAAACTCCCATAGGTCCATT | |||
| P5CS | Δ1-pyrroline-5-carboxylate synthase | F:CTTCATCAATCCCAGAAAGAGTT; R:ATGTCCAAGAACTGGAATTTTTGTG | 173 |
| OAT | Ornithine δ-aminotransferase | F:GAAGTCCACCCTGATGTCGT R:AAATGTGCTTCCATGCTCCC | 126 |
| PHD | Proline hydroxylase | F:TTTGATTATGTGCCGGAAGC R:TCTTCGGACAACCATCTCG | 242 |
| GAD | Glutamic acid decarboxylase | F:CAAGCAGACTGGATGGGATA R:CAATGGCAACCTAAAGTCCC | 106 |
| Treatments | geNorm | NormFinder | Bestkeeper | RefFinder | Rank | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Stability | Gene | Stability | Gene | CV | SD | Gene | Value | ||
| Leaf | PGK | 0.057 | PPP2R5 | 0.052 | CYP | 0.04 | 0.17 | CYP | 1.41 | 1 |
| CYP | 0.057 | CYP | 0.079 | PGK | 0.07 | 0.26 | TUA | 2.00 | 2 | |
| TUA | 0.092 | TUA | 0.080 | TUA | 0.07 | 0.28 | PGK | 2.51 | 3 | |
| PPP2R5 | 0.161 | PGK | 0.115 | PPP2R5 | 0.12 | 0.46 | PPP2R5 | 3.00 | 4 | |
| ACT | 0.198 | ACT | 0.124 | TUB | 0.18 | 0.59 | ACT | 4.95 | 5 | |
| TUB | 0.213 | TUB | 0.152 | ACT | 0.19 | 0.70 | TUB | 5.73 | 6 | |
| PPP2R3 | 0.220 | PPP2R3 | 0.175 | PPP2R3 | 0.19 | 0.70 | PPP2R3 | 7.00 | 7 | |
| PPP2R2 | 0.242 | PPP2R2 | 0.184 | PPP2R2 | 0.21 | 0.74 | PPP2R2 | 8.00 | 8 | |
| EF-1α | 0.289 | EF-1ɑ | 0.300 | EF-1α | 0.27 | 1.08 | EF-1α | 9.00 | 9 | |
| Stem | PGK | 0.027 | PPP2R3 | 0.009 | PPP2R5 | 0.00 | 0.02 | PPP2R3 | 1.41 | 1 |
| PPP2R3 | 0.027 | PGK | 0.009 | PPP2R3 | 0.03 | 0.10 | PGK | 1.73 | 2 | |
| PPP2R5 | 0.037 | PPP2R5 | 0.010 | PGK | 0.05 | 0.20 | PPP2R5 | 2.06 | 3 | |
| CYP | 0.059 | ACT | 0.031 | CYP | 0.05 | 0.20 | CYP | 4.23 | 4 | |
| ACT | 0.088 | CYP | 0.048 | ACT | 0.07 | 0.27 | ACT | 4.73 | 5 | |
| PPP2R2 | 0.124 | PPP2R2 | 0.126 | PPP2R2 | 0.14 | 0.50 | PPP2R2 | 6.00 | 6 | |
| TUA | 0.159 | TUA | 0.198 | TUA | 0.19 | 0.76 | TUA | 7.00 | 7 | |
| EF-1α | 0.223 | EF-1α | 0.284 | TUB | 0.26 | 0.88 | EF-1α | 8.24 | 8 | |
| TUB | 0.271 | TUB | 0.289 | EF-1α | 0.28 | 1.05 | TUB | 8.74 | 9 | |
| Root | EF-1α | 0.025 | CYP | 0.004 | ACT | 0.01 | 0.04 | ACT | 1.86 | 1 |
| TUA | 0.025 | TUA | 0.008 | TUA | 0.02 | 0.06 | TUA | 2.11 | 2 | |
| ACT | 0.031 | EF-1α | 0.009 | EF-1α | 0.02 | 0.09 | EF-1α | 2.28 | 3 | |
| CYP | 0.047 | ACT | 0.011 | CYP | 0.04 | 0.17 | CYP | 2.38 | 4 | |
| PGK | 0.052 | PGK | 0.018 | PGK | 0.05 | 0.19 | PGK | 4.73 | 5 | |
| TUB | 0.080 | TUB | 0.102 | PPP2R5 | 0.11 | 0.43 | TUB | 6.24 | 6 | |
| PPP2R5 | 0.099 | PPP2R5 | 0.103 | TUB | 0.12 | 0.46 | PPP2R5 | 6.74 | 7 | |
| PPP2R2 | 0.119 | PPP2R2 | 0.121 | PPP2R2 | 0.12 | 0.43 | PPP2R2 | 8.00 | 8 | |
| PPP2R3 | 0.153 | PPP2R3 | 0.182 | PPP2R3 | 0.17 | 0.59 | PPP2R3 | 9.00 | 9 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Zhang, T.; Jiang, L.; Fan, G. Evaluation of Suitable Reference Genes for Quantitative Real-Time PCR in Various Tissues of Apocynum venetum. Genes 2024, 15, 231. https://doi.org/10.3390/genes15020231
Li X, Zhang T, Jiang L, Fan G. Evaluation of Suitable Reference Genes for Quantitative Real-Time PCR in Various Tissues of Apocynum venetum. Genes. 2024; 15(2):231. https://doi.org/10.3390/genes15020231
Chicago/Turabian StyleLi, Xiaoshuang, Tingting Zhang, Li Jiang, and Guizhi Fan. 2024. "Evaluation of Suitable Reference Genes for Quantitative Real-Time PCR in Various Tissues of Apocynum venetum" Genes 15, no. 2: 231. https://doi.org/10.3390/genes15020231
APA StyleLi, X., Zhang, T., Jiang, L., & Fan, G. (2024). Evaluation of Suitable Reference Genes for Quantitative Real-Time PCR in Various Tissues of Apocynum venetum. Genes, 15(2), 231. https://doi.org/10.3390/genes15020231





