Transcription Factor Regulation of Gene Expression Network by ZNF385D and HAND2 in Carotid Atherosclerosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Samples and Transcriptomic Data
2.2. Transcriptome-Wide Association Study (TWAS)
2.3. TF Regulatory Network Inference
2.4. TF Regulatory Network Analysis
2.5. Functional Annotation
3. Results
3.1. Global Differential Expression Analysis
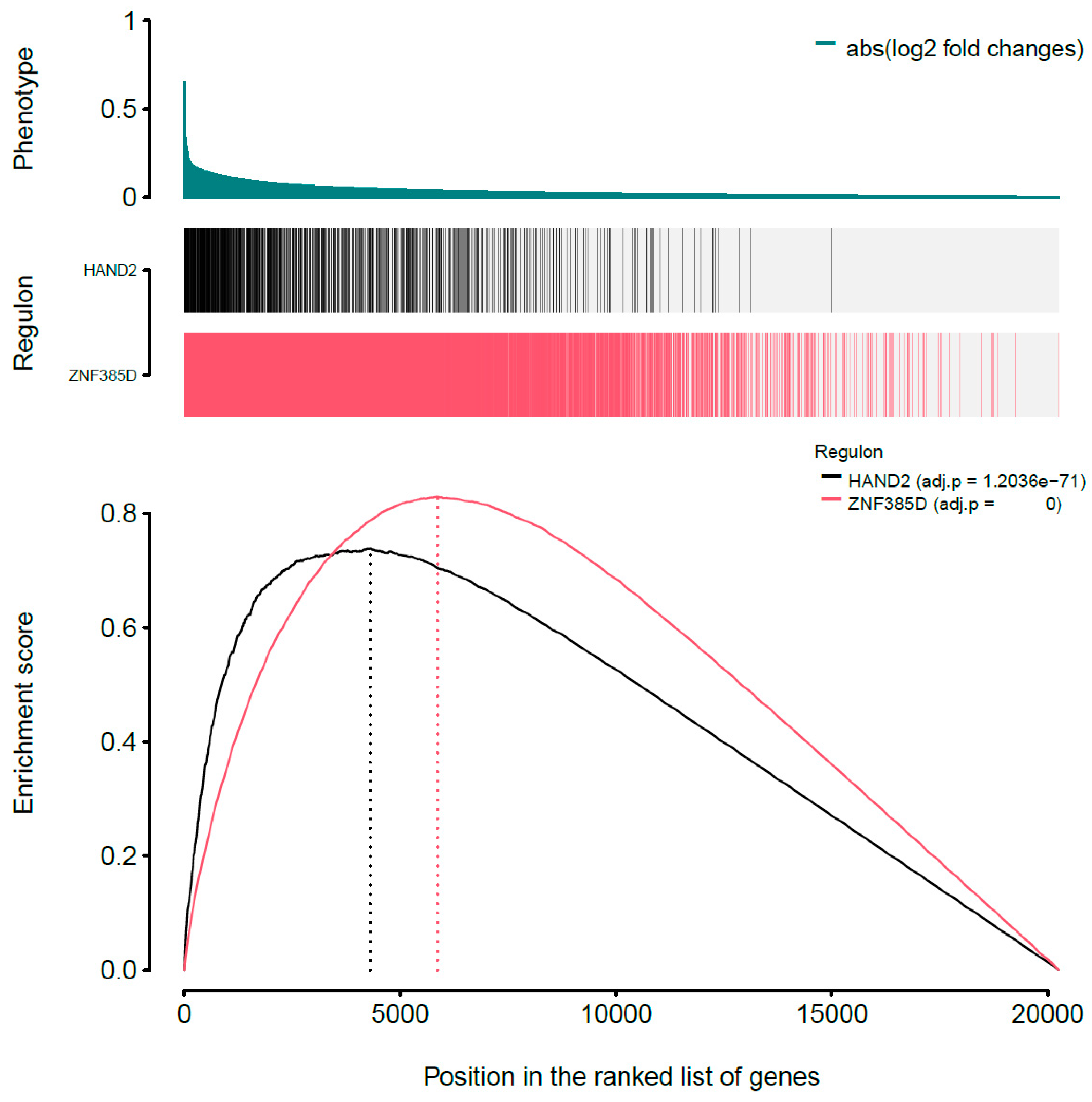
3.2. Inference of Transcriptional Network
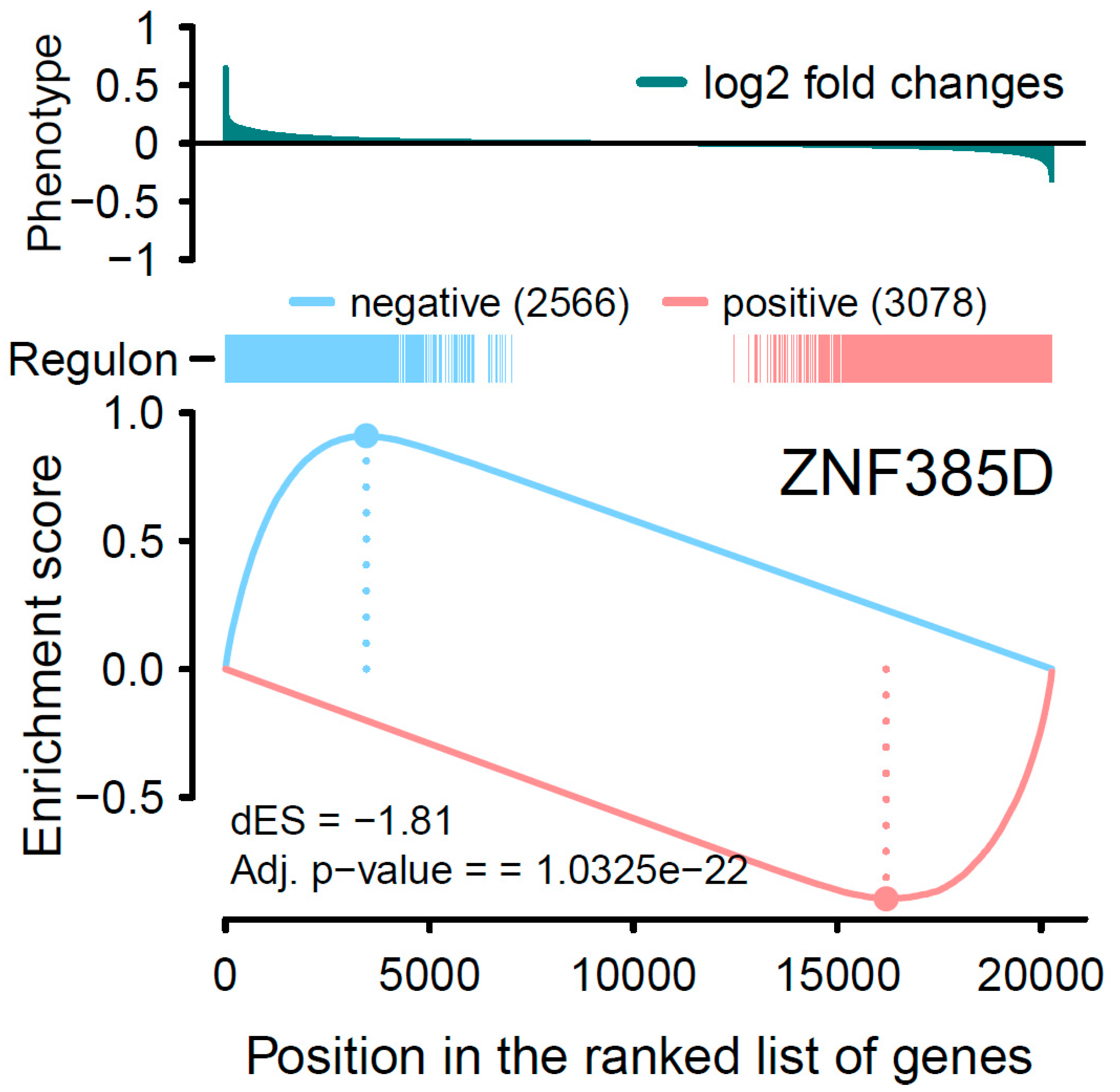
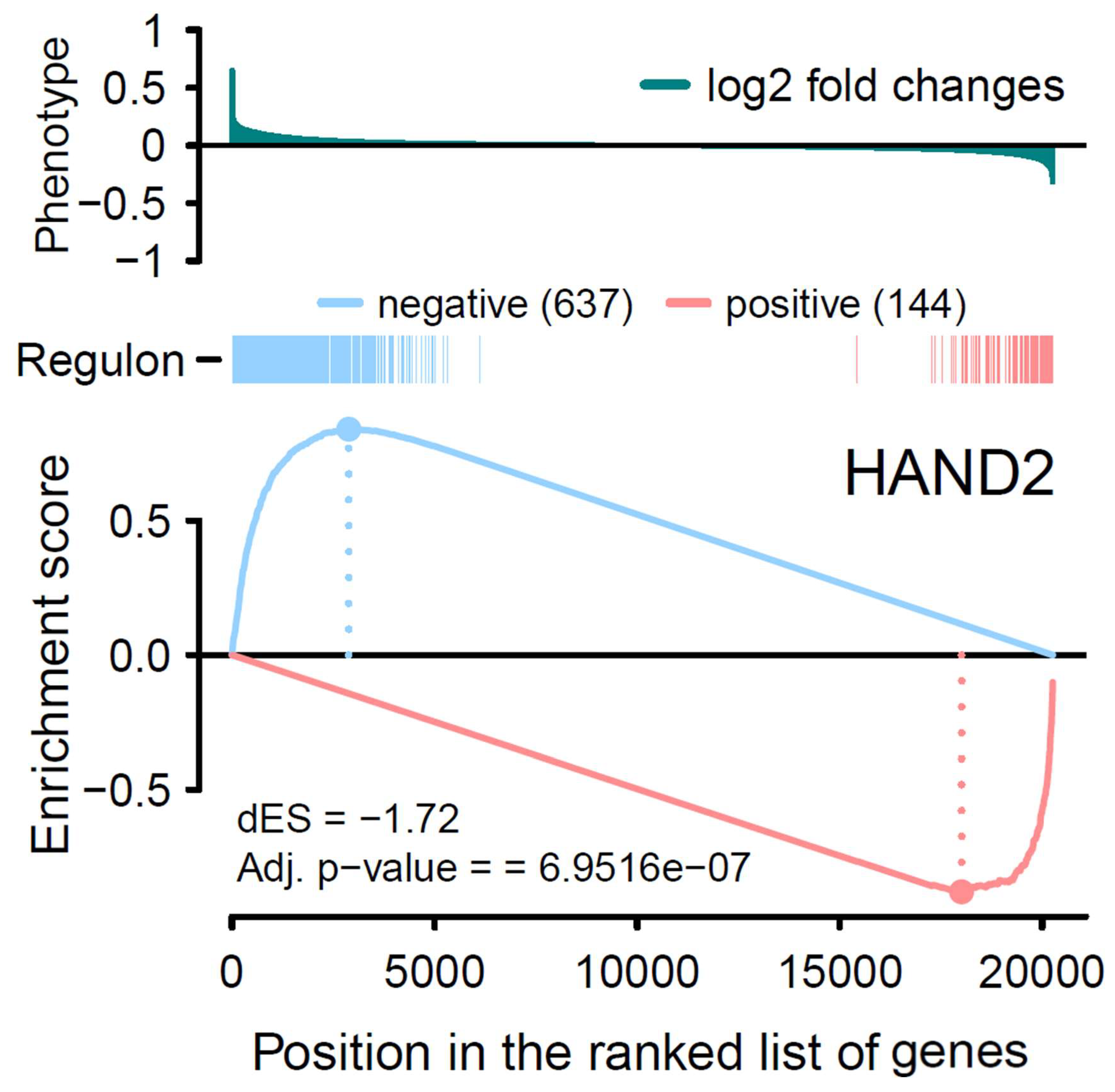
3.3. Analysis of Transcriptional Network
3.4. Enriched KEGG Pathways by TF Target Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gardener, H.; Caunca, M.R.; Dong, C.; Cheung, Y.K.; Elkind, M.S.V.; Sacco, R.L.; Rundek, T.; Wright, C.B. Ultrasound Markers of Carotid Atherosclerosis and Cognition: The Northern Manhattan Study. Stroke 2017, 48, 1855–1861. [Google Scholar] [CrossRef]
- Park, J.; Park, J.H.; Park, H. Association Between Carotid Artery Intima-Media Thickness and Combinations of Mild Cognitive Impairment and Pre-Frailty in Older Adults. Int. J. Environ. Res. Public. Health 2019, 16, 2978. [Google Scholar] [CrossRef] [PubMed]
- Della-Morte, D.; Dong, C.; Markert, M.S.; Elkind, M.S.V.; Sacco, R.L.; Wright, C.B.; Rundek, T. Carotid Intima-Media Thickness Is Associated With White Matter Hyperintensities: The Northern Manhattan Study. Stroke 2018, 45, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Marshall, D.; Elaine, W.; Vernalis, M. The effect of a one-year lifestyle intervention program on carotid intima media thickness. Mil. Med. 2011, 176, 798–804. [Google Scholar] [CrossRef] [PubMed]
- Akbari-Sedigh, A.; Asghari, G.; Yuzbashian, E.; Dehghan, P.; Imani, H.; Mirmiran, P. Association of dietary pattern withcarotid intima media thickness among children with overweight or obesity. Diabetol. Metab. Syndr. 2019, 11, 77. [Google Scholar] [CrossRef] [PubMed]
- Juo, S.H.; Lin, H.F.; Rundek, T.; Sabala, E.A.; Boden-Albala, B.; Park, N.; Lan, M.Y.; Sacco, R.L. Genetic and environmental contributions to carotid intima-media thickness and obesity phenotypes in the Northern Manhattan Family Study. Stroke 2004, 35, 2243–2247. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Cheema, F.A.; Bremner, J.D.; Goldberg, J.; Su, S.; Snieder, H.; Maisano, C.; Jones, L.; Javed, F.; Murrah, N.; et al. Heritability of carotid intima-media thickness: A twin study. Atherosclerosis 2008, 197, 814–820. [Google Scholar] [CrossRef] [PubMed]
- Franceschini, N.; Giambartolomei, C.; de Vries, P.S.; Finan, C.; Bis, J.C.; Huntley, R.P.; Lovering, R.C.; Tajuddin, S.M.; Winkler, T.W.; Graff, M.; et al. GWAS and colocalization analyses implicate carotid intima-media thickness and carotid plaque loci in cardiovascular outcomes. Nat. Commun. 2018, 9, 5141. [Google Scholar] [CrossRef]
- Yeung, M.W.; Wang, S.; van de Vegte, Y.J.; Borisov, O.; van Setten, J.; Snieder, H.; Verweij, N.; Said, M.A.; van der Harst, P. Twenty-Five Novel Loci for Carotid Intima-Media Thickness: A Genome-Wide Association Study in >45 000 Individuals and Meta-Analysis of >100 000 Individuals. Arter. Thromb. Vasc. Biol. 2022, 42, 484–501. [Google Scholar] [CrossRef]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The Human Transcription Factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef]
- Bushweller, J.H. Targeting transcription factors in cancer—from undruggable to reality. Nat. Rev. Cancer 2019, 19, 611–624. [Google Scholar] [CrossRef] [PubMed]
- Laurent, F.; Girdziusaite, A.; Gamart, J.; Barozzi, I.; Osterwalder, M.; Akiyama, J.A.; Lincoln, J.; Lopez-Rios, J.; Visel, A.; Zuniga, A.; et al. HAND2 Target Gene Regulatory Networks Control Atrioventricular Canal and Cardiac Valve Development. Cell Rep. 2017, 19, 1602–1613. [Google Scholar] [CrossRef] [PubMed]
- Videira, R.F.; Koop, A.M.C.; Ottaviani, L.; Poels, E.M.; Kocken, J.M.M.; Dos Remedios, C.; Mendes-Ferreira, P.; Van De Kolk, K.W.; Sarvaas, G.J.D.M.; Lourenço, A.; et al. The adult heart requires baseline expression of the transcription factor Hand2 to withstand right ventricular pressure overload. Cardiovasc. Res. 2022, 118, 2688–2702. [Google Scholar] [CrossRef]
- Tabassum, R.; Project, F.; Rämö, J.T.; Ripatti, P.; Koskela, J.; Kurki, M.; Karjalainen, J.; Palta, P.; Hassan, S.; Nunez-Fontarnau, J.; et al. Genetic architecture of human plasma lipidome and its link to cardiovascular disease. Nat. Commun. 2019, 10, 4329. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Aragam, N.; Li, X.; Villla, E.C.; Wang, L.; Briones, D.; Petty, L.; Posada, Y.; Arana, T.B.; Cruz, G.; et al. BCL9 and C9orf5 are associated with negative symptoms in schizophrenia: Meta-analysis of two genome-wide association studies. PLoS ONE 2013, 8, e51674. [Google Scholar] [CrossRef]
- Yohannes, A.M.; Alexopoulos, G.S. Depression and anxiety in patients with COPD. Eur. Respir. Rev. 2014, 23, 345–349. [Google Scholar] [CrossRef]
- Fletcher, M.N.; Castro, M.A.; Wang, X.; de Santiago, I.; O’Reilly, M.; Chin, S.-F.; Rueda, O.M.; Caldas, C.; Ponder, B.A.J.; Markowetz, F.; et al. Master regulators of FGFR2 signalling and breast cancer risk. Nat. Commun. 2013, 4, 2464. [Google Scholar] [CrossRef]
- Ayari, H.; Bricca, G. Identification of two genes potentially associated in iron-heme homeostasis in human carotid plaque using microarray analysis. J. Biosci. 2013, 38, 311–315. [Google Scholar] [CrossRef]
- Kinney, J.B.; Atwal, G.S. Equitability, mutual information, and the maximal information coefficient. Proc. Natl. Acad. Sci. USA 2014, 111, 3354–3359. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Flynn, E.D.; Tsu, A.L.; Kasela, S.; Kim-Hellmuth, S.; Aguet, F.; Ardlie, K.G.; Bussemaker, H.J.; Mohammadi, P.; Lappalainen, T. Transcription factor regulation of eQTL activity across individuals and tissues. PLOS Genet. 2022, 18, e1009719. [Google Scholar] [CrossRef] [PubMed]
- Holdt, L.M.; Teupser, D. Genetic background of atherosclerosis and its risk factors. In The ESC Textbook of Preventive Cardiology; ESC Textbook; Gielen, S., Ed.; Oxford University Press (OUP): Oxford, UK, 2015. [Google Scholar] [CrossRef]
- Björck, H.M.; Renner, J.; Maleki, S.; Nilsson, S.F.E.; Kihlberg, J.; Folkersen, L.; Karlsson, M.; Ebbers, T.; Eriksson, P.; Länne, T. Characterization of shear-sensitive genes in the normal rat aorta identifies Hand2 as a major flow-responsive transcription factor. PLoS ONE 2012, 7, e52227. [Google Scholar] [CrossRef]
- Rodrigues, A.C.Z.; Messi, M.L.; Wang, Z.; Bonilla, H.J.; Freeman, W.M.; Delbono, O. Long-term, induced expression of Hand2 in peripheral sympathetic neurons ameliorates sarcopenia in geriatric mice. J. Cachex Sarcopenia Muscle 2021, 12, 1908–1924. [Google Scholar] [CrossRef]
- Abhashi, S.A.; Kryeziu, F.U.; Nazreku, F.D. Increased carotid intima-media thickness associated with high hs-CRP levels is a predictor of unstable coronary artery disease. Cardiovasc. J. Afr. 2013, 24, 270–273. [Google Scholar] [CrossRef]
- Liao, H.; Li, Z.; Zheng, D.; Liu, J.; Liu, Y.; Xiao, C.; Wang, H. Increased Hs-CRP/adiponectin ratio is associated with increase carotid intima-media thickness. Lipids Heal. Dis. 2014, 13, 120. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Du, X.; Li, J.; Ma, L.; Zhang, S.; Wu, S.; Ji, C.; Shi, J.; Guo, R. Correlation between serum high sensitivity C-reactive protein and carotid intima-media thickness. Clin. Med. China 2021, 12, 26–30. [Google Scholar] [CrossRef]
- Kivimäki, M.; Lawlor, D.A.; Smith, G.D.; Kumari, M.; Donald, A.; Britton, A.; Casas, J.P.; Shah, T.; Brunner, E.; Timpson, N.J.; et al. Does high C-reactive protein concentration increase atherosclerosis? The Whitehall II Study. PLoS ONE 2008, 3, e3013. [Google Scholar] [CrossRef]
- Çırakoğlu, Ö.F.; Yılmaz, A.S. Systemic immune-inflammation index is associated with increased carotid intima-media thickness in hypertensive patients. Clin. Exp. Hypertens. 2021, 43, 565–571. [Google Scholar] [CrossRef]
- van Zanten, J.J.V.; Kitas, G.D. Inflammation, carotid intima-media thickness and atherosclerosis in rheumatoid arthritis. Arthritis Res. Ther. 2008, 10, 102. [Google Scholar] [CrossRef][Green Version]
- Ross, A.C.; Rizk, N.; O’riordan, M.A.; Dogra, V.; El-Bejjani, D.; Storer, N.; Harrill, D.; Tungsiripat, M.; Adell, J.; McComsey, G.A. Relationship between inflammatory markers, endothelial activation markers, and carotid intima-media thickness in HIV-infected patients receiving antiretroviral therapy. Clin. Infect. Dis. 2009, 49, 1119–1127. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Camba, A.; Carrillo-Palau, M.; Ramos, L.; Alvarez-Buylla, N.H.; Alonso-Abreu, I.; Hernández-Pérez, A.; Vela, M.; Arranz, L.; Hernández-Guerra, M.; González-Gay, M.; et al. Carotid Plaque Assessment Reclassifies Patients with Inflammatory Bowel Disease into Very-High Cardiovascular Risk. J. Clin. Med. 2021, 10, 1671. [Google Scholar] [CrossRef] [PubMed]
- Alfaddagh, A.; Martin, S.S.; Leucker, T.M.; Michos, E.D.; Blaha, M.J.; Lowenstein, C.J.; Jones, S.R.; Toth, P.P. Inflammation and cardiovascular disease: From mechanisms to therapeutics. Am. J. Prev. Cardiol. 2020, 4, 100130. [Google Scholar] [CrossRef] [PubMed]


| ZNF38D5 | HAND2 | |
|---|---|---|
| Network inference | ||
| Regulon size | 5644 | 781 |
| Positive regulation | 3078 | 144 |
| Negative regulation | 2566 | 637 |
| Enrichment analysis | ||
| GSEA one-sided | ||
| ES * | 0.83 | 0.74 |
| p value | <2.26 × 10−308 | 1.20 × 10−71 |
| GSEA two-sided | ||
| Positive enrichment | ||
| ESpositive | −0.90 | −0.88 |
| p value | 1.18 × 10−20 | 2.64 × 10−4 |
| Negative enrichment | ||
| ESnegative | 0.91 | 0.84 |
| p value | 3.03 × 10−7 | 4.4 × 10−4 |
| Differential | ||
| ∆ES * | −1.81 | −1.72 |
| p value | 5.16 × 10−23 | 6.95 × 10−7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, M.; Andersen, L.J.; Bruun, N.E.; Lindholm, M.G.; Tan, Q.; Snoer, M. Transcription Factor Regulation of Gene Expression Network by ZNF385D and HAND2 in Carotid Atherosclerosis. Genes 2024, 15, 213. https://doi.org/10.3390/genes15020213
Tan M, Andersen LJ, Bruun NE, Lindholm MG, Tan Q, Snoer M. Transcription Factor Regulation of Gene Expression Network by ZNF385D and HAND2 in Carotid Atherosclerosis. Genes. 2024; 15(2):213. https://doi.org/10.3390/genes15020213
Chicago/Turabian StyleTan, Ming, Lars Juel Andersen, Niels Eske Bruun, Matias Greve Lindholm, Qihua Tan, and Martin Snoer. 2024. "Transcription Factor Regulation of Gene Expression Network by ZNF385D and HAND2 in Carotid Atherosclerosis" Genes 15, no. 2: 213. https://doi.org/10.3390/genes15020213
APA StyleTan, M., Andersen, L. J., Bruun, N. E., Lindholm, M. G., Tan, Q., & Snoer, M. (2024). Transcription Factor Regulation of Gene Expression Network by ZNF385D and HAND2 in Carotid Atherosclerosis. Genes, 15(2), 213. https://doi.org/10.3390/genes15020213






