Analysis of Transcriptomic Differences in the Ovaries of High- and Low-Laying Ducks
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. Serum Physiological and Biochemical Index Testing
2.3. RNA Extraction, Library Preparation, and Illumina Hiseq Sequencing
2.4. RNA-Seq Data Processing and Analysis
2.5. Gene Ontology and KEGG Pathway Analysis of DEGs
2.6. Protein–Protein Interaction Network Analysis and Module Selection
2.7. Quantitative Real-Time PCR Analysis
2.8. Statistical Analysis
3. Results
3.1. Egg Quality Differences between the HEP and LEP Groups
3.2. Serum Physiological and Biochemical Differences between the HEP and LEP Groups
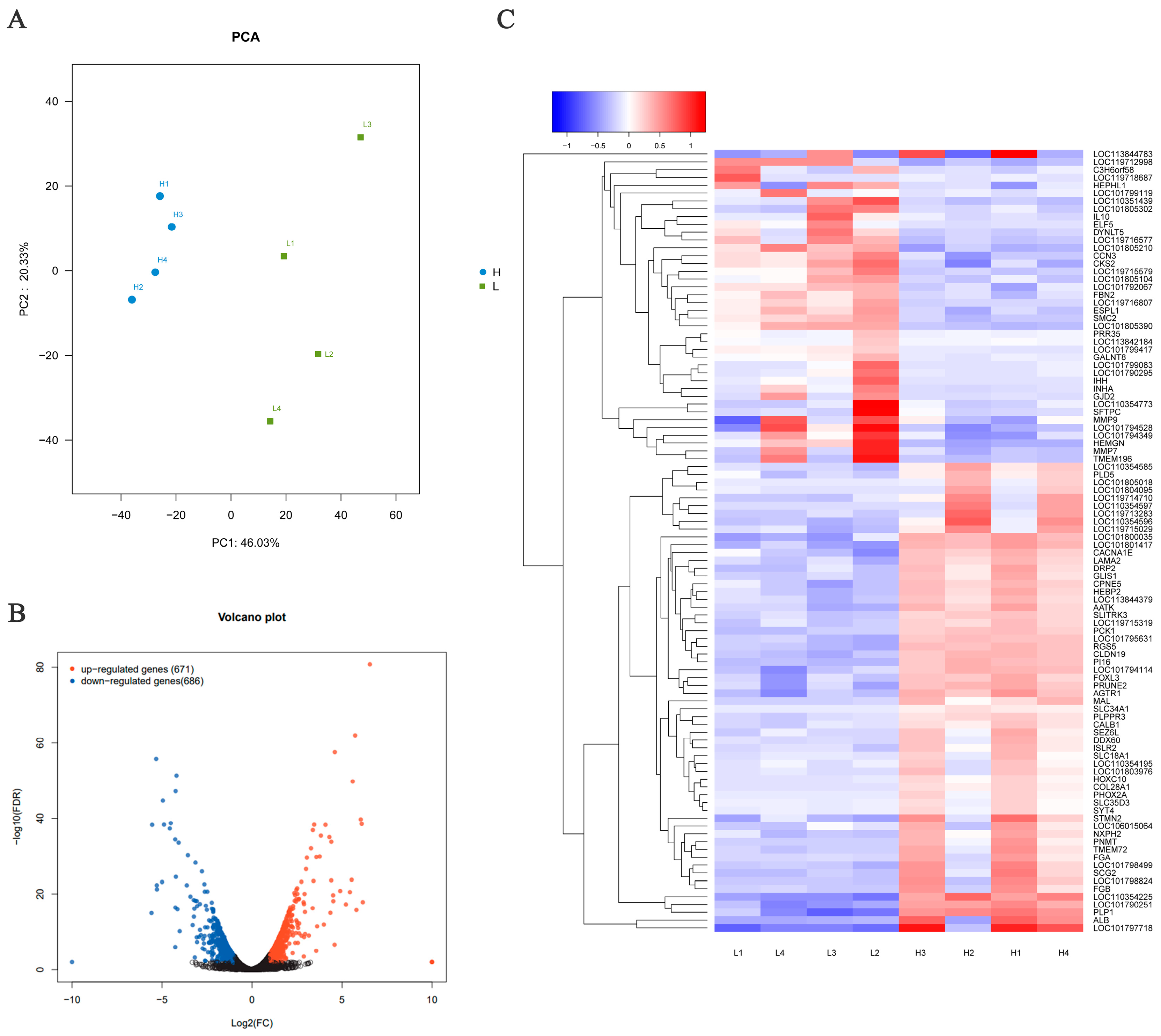
3.3. Transcriptome Alignment and Mapping Statistics
3.4. Differentially Expressed Genes (DEG) between the HEP and LEP Groups
3.5. Gene Ontology (GO) and KEGG Analyses for DEGs
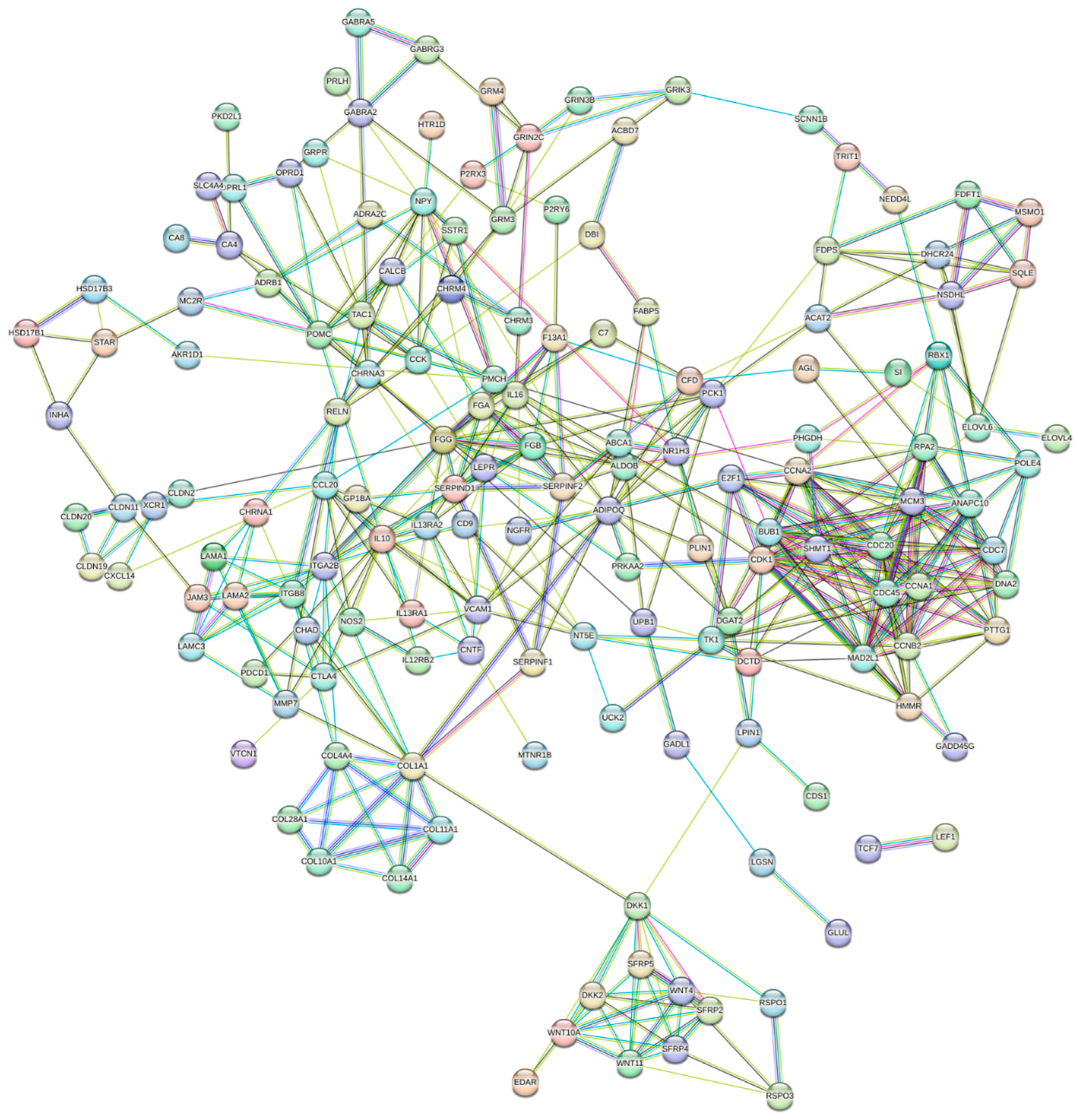
3.6. Identification of Hub Genes and Pathways through PPI Network Analysis of DEGs
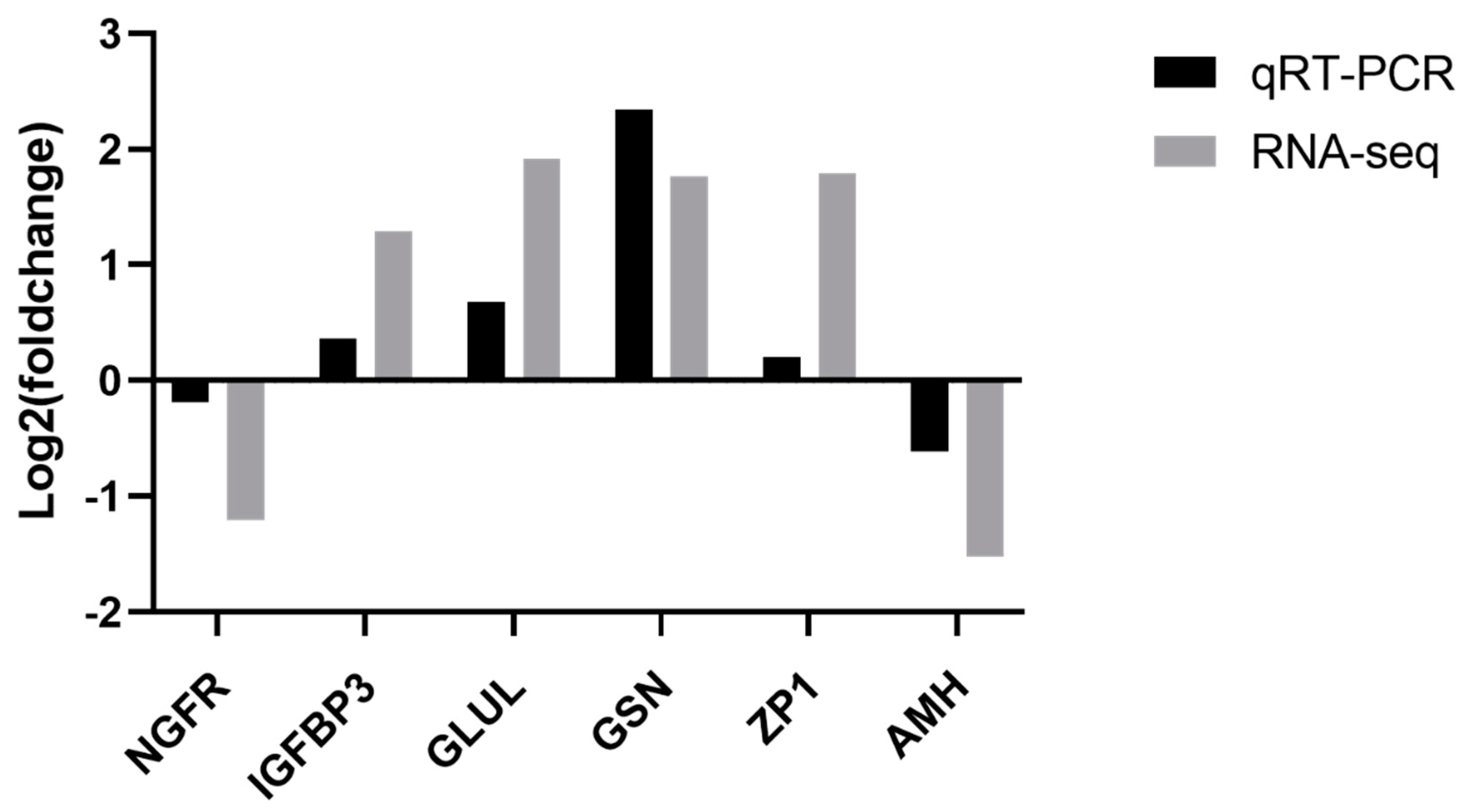
3.7. Validation of RNA-Seq Results by qRT-PCR
4. Discussion
4.1. Differences between Egg Quality in High- and Low-Egg-Production Shan Ma Ducks
4.2. Blood Physiological and Biochemical Indicators
4.3. Analysis of DEGs
4.4. Functional Classification Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tao, Z.; Song, W.; Zhu, C.; Xu, W.; Liu, H.; Zhang, S.; Huifang, L. Comparative transcriptomic analysis of high and low egg-producing duck ovaries. Poult. Sci. 2017, 96, 4378–4388. [Google Scholar] [CrossRef]
- Bhavana, K.; Foote, D.J.; Srikanth, K.; Balakrishnan, C.N.; Prabhu, V.R.; Sankaralingam, S.; Singha, H.S.; Gopalakrishnan, A.; Nagarajan, M. Comparative transcriptome analysis of Indian domestic duck reveals candidate genes associated with egg production. Sci. Rep. 2022, 12, 10943. [Google Scholar] [CrossRef] [PubMed]
- Bao, X.; Song, Y.; Li, T.; Zhang, S.; Huang, L.; Zhang, S.; Cao, J.; Liu, X.; Zhang, J. Comparative Transcriptome Profiling of Ovary Tissue between Black Muscovy Duck and White Muscovy Duck with High- and Low-Egg Production. Genes 2020, 12, 57. [Google Scholar] [CrossRef] [PubMed]
- Mu, R.; Yu, Y.Y.; Gegen, T.; Wen, D.; Wang, F.; Chen, Z.; Xu, W.B. Transcriptome analysis of ovary tissues from low- and high-yielding Changshun green-shell laying hens. BMC Genom. 2021, 22, 349. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Q.; Hu, S.; Wang, G.; Hu, J.; Zhang, J.; Li, L.; Hu, B.; He, H.; Liu, H.; Xia, L.; et al. Comparative Transcriptome Analysis Suggests Key Roles for 5-Hydroxytryptamlne Receptors in Control of Goose Egg Production. Genes 2020, 11, 455. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Chen, L.; Han, K.; Zhang, X.; Zhang, G.; Dai, G.; Wang, J.; Xie, K. Transcriptome analysis of ovary in relatively greater and lesser egg producing Jinghai Yellow Chicken. Anim. Reprod. Sci. 2019, 208, 106114. [Google Scholar] [CrossRef]
- Zhu, Z.; Miao, Z.; Chen, H.; Xin, Q.; Li, L.; Lin, R.; Huang, Q.; Zheng, N. Ovarian transcriptomic analysis of Shan Ma ducks at peak and late stages of egg production. Asian-Australas J. Anim. Sci. 2017, 30, 1215–1224. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.L.; Chen, H.P.; Rouvier, R.; Marie-Etancelin, C. Genetic parameters of body weight, egg production, and shell quality traits in the Shan Ma laying duck (Anas platyrhynchos). Poult. Sci. 2016, 95, 2514–2519. [Google Scholar] [CrossRef] [PubMed]
- Kostaman, T.; Sopiyana, S. The differences in egg quality of white leghorn and naked neck chicken. In International Seminar on Livestock Production and Veterinary Technology; IAARD Press: Jakarta, Indonesian, 2016; pp. 365–369. [Google Scholar]
- Wolc, A.; Arango, J.; Settar, P.; O’Sullivan, N.P.; Olori, V.E.; White, I.M.; Hill, W.G.; Dekkers, J.C. Genetic parameters of egg defects and egg quality in layer chickens. Poult. Sci. 2012, 91, 1292–1298. [Google Scholar] [CrossRef]
- Kowalska, E.; Kucharska-Gaca, J.; Kuźniacka, J.; Lewko, L.; Gornowicz, E.; Biesek, J.; Adamski, M. Egg quality depending on the diet with different sources of protein and age of the hens. Sci. Rep. 2021, 11, 2638. [Google Scholar] [CrossRef]
- Nasr, M.A.F.; Mohammed, H.; Hassan, R.A.; Swelum, A.A.; Saadeldin, I.M. Does light intensity affect the behavior, welfare, performance, meat quality, amino acid profile, and egg quality of Japanese quails? Poult. Sci. 2019, 98, 3093–3102. [Google Scholar] [CrossRef]
- Kumar, M.; Dahiya, S.P.; Ratwan, P.; Sheoran, N.; Kumar, S.; Kumar, N. Assessment of egg quality and biochemical parameters of Aseel and Kadaknath indigenous chicken breeds of India under backyard poultry farming. Poult. Sci. 2022, 101, 101589. [Google Scholar] [CrossRef]
- Onbasilar, E.E.; Erdem, E.; Poyraz, O.; Yalçin, S. Effects of hen production cycle and egg weight on egg quality and composition, hatchability, duckling quality, and first-week body weight in Pekin ducks. Poult. Sci. 2011, 90, 2642–2647. [Google Scholar] [CrossRef]
- Yao, B.N.; Liao, F.Y.; Yang, J.Y.; Liu, A.; Wang, J.; Zhu, B.G.; Feng, G.; Yang, S.L. Effect of sea buckthorn extract on production performance, serum biochemical indexes, egg quality, and cholesterol deposition of laying ducks. Front. Vet. Sci. 2023, 10, 1127117. [Google Scholar] [CrossRef]
- Chang, C.; Lahti, T.; Tanaka, T.; Nickerson, M.T. Egg proteins: Fractionation, bioactive peptides and allergenicity. J. Sci. Food Agric. 2018, 98, 5547–5558. [Google Scholar] [CrossRef] [PubMed]
- Weishaar, R.; Wellmann, R.; Camarinha-Silva, A.; Rodehutscord, M.; Bennewitz, J. Selecting the hologenome to breed for an improved feed efficiency in pigs—A novel selection index. J. Anim. Breed. Genet. 2020, 137, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Kumar, S.; Rahim, A.; Kokate, L.; Mishra, A. Genetic analysis of body conformation and feed efficiency characteristics in a selected line of Rhode Island Red chicken. Asian J. Anim. Sci. 2015, 9, 434–440. [Google Scholar] [CrossRef]
- Brady, K.; Porter, T.E.; Liu, H.C.; Long, J.A. Characterization of the hypothalamo-pituitary-gonadal axis in low and high egg producing turkey hens. Poult. Sci. 2020, 99, 1163–1173. [Google Scholar] [CrossRef] [PubMed]
- Etches, R.J.; Cunningham, F.J. The interrelationship between progesterone and luteinizing hormone during the ovulation cycle of the hen (Gallus domesticus). J. Endocrinol. 1976, 71, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Zhang, L.; Qi, Q.; Li, J.; Yan, F.; Hou, J. Growth hormone treatment improves the development of follicles and oocytes in prepubertal lambs. J. Ovarian Res. 2023, 16, 132. [Google Scholar] [CrossRef]
- Vasilatos-Younken, R.; Wang, X.H.; Zhou, Y.; Day, J.R.; McMurtry, J.P.; Rosebrough, R.W.; Decuypere, E.; Buys, N.; Darras, V.; Beard, J.L.; et al. New insights into the mechanism and actions of growth hormone (GH) in poultry. Domest. Anim. Endocrinol. 1999, 17, 181–190. [Google Scholar] [CrossRef]
- Mohammadi, H.; Ansari-Pirsaraei, Z. Changes in some blood parameters and production performance of old laying hens due to growth hormone and testosterone injection. J. Anim. Physiol. Anim. Nutr. 2014, 98, 483–490. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, Y.; Iwashita, M.; Karube, M.; Oda, T.; Akiba, M.; Shiokawa, S.; Ando, M.; Yoshinaga, A.; Nakamura, Y. Growth hormone stimulates follicular development by stimulating ovarian production of insulin-like growth factor-I. Endocrinology 1994, 135, 887–894. [Google Scholar] [CrossRef]
- Wang, P.; Gong, Y.; Li, D.; Zhao, X.; Zhang, Y.; Zhang, J.; Geng, X.; Zhang, X.; Tian, Y.; Li, W.; et al. Effect of induced molting on ovarian function remodeling in laying hens. Poult. Sci. 2023, 102, 102820. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, Y.; Tamura, T. Effects of estradiol administration on the follicular tissue of hypophysectomized hens. Poult. Sci. 1986, 65, 1808–1810. [Google Scholar] [CrossRef] [PubMed]
- Dorrington, J.; Gore-Langton, R.E. Prolactin inhibits oestrogen synthesis in the ovary. Nature 1981, 290, 600–602. [Google Scholar] [CrossRef]
- Laborde, C.J.; Chapa, A.M.; Burleigh, D.W.; Salgado, D.J.; Fernandez, J.M. Effects of processing and storage on the measurement of nitrogenous compounds in ovine blood. Small Rumin. Res. 1995, 17, 159–166. [Google Scholar] [CrossRef]
- Huang, Z.; Zhong, Z.; Xiao, T. Correlation Analysis on Egg-laying Performance and Blood Biochemical Parameters in Shanma Duck. Chin. Agric. Sci. Bull. 2011, 27, 13–17. [Google Scholar]
- Wang, Z.W. Effect of dietary nutrient levels on growth performance and serum parameters for northeast geese (1~28 d). J. Nucl. Agric. Sci. 2009, 16, 27–31. [Google Scholar]
- Wang, H.H.; Garruti, G.; Liu, M.; Portincasa, P.; Wang, D.Q. Cholesterol and Lipoprotein Metabolism and Atherosclerosis: Recent Advances in reverse Cholesterol Transport. Ann. Hepatol. 2017, 16, s27–s42. [Google Scholar] [CrossRef]
- Kang, W.C.; Ang, L.I.; Zhu, Z.N. Relationships between blood biochemical indices and egg-laying performances in muscovy ducks. J. Fujian Agric. Univ. 2001, 30, 510–513. [Google Scholar]
- McDonald, J.K. Role of neuropeptide Y in reproductive function. Ann. N. Y. Acad. Sci. 1990, 611, 258–272. [Google Scholar] [CrossRef] [PubMed]
- Barb, C.R.; Kraeling, R.R.; Rampacek, G.B.; Hausman, G.J. The role of neuropeptide Y and interaction with leptin in regulating feed intake and luteinizing hormone and growth hormone secretion in the pig. Reproduction 2006, 131, 1127–1135. [Google Scholar] [CrossRef] [PubMed]
- Urata, Y.; Salehi, R.; Lima, P.D.A.; Osuga, Y.; Tsang, B.K. Neuropeptide Y regulates proliferation and apoptosis in granulosa cells in a follicular stage-dependent manner. J. Ovarian Res. 2020, 13, 5. [Google Scholar] [CrossRef] [PubMed]
- Catania, M.G.; Mischel, P.S.; Vinters, H.V. Hamartin and tuberin interaction with the G2/M cyclin-dependent kinase CDK1 and its regulatory cyclins A and B. J. Neuropathol. Exp. Neurol. 2001, 60, 711–723. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Bird, R.C. Selective induction of cell cycle regulatory genes cdk1 (p34cdc2), cyclins A/B, and the tumor suppressor gene Rb in transformed cells by okadaic acid. J. Cell. Physiol. 1995, 164, 424–433. [Google Scholar] [CrossRef] [PubMed]
- Song, P.; Chen, X.; Zhang, P.; Zhou, Y.; Zhou, R. miR-200b/MYBL2/CDK1 suppresses proliferation and induces senescence through cell cycle arrest in ovine granulosa cells. Theriogenology 2023, 207, 19–30. [Google Scholar] [CrossRef]
- Yang, W.; Chen, X.; Liu, Z.; Zhao, Y.; Chen, Y.; Geng, Z. Integrated transcriptome and proteome revealed that the declined expression of cell cycle-related genes associated with follicular atresia in geese. BMC Genom. 2023, 24, 24. [Google Scholar] [CrossRef]
- Knezevic, D.; Brash, D.E. Role of E2F1 in apoptosis: A case study in feedback loops. Cell Cycle 2004, 3, 729–732. [Google Scholar] [CrossRef]
- Inoshita, S.; Terada, Y.; Nakashima, O.; Kuwahara, M.; Sasaki, S.; Marumo, F. Regulation of the G1/S transition phase in mesangial cells by E2F1. Kidney Int. 1999, 56, 1238–1241. [Google Scholar] [CrossRef]
- Avellino, R.; Romano, S.; Parasole, R.; Bisogni, R.; Lamberti, A.; Poggi, V.; Venuta, S.; Romano, M.F. Rapamycin stimulates apoptosis of childhood acute lymphoblastic leukemia cells. Blood 2005, 106, 1400–1406. [Google Scholar] [CrossRef] [PubMed]
- Richani, D.; Gilchrist, R.B. The epidermal growth factor network: Role in oocyte growth, maturation and developmental competence. Hum. Reprod. Update 2018, 24, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, M.; Conti, M. G-protein-coupled receptor signaling and the EGF network in endocrine systems. Trends Endocrinol. Metab. 2005, 16, 320–326. [Google Scholar] [CrossRef]
- Park, J.Y.; Su, Y.Q.; Ariga, M.; Law, E.; Jin, S.L.; Conti, M. EGF-like growth factors as mediators of LH action in the ovulatory follicle. Science 2004, 303, 682–684. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Xiao, C.; Deng, J.; Yang, Z.; Zou, L.; Du, W.; Li, S.; Huo, X.; Zeng, L.; Yang, X. Transcriptome analysis reveals key genes and pathways associated with egg production in Nandan-Yao domestic chicken. Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 40, 100889. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhao, X.; Chen, L.; Wang, J.; Duan, Y.; Li, H.; Lu, L. Transcriptomic Analyses of the Hypothalamic-Pituitary-Gonadal Axis Identify Candidate Genes Related to Egg Production in Xinjiang Yili Geese. Animals 2020, 10, 90. [Google Scholar] [CrossRef] [PubMed]
- Abdal Dayem, A.; Lee, S.; Choi, H.Y.; Cho, S.G. The Impact of Adhesion Molecules on the In Vitro Culture and Differentiation of Stem Cells. Biotechnol. J. 2018, 13, 1700575. [Google Scholar] [CrossRef] [PubMed]
- Thomas, G.J.; Speight, P.M. Cell adhesion molecules and oral cancer. Crit. Rev. Oral Biol. Med. 2001, 12, 479–498. [Google Scholar] [CrossRef]
- Schmidt, E.P.; Kuebler, W.M.; Lee, W.L.; Downey, G.P. Adhesion Molecules: Master Controllers of the Circulatory System. Compr. Physiol. 2016, 6, 945–973. [Google Scholar]
- Yin, M.; Li, C.; Jiang, J.; Le, J.; Luo, B.; Yang, F.; Fang, Y.; Yang, M.; Deng, Z.; Ni, W.; et al. Cell adhesion molecule-mediated therapeutic strategies in atherosclerosis: From a biological basis and molecular mechanism to drug delivery nanosystems. Biochem. Pharmacol. 2021, 186, 114471. [Google Scholar] [CrossRef]
- Hynes, R.O. The extracellular matrix: Not just pretty fibrils. Science 2009, 326, 1216–1219. [Google Scholar] [CrossRef] [PubMed]
- Järveläinen, H.; Sainio, A.; Koulu, M.; Wight, T.N.; Penttinen, R. Extracellular matrix molecules: Potential targets in pharmacotherapy. Pharmacol. Rev. 2009, 61, 198–223. [Google Scholar] [CrossRef] [PubMed]
- Gilchrist, R.B.; Ritter, L.J.; Armstrong, D.T. Oocyte-somatic cell interactions during follicle development in mammals. Anim. Reprod. Sci. 2004, 82–83, 431–446. [Google Scholar] [CrossRef] [PubMed]


| Item | LEP | HEP | p-Value |
|---|---|---|---|
| Egg production | 184.33 ± 4.55 | 215.50 ± 13.52 ** | 0.002 |
| Eggshell thickness (mm) | 0.44 ± 0.02 | 0.42 ± 0.04 | 0.069 |
| Egg weight (g) | 69.53 ± 7.43 | 78.55 ± 7.43 ** | 0.008 |
| Ovary weight (g) | 2.58 ± 0.95 | 3.03 ± 1.08 | 0.784 |
| Egg shape index | 1.31 ± 0.06 | 1.32 ± 0.06 | 0.279 |
| Egg yolk color | 11.25 ± 0.45 | 11.36 ± 0.50 | 0.909 |
| Eggshell strength (kg/cm2) | 4.74 ± 0.37 | 4.77 ± 0.79 | 0.151 |
| Hastelloy unit | 63.87 ± 17.86 | 69.93 ± 12.36 | 0.002 |
| Item | LEP | HEP | p-Value |
|---|---|---|---|
| Follicle-stimulating hormone (mIU/mL) | 1.77 ± 0.26 | 3.07 ± 0.19 ** | 0.000 |
| Luteinizing hormone (mIU/mL) | 5.64 ± 0.83 | 8.22 ± 1.44 * | 0.021 |
| Estradiol (pg/mL) | 31.55 ± 6.14 | 47.20 ± 5.45 ** | 0.009 |
| Progesterone (ng/mL) | 12.12 ± 0.92 | 8.26 ± 0.90 ** | 0.001 |
| Growth hormone (ng/mL) | 4.70 ± 0.72 | 7.06 ± 0.56 * | 0.002 |
| Prolactin (μIU/mL) | 99.80 ± 2.86 | 87.12 ± 6.46 ** | 0.012 |
| Triiodothyronine (ng/mL) | 0.73 ± 0.17 | 1.06 ± 0.09 * | 0.015 |
| Thyroxine (ng/mL) | 9.14 ± 0.64 | 12.71 ± 1.09 ** | 0.001 |
| Total protein (g/L) | 37.51 ± 5.99 | 57.55 ± 7.35 ** | 0.006 |
| Albumin (g/L) | 13.15 ± 1.01 | 15.81 ± 0.88 ** | 0.007 |
| Globulin (g/L) | 32.03 ± 1.25 | 36.24 ± 2.49 * | 0.023 |
| Total cholesterol (mmol/L) | 1.98 ± 0.10 | 3.65 ± 0.38 ** | 0.000 |
| Triglycerides (mmol/L) | 9.30 ± 1.95 | 17.49 ± 2.75 ** | 0.003 |
| High-density lipoprotein (mmol/L) | 1.74 ± 0.50 | 0.90 ± 0.14 * | 0.019 |
| Low-density lipoprotein (mmol/L) | 0.88 ± 0.19 | 1.16 ± 0.11 * | 0.045 |
| Aspartate transaminase (U/L) | 48.90 ± 7.19 | 115.96 ± 56.41 | 0.056 |
| Alanine transaminase (U/L) | 60.99 ± 5.25 | 82.22 ± 6.71 ** | 0.002 |
| Gene ID | Read Count (LEP) | Read Count (HEP) | Log2 Fold-Change (HEP/LEP) | Up/Down (HEP/LEP) | p-Value | Gene Description |
|---|---|---|---|---|---|---|
| LOC101797718 | 0.58 | 54.78 | 6.55 | up | 0.00 | Apovitellenin-1-like precursor |
| LOC101805018 | 0.00 | 0.86 | 9.76 | up | 0.00 | Prostatic acid phosphatase-like |
| MAL | 0.03 | 2.03 | 6.16 | up | 0.00 | Myelin and lymphocyte protein |
| LOC110354597 | 0.00 | 2.13 | 11.05 | up | 0.00 | Cygnin |
| FGA | 0.03 | 2.12 | 6.10 | up | 0.00 | Fibrinogen alpha chain |
| FGB | 0.05 | 3.07 | 6.04 | up | 0.00 | Fibrinogen beta chain |
| ALB | 0.20 | 10.51 | 5.73 | up | 0.00 | Serum albumin precursor |
| LOC101793493 | 0.00 | 1.46 | 10.51 | up | 0.00 | Fatty acid-binding protein, liver |
| PHOX2A | 0.01 | 0.79 | 5.80 | up | 0.00 | Paired mesoderm homeobox Protein 2A |
| PNMT | 0.00 | 2.20 | 11.10 | up | 0.00 | Phenylethanolamine N-methyltransferase |
| LOC110351439 | 5.40 | 0.14 | −5.28 | down | 0.00 | Histone H3 |
| LOC101799083 | 1.67 | 0.03 | −5.58 | down | 0.00 | Trefoil factor 2 |
| LOC119716807 | 2.07 | 0.06 | −5.00 | down | 0.00 | Putative short transient receptor potential channel 2-like protein |
| TMEM196 | 9.33 | 0.23 | −5.32 | down | 0.00 | Transmembrane protein 196 isoform X3 |
| GJD2 | 2.05 | 0.07 | −4.95 | down | 0.00 | Gap junction delta-2 protein |
| IHH | 1.96 | 0.04 | −5.55 | down | 0.00 | Indian hedgehog protein |
| LOC119718687 | 1.86 | 0.05 | −5.29 | down | 0.00 | Butyrophilin subfamily 1 member A1-like |
| SFTPC | 6.33 | 0.21 | −4.89 | down | 0.00 | Pulmonary surfactant-associated protein C isoform X1 |
| LOC110354773 | 7.78 | 0.33 | −4.57 | down | 0.00 | Pulmonary surfactant-associated protein C-like |
| LOC119715269 | 0.40 | 0.00 | −8.66 | down | 0.00 | Trypsin-3-like |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, Y.; Guo, R.; Zeng, T.; Sun, H.; Tian, Y.; Han, X.; Cao, Y.; Xu, L.; Duan, M.; Lu, L.; et al. Analysis of Transcriptomic Differences in the Ovaries of High- and Low-Laying Ducks. Genes 2024, 15, 181. https://doi.org/10.3390/genes15020181
Chang Y, Guo R, Zeng T, Sun H, Tian Y, Han X, Cao Y, Xu L, Duan M, Lu L, et al. Analysis of Transcriptomic Differences in the Ovaries of High- and Low-Laying Ducks. Genes. 2024; 15(2):181. https://doi.org/10.3390/genes15020181
Chicago/Turabian StyleChang, Yuguang, Rongbing Guo, Tao Zeng, Hanxue Sun, Yong Tian, Xue Han, Yongqing Cao, Ligen Xu, Mingcai Duan, Lizhi Lu, and et al. 2024. "Analysis of Transcriptomic Differences in the Ovaries of High- and Low-Laying Ducks" Genes 15, no. 2: 181. https://doi.org/10.3390/genes15020181
APA StyleChang, Y., Guo, R., Zeng, T., Sun, H., Tian, Y., Han, X., Cao, Y., Xu, L., Duan, M., Lu, L., & Chen, L. (2024). Analysis of Transcriptomic Differences in the Ovaries of High- and Low-Laying Ducks. Genes, 15(2), 181. https://doi.org/10.3390/genes15020181





