The Intriguing Mystery of RPA Phosphorylation in DNA Double-Strand Break Repair
Abstract
1. Introduction
2. Structure of RPA
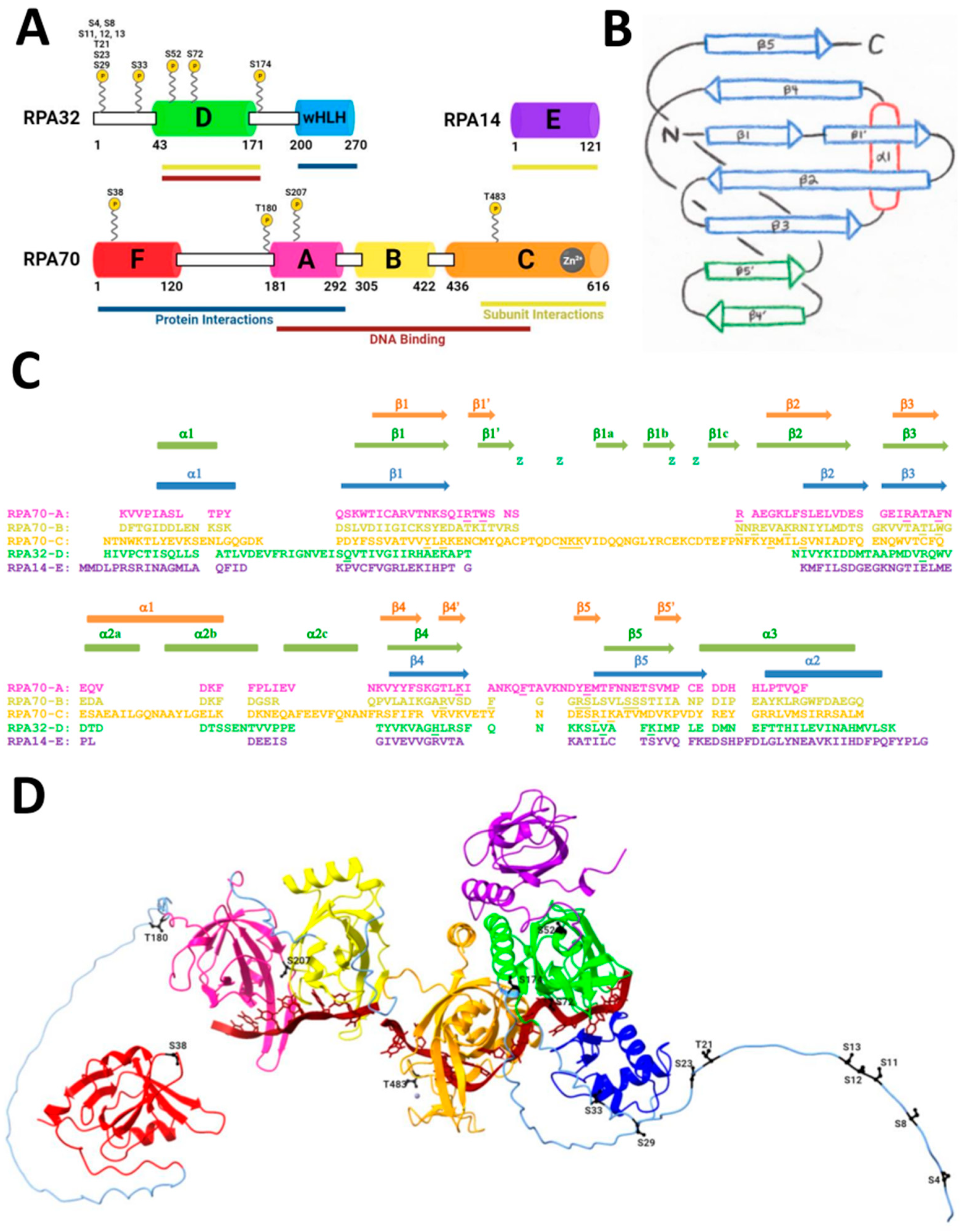
3. The Function of RPA in DNA Double-Strand Break Repair
4. ssDNA and Protein Interactions involving RPA
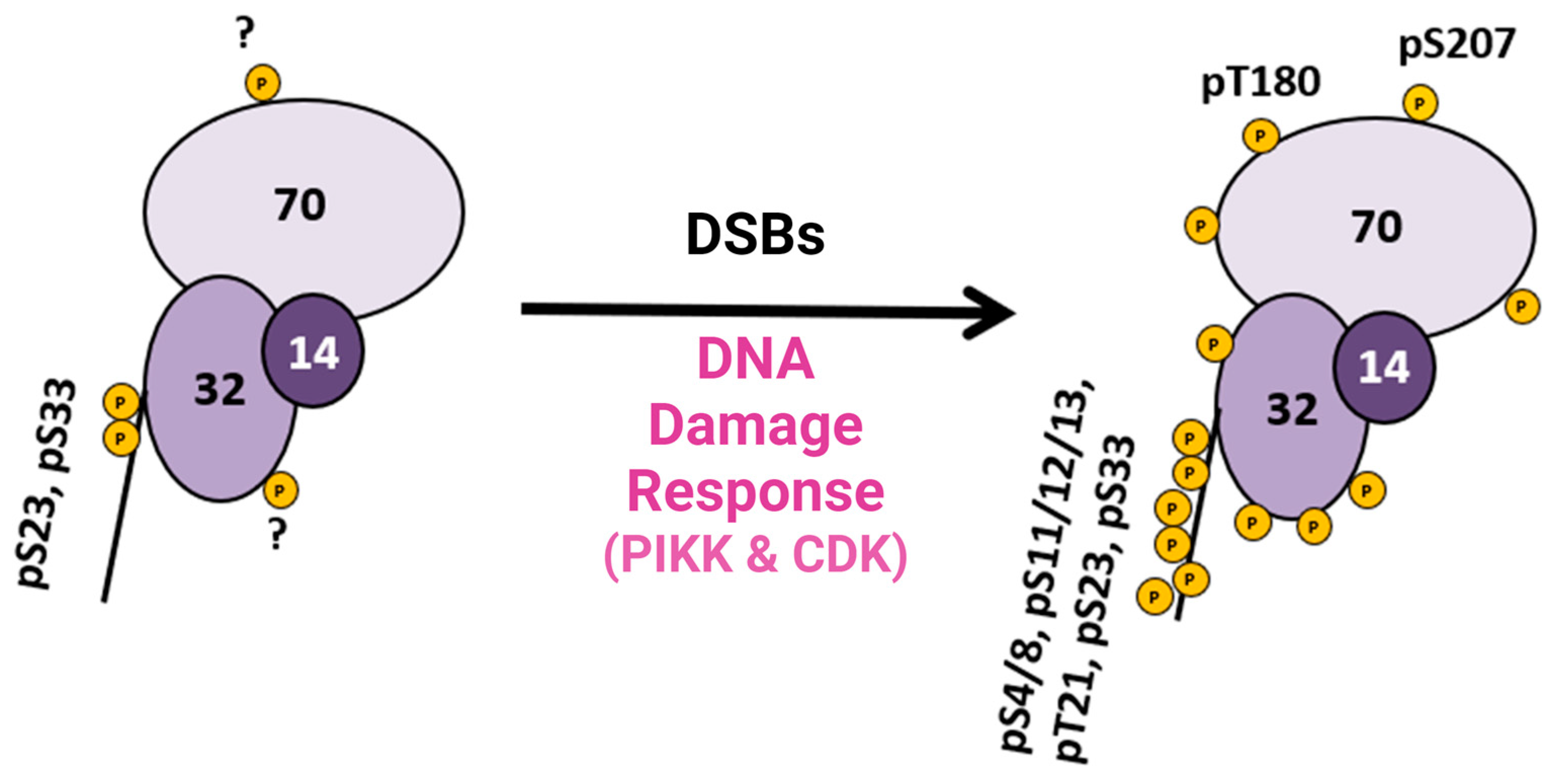
5. PTMs That Regulate RPA in DNA Metabolism
6. Future Directions for RPA Research
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bastin-Shanower, S.A.; Brill, S.J. Functional analysis of the four DNA binding domains of replication protein A. The role of RPA2 in ssDNA binding. J. Biol. Chem. 2001, 276, 36446–36453. [Google Scholar] [CrossRef]
- Wold, M.S.; Weinberg, D.H.; Virshup, D.M.; Li, J.J.; Kelly, T.J. Identification of cellular proteins required for simian virus 40 DNA replication. J. Biol. Chem. 1989, 264, 2801–2809. [Google Scholar] [CrossRef]
- Wold, M.S.; McMacken, R. Regulation of expression of the Escherichia coli dnaG gene and amplification of the dnaG primase. Proc. Natl. Acad. Sci. USA 1982, 79, 4907–4911. [Google Scholar] [CrossRef]
- Wold, M.S.; Mallory, J.B.; Roberts, J.D.; LeBowitz, J.H.; McMacken, R. Initiation of bacteriophage lambda DNA replication in vitro with purified lambda replication proteins. Proc. Natl. Acad. Sci. USA 1982, 79, 6176–6180. [Google Scholar] [CrossRef] [PubMed]
- Wold, M.S.; Li, J.J.; Kelly, T.J. Initiation of simian virus 40 DNA replication in vitro: Large-tumor-antigen- and origin-dependent unwinding of the template. Proc. Natl. Acad. Sci. USA 1987, 84, 3643–3647. [Google Scholar] [CrossRef] [PubMed]
- Kelly, T.J.; Rosenfeld, P.J.; Wides, R.J.; O’Neill, E.A.; Li, J.J.; Wold, M.S. Replication of adenovirus and SV40 chromosomes in vitro. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1987, 317, 429–438. [Google Scholar] [CrossRef]
- Wold, M.S.; Kelly, T. Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc. Natl. Acad. Sci. USA 1988, 85, 2523–2527. [Google Scholar] [CrossRef]
- Erdile, L.F.; Wold, M.S.; Kelly, T.J. The primary structure of the 32-kDa subunit of human replication protein A. J. Biol. Chem. 1990, 265, 3177–3182. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Snyder, R.O.; Wold, M.S. Binding properties of replication protein A from human and yeast cells. Mol. Cell Biol. 1992, 12, 3050–3059. [Google Scholar] [CrossRef]
- Henricksen, L.A.; Umbricht, C.B.; Wold, M.S. Recombinant replication protein A: Expression, complex formation, and functional characterization. J. Biol. Chem. 1994, 269, 11121–11132. [Google Scholar] [CrossRef]
- Henricksen, L.A.; Wold, M.S. Replication protein A mutants lacking phosphorylation sites for p34cdc2 kinase support DNA replication. J. Biol. Chem. 1994, 269, 24203–24208. [Google Scholar] [CrossRef]
- Gomes, X.V.; Wold, M.S. Structural analysis of human replication protein A. Mapping functional domains of the 70-kDa subunit. J. Biol. Chem. 1995, 270, 4534–4543. [Google Scholar] [CrossRef]
- He, Z.; Henricksen, L.A.; Wold, M.S.; Ingles, C.J. RPA involvement in the damage-recognition and incision steps of nucleotide excision repair. Nature 1995, 374, 566–569. [Google Scholar] [CrossRef]
- Gomes, X.V.; Henricksen, L.A.; Wold, M.S. Proteolytic mapping of human replication protein A: Evidence for multiple structural domains and a conformational change upon interaction with single-stranded DNA. Biochemistry 1996, 35, 5586–5595. [Google Scholar] [CrossRef]
- Henricksen, L.A.; Carter, T.; Dutta, A.; Wold, M.S. Phosphorylation of human replication protein A by the DNA-dependent protein kinase is involved in the modulation of DNA replication. Nucleic Acids Res. 1996, 24, 3107–3112. [Google Scholar] [CrossRef]
- Wold, M.S. Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997, 66, 61–92. [Google Scholar] [CrossRef]
- Sibenaller, Z.A.; Sorensen, B.R.; Wold, M.S. The 32- and 14-kilodalton subunits of replication protein A are responsible for species-specific interactions with single-stranded DNA. Biochemistry 1998, 37, 12496–12506. [Google Scholar] [CrossRef] [PubMed]
- Shao, R.G.; Cao, C.X.; Zhang, H.; Kohn, K.W.; Wold, M.S.; Pommier, Y. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J. 1999, 18, 1397–1406. [Google Scholar] [CrossRef] [PubMed]
- Walther, A.P.; Gomes, X.V.; Lao, Y.; Lee, C.G.; Wold, M.S. Replication protein A interactions with DNA. 1. Functions of the DNA-binding and zinc-finger domains of the 70-kDa subunit. Biochemistry 1999, 38, 3963–3973. [Google Scholar] [CrossRef] [PubMed]
- Buchko, G.W.; Daughdrill, G.W.; de Lorimier, R.; Rao, B.K.; Isern, N.G.; Lingbeck, J.M.; Taylor, J.S.; Wold, M.S.; Gochin, M.; Spicer, L.D.; et al. Interactions of human nucleotide excision repair protein XPA with DNA and RPA70 Delta C327: Chemical shift mapping and 15N NMR relaxation studies. Biochemistry 1999, 38, 15116–15128. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D.; Dhar, K.; Wahl, J.K.; Wold, M.S.; Borgstahl, G.E. Analysis of the human replication protein A:Rad52 complex: Evidence for crosstalk between RPA32, RPA70, Rad52 and DNA. J. Mol. Biol. 2002, 321, 133–148. [Google Scholar] [CrossRef]
- Binz, S.K.; Lao, Y.; Lowry, D.F.; Wold, M.S. The phosphorylation domain of the 32-kDa subunit of replication protein A (RPA) modulates RPA-DNA interactions. Evidence for an intersubunit interaction. J. Biol. Chem. 2003, 278, 35584–35591. [Google Scholar] [CrossRef] [PubMed]
- Binz, S.K.; Sheehan, A.M.; Wold, M.S. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair 2004, 3, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Mason, A.C.; Haring, S.J.; Pryor, J.M.; Staloch, C.A.; Gan, T.F.; Wold, M.S. An alternative form of replication protein a prevents viral replication in vitro. J. Biol. Chem. 2009, 284, 5324–5331. [Google Scholar] [CrossRef] [PubMed]
- Kemp, M.G.; Mason, A.C.; Carreira, A.; Reardon, J.T.; Haring, S.J.; Borgstahl, G.E.; Kowalczykowski, S.C.; Sancar, A.; Wold, M.S. An alternative form of replication protein a expressed in normal human tissues supports DNA repair. J. Biol. Chem. 2010, 285, 4788–4797. [Google Scholar] [CrossRef] [PubMed]
- Hass, C.S.; Gakhar, L.; Wold, M.S. Functional characterization of a cancer causing mutation in human replication protein A. Mol. Cancer Res. 2010, 8, 1017–1026. [Google Scholar] [CrossRef] [PubMed]
- Hass, C.S.; Lam, K.; Wold, M.S. Repair-specific functions of replication protein A. J. Biol. Chem. 2012, 287, 3908–3918. [Google Scholar] [CrossRef] [PubMed]
- Hass, C.S.; Chen, R.; Wold, M.S. Detection of posttranslational modifications of replication protein A. Methods Mol. Biol. 2012, 922, 193–204. [Google Scholar] [CrossRef]
- Chen, R.; Wold, M.S. Replication protein A: Single-stranded DNA’s first responder: Dynamic DNA-interactions allow replication protein A to direct single-strand DNA intermediates into different pathways for synthesis or repair. Bioessays 2014, 36, 1156–1161. [Google Scholar] [CrossRef]
- Chen, R.; Subramanyam, S.; Elcock, A.H.; Spies, M.; Wold, M.S. Dynamic binding of replication protein a is required for DNA repair. Nucleic Acids Res. 2016, 44, 5758–5772. [Google Scholar] [CrossRef]
- Bain, F.E.; Fischer, L.A.; Chen, R.; Wold, M.S. Single-Molecule Analysis of Replication Protein A-DNA Interactions. Methods Enzymol. 2018, 600, 439–461. [Google Scholar] [CrossRef] [PubMed]
- Pokhrel, N.; Caldwell, C.C.; Corless, E.I.; Tillison, E.A.; Tibbs, J.; Jocic, N.; Tabei, S.M.A.; Wold, M.S.; Spies, M.; Antony, E. Dynamics and selective remodeling of the DNA-binding domains of RPA. Nat. Struct. Mol. Biol. 2019, 26, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Gall-Duncan, T.; Luo, J.; Jurkovic, C.M.; Fischer, L.A.; Fujita, K.; Deshmukh, A.L.; Harding, R.J.; Tran, S.; Mehkary, M.; Li, V.; et al. Antagonistic roles of canonical and Alternative-RPA in disease-associated tandem CAG repeat instability. Cell 2023, 186, 4898–4919.e25. [Google Scholar] [CrossRef] [PubMed]
- Wyka, I.M.; Dhar, K.; Binz, S.K.; Wold, M.S. Replication protein A interactions with DNA: Differential binding of the core domains and analysis of the DNA interaction surface. Biochemistry 2003, 42, 12909–12918. [Google Scholar] [CrossRef]
- Caldwell, C.C.; Spies, M. Dynamic elements of replication protein A at the crossroads of DNA replication, recombination, and repair. Crit. Rev. Biochem. Mol. Biol. 2020, 55, 482–507. [Google Scholar] [CrossRef]
- He, H.; Wang, J.; Liu, T. UV-Induced RPA1 Acetylation Promotes Nucleotide Excision Repair. Cell Rep. 2017, 20, 2010–2025. [Google Scholar] [CrossRef]
- Xia, D.; Zhu, X.; Wang, Y.; Gong, P.; Su, H.S.; Xu, X. Implications of ubiquitination and the maintenance of replication fork stability in cancer therapy. Biosci. Rep. 2023, 43, BSR20222591. [Google Scholar] [CrossRef]
- Dou, H.; Huang, C.; Singh, M.; Carpenter, P.B.; Yeh, E.T. Regulation of DNA repair through deSUMOylation and SUMOylation of replication protein A complex. Mol. Cell 2010, 39, 333–345. [Google Scholar] [CrossRef]
- Din, S.; Brill, S.J.; Fairman, M.P.; Stillman, B. Cell-cycle-regulated phosphorylation of DNA replication factor A from human and yeast cells. Genes. Dev. 1990, 4, 968–977. [Google Scholar] [CrossRef]
- Borgstahl, G.E.; Brader, K.; Mosel, A.; Liu, S.; Kremmer, E.; Goettsch, K.A.; Kolar, C.; Nasheuer, H.P.; Oakley, G.G. Interplay of DNA damage and cell cycle signaling at the level of human replication protein A. DNA Repair 2014, 21, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Brush, G.S.; Kelly, T.J. Phosphorylation of the replication protein A large subunit in the Saccharomyces cerevisiae checkpoint response. Nucleic Acids Res. 2000, 28, 3725–3732. [Google Scholar] [CrossRef]
- Trovesi, C.; Manfrini, N.; Falcettoni, M.; Longhese, M.P. Regulation of the DNA damage response by cyclin-dependent kinases. J. Mol. Biol. 2013, 425, 4756–4766. [Google Scholar] [CrossRef]
- Block, W.D.; Yu, Y.; Lees-Miller, S.P. Phosphatidyl inositol 3-kinase-like serine/threonine protein kinases (PIKKs) are required for DNA damage-induced phosphorylation of the 32 kDa subunit of replication protein A at threonine 21. Nucleic Acids Res. 2004, 32, 997–1005. [Google Scholar] [CrossRef]
- Priya, B.; Ravi, S.; Kirubakaran, S. Targeting ATM and ATR for cancer therapeutics: Inhibitors in clinic. Drug Discov. Today 2023, 28, 103662. [Google Scholar] [CrossRef]
- Stillman, B.; Bell, S.P.; Dutta, A.; Marahrens, Y. DNA replication and the cell cycle. Ciba Found. Symp. 1992, 170, 147–156; discussion 156–160. [Google Scholar] [CrossRef] [PubMed]
- Sancar, A. DNA excision repair. Annu. Rev. Biochem. 1996, 65, 43–81. [Google Scholar] [CrossRef] [PubMed]
- Lohrer, H.D. Regulation of the cell cycle following DNA damage in normal and Ataxia telangiectasia cells. Experientia 1996, 52, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Iftode, C.; Daniely, Y.; Borowiec, J.A. Replication protein A (RPA): The eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 1999, 34, 141–180. [Google Scholar] [CrossRef] [PubMed]
- Bochkarev, A.; Bochkareva, E. From RPA to BRCA2: Lessons from single-stranded DNA binding by the OB-fold. Curr. Opin. Struct. Biol. 2004, 14, 36–42. [Google Scholar] [CrossRef]
- Zou, Y.; Liu, Y.; Wu, X.; Shell, S.M. Functions of human replication protein A (RPA): From DNA replication to DNA damage and stress responses. J. Cell Physiol. 2006, 208, 267–273. [Google Scholar] [CrossRef]
- Fanning, E.; Klimovich, V.; Nager, A.R. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006, 34, 4126–4137. [Google Scholar] [CrossRef]
- Oakley, G.G.; Patrick, S.M. Replication protein A: Directing traffic at the intersection of replication and repair. Front. Biosci. (Landmark Ed.) 2010, 15, 883–900. [Google Scholar] [CrossRef]
- Prakash, A.; Borgstahl, G.E. The structure and function of replication protein A in DNA replication. Subcell. Biochem. 2012, 62, 171–196. [Google Scholar] [CrossRef]
- Marechal, A.; Zou, L. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 2015, 25, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Huang, J. Replication protein A and more: Single-stranded DNA-binding proteins in eukaryotic cells. Acta Biochim Biophys Sin 2016, 48, 665–670. [Google Scholar] [CrossRef] [PubMed]
- Byrne, B.M.; Oakley, G.G. Replication protein A, the laxative that keeps DNA regular: The importance of RPA phosphorylation in maintaining genome stability. Semin. Cell Dev. Biol. 2019, 86, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Anindya, R. Single-stranded DNA damage: Protecting the single-stranded DNA from chemical attack. DNA Repair 2020, 87, 102804. [Google Scholar] [CrossRef] [PubMed]
- Rechkunova, N.I.; Lavrik, O.I. Photoreactive DNA as a Tool to Study Replication Protein A Functioning in DNA Replication and Repair. Photochem. Photobiol. 2020, 96, 440–449. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.; Chowdhury, A.B.; Kumar, M.; Chakraborty, S. Revisiting regulatory roles of replication protein A in plant DNA metabolism. Planta 2021, 253, 130. [Google Scholar] [CrossRef] [PubMed]
- Barbour, A.T.; Wuttke, D.S. RPA-like single-stranded DNA-binding protein complexes including CST serve as specialized processivity factors for polymerases. Curr. Opin. Struct. Biol. 2023, 81, 102611. [Google Scholar] [CrossRef]
- Kusi-Appauh, N.; Ralph, S.F.; van Oijen, A.M.; Spenkelink, L.M. Understanding G-Quadruplex Biology and Stability Using Single-Molecule Techniques. J. Phys. Chem. B 2023, 127, 5521–5540. [Google Scholar] [CrossRef] [PubMed]
- Nasheuer, H.P.; Meaney, A.M.; Hulshoff, T.; Thiele, I.; Onwubiko, N.O. Replication Protein A, the Main Eukaryotic Single-Stranded DNA Binding Protein, a Focal Point in Cellular DNA Metabolism. Int. J. Mol. Sci. 2024, 25, 588. [Google Scholar] [CrossRef] [PubMed]
- Bochkareva, E.; Belegu, V.; Korolev, S.; Bochkarev, A. Structure of the major single-stranded DNA-binding domain of replication protein A suggests a dynamic mechanism for DNA binding. EMBO J. 2001, 20, 612–618. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Lin, L.J.; Sriratana, P.; Coleman, K.; Ha, T.; Spies, M.; Cann, I.K. Engineering of functional replication protein a homologs based on insights into the evolution of oligonucleotide/oligosaccharide-binding folds. J. Bacteriol. 2008, 190, 5766–5780. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, C.A.H.; Morea, E.G.O.; Cano, M.I.N. RPA-1 from Leishmania sp.: Recombinant Protein Expression and Purification, Molecular Modeling, and Molecular Dynamics Simulations Protocols. Methods Mol. Biol. 2021, 2281, 169–191. [Google Scholar] [CrossRef] [PubMed]
- Bochkarev, A.; Pfuetzner, R.A.; Edwards, A.M.; Frappier, L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature 1997, 385, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Pavletich, N.P. Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes. Dev. 2012, 26, 2337–2347. [Google Scholar] [CrossRef]
- Jacobs, D.M.; Lipton, A.S.; Isern, N.G.; Daughdrill, G.W.; Lowry, D.F.; Gomes, X.; Wold, M.S. Human replication protein A: Global fold of the N-terminal RPA-70 domain reveals a basic cleft and flexible C-terminal linker. J. Biomol. NMR 1999, 14, 321–331. [Google Scholar] [CrossRef]
- Mer, G.; Bochkarev, A.; Gupta, R.; Bochkareva, E.; Frappier, L.; Ingles, C.J.; Edwards, A.M.; Chazin, W.J. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell 2000, 103, 449–456. [Google Scholar] [CrossRef]
- Zhou, C.; Pourmal, S.; Pavletich, N.P. Dna2 nuclease-helicase structure, mechanism and regulation by Rpa. Elife 2015, 4, e09832. [Google Scholar] [CrossRef]
- Bochkareva, E.; Korolev, S.; Lees-Miller, S.P.; Bochkarev, A. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J. 2002, 21, 1855–1863. [Google Scholar] [CrossRef]
- Bochkareva, E.; Kaustov, L.; Ayed, A.; Yi, G.S.; Lu, Y.; Pineda-Lucena, A.; Liao, J.C.; Okorokov, A.L.; Milner, J.; Arrowsmith, C.H.; et al. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc. Natl. Acad. Sci. USA 2005, 102, 15412–15417. [Google Scholar] [CrossRef]
- Frank, A.O.; Feldkamp, M.D.; Kennedy, J.P.; Waterson, A.G.; Pelz, N.F.; Patrone, J.D.; Vangamudi, B.; Camper, D.V.; Rossanese, O.W.; Chazin, W.J.; et al. Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach. J. Med. Chem. 2013, 56, 9242–9250. [Google Scholar] [CrossRef]
- Guilliam, T.A.; Brissett, N.C.; Ehlinger, A.; Keen, B.A.; Kolesar, P.; Taylor, E.M.; Bailey, L.J.; Lindsay, H.D.; Chazin, W.J.; Doherty, A.J. Molecular basis for PrimPol recruitment to replication forks by RPA. Nat. Commun. 2017, 8, 15222. [Google Scholar] [CrossRef]
- Deng, X.; Habel, J.E.; Kabaleeswaran, V.; Snell, E.H.; Wold, M.S.; Borgstahl, G.E. Structure of the full-length human RPA14/32 complex gives insights into the mechanism of DNA binding and complex formation. J. Mol. Biol. 2007, 374, 865–876. [Google Scholar] [CrossRef]
- Wu, Y.; Fu, W.; Zang, N.; Zhou, C. Structural characterization of human RPA70N association with DNA damage response proteins. Elife 2023, 12, e81639. [Google Scholar] [CrossRef]
- Patrone, J.D.; Pelz, N.F.; Bates, B.S.; Souza-Fagundes, E.M.; Vangamudi, B.; Camper, D.V.; Kuznetsov, A.G.; Browning, C.F.; Feldkamp, M.D.; Frank, A.O.; et al. Identification and Optimization of Anthranilic Acid Based Inhibitors of Replication Protein A. ChemMedChem 2016, 11, 893–899. [Google Scholar] [CrossRef] [PubMed]
- Waterson, A.G.; Kennedy, J.P.; Patrone, J.D.; Pelz, N.F.; Feldkamp, M.D.; Frank, A.O.; Vangamudi, B.; Souza-Fagundes, E.M.; Rossanese, O.W.; Chazin, W.J.; et al. Diphenylpyrazoles as replication protein a inhibitors. ACS Med. Chem. Lett. 2015, 6, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Feldkamp, M.D.; Frank, A.O.; Kennedy, J.P.; Patrone, J.D.; Vangamudi, B.; Waterson, A.G.; Fesik, S.W.; Chazin, W.J. Surface reengineering of RPA70N enables cocrystallization with an inhibitor of the replication protein A interaction motif of ATR interacting protein. Biochemistry 2013, 52, 6515–6524. [Google Scholar] [CrossRef] [PubMed]
- Patrone, J.D.; Kennedy, J.P.; Frank, A.O.; Feldkamp, M.D.; Vangamudi, B.; Pelz, N.F.; Rossanese, O.W.; Waterson, A.G.; Chazin, W.J.; Fesik, S.W. Discovery of Protein-Protein Interaction Inhibitors of Replication Protein A. ACS Med. Chem. Lett. 2013, 4, 601–605. [Google Scholar] [CrossRef] [PubMed]
- Bochkarev, A.; Bochkareva, E.; Frappier, L.; Edwards, A.M. The crystal structure of the complex of replication protein A subunits RPA32 and RPA14 reveals a mechanism for single-stranded DNA binding. EMBO J. 1999, 18, 4498–4504. [Google Scholar] [CrossRef] [PubMed]
- Feldkamp, M.D.; Mason, A.C.; Eichman, B.F.; Chazin, W.J. Structural analysis of replication protein A recruitment of the DNA damage response protein SMARCAL1. Biochemistry 2014, 53, 3052–3061. [Google Scholar] [CrossRef] [PubMed]
- Madru, C.; Martinez-Carranza, M.; Laurent, S.; Alberti, A.C.; Chevreuil, M.; Raynal, B.; Haouz, A.; Le Meur, R.A.; Delarue, M.; Henneke, G.; et al. DNA-binding mechanism and evolution of replication protein A. Nat. Commun. 2023, 14, 2326. [Google Scholar] [CrossRef] [PubMed]
- Brosey, C.A.; Soss, S.E.; Brooks, S.; Yan, C.; Ivanov, I.; Dorai, K.; Chazin, W.J. Functional dynamics in replication protein A DNA binding and protein recruitment domains. Structure 2015, 23, 1028–1038. [Google Scholar] [CrossRef] [PubMed]
- Pretto, D.I.; Tsutakawa, S.; Brosey, C.A.; Castillo, A.; Chagot, M.E.; Smith, J.A.; Tainer, J.A.; Chazin, W.J. Structural dynamics and single-stranded DNA binding activity of the three N-terminal domains of the large subunit of replication protein A from small angle X-ray scattering. Biochemistry 2010, 49, 2880–2889. [Google Scholar] [CrossRef] [PubMed]
- Powers, K.T.; Gildenberg, M.S.; Washington, M.T. Modeling Conformationally Flexible Proteins With X-ray Scattering and Molecular Simulations. Comput. Struct. Biotechnol. J. 2019, 17, 570–578. [Google Scholar] [CrossRef] [PubMed]
- Dutta, A.; Din, S.; Brill, S.J.; Stillman, B. Phosphorylation of replication protein A: A role for cdc2 kinase in G1/S regulation. Cold Spring Harb. Symp. Quant. Biol. 1991, 56, 315–324. [Google Scholar] [CrossRef]
- Murti, K.G.; He, D.C.; Brinkley, B.R.; Scott, R.; Lee, S.H. Dynamics of human replication protein A subunit distribution and partitioning in the cell cycle. Exp. Cell Res. 1996, 223, 279–289. [Google Scholar] [CrossRef]
- Thoma, B.S.; Vasquez, K.M. Critical DNA damage recognition functions of XPC-hHR23B and XPA-RPA in nucleotide excision repair. Mol. Carcinog. 2003, 38, 1–13. [Google Scholar] [CrossRef]
- Zhao, M.; Geng, R.; Guo, X.; Yuan, R.; Zhou, X.; Zhong, Y.; Huo, Y.; Zhou, M.; Shen, Q.; Li, Y.; et al. PCAF/GCN5-Mediated Acetylation of RPA1 Promotes Nucleotide Excision Repair. Cell Rep. 2017, 20, 1997–2009. [Google Scholar] [CrossRef]
- Hayran, A.B.; Liabakk, N.B.; Aas, P.A.; Kusnierczyk, A.; Vagbo, C.B.; Sarno, A.; Iveland, T.S.; Chawla, K.; Zahn, A.; Di Noia, J.M.; et al. RPA guides UNG to uracil in ssDNA to facilitate antibody class switching and repair of mutagenic uracil at the replication fork. Nucleic Acids Res. 2024, 52, 784–800. [Google Scholar] [CrossRef]
- Gavande, N.S.; VanderVere-Carozza, P.S.; Hinshaw, H.D.; Jalal, S.I.; Sears, C.R.; Pawelczak, K.S.; Turchi, J.J. DNA repair targeted therapy: The past or future of cancer treatment? Pharmacol. Ther. 2016, 160, 65–83. [Google Scholar] [CrossRef]
- Deng, X.; Prakash, A.; Dhar, K.; Baia, G.S.; Kolar, C.; Oakley, G.G.; Borgstahl, G.E. Human replication protein A-Rad52-single-stranded DNA complex: Stoichiometry and evidence for strand transfer regulation by phosphorylation. Biochemistry 2009, 48, 6633–6643. [Google Scholar] [CrossRef]
- Wright, W.D.; Shah, S.S.; Heyer, W.D. Homologous recombination and the repair of DNA double-strand breaks. J. Biol. Chem. 2018, 293, 10524–10535. [Google Scholar] [CrossRef] [PubMed]
- Creeden, J.F.; Nanavaty, N.S.; Einloth, K.R.; Gillman, C.E.; Stanbery, L.; Hamouda, D.M.; Dworkin, L.; Nemunaitis, J. Homologous recombination proficiency in ovarian and breast cancer patients. BMC Cancer 2021, 21, 1154. [Google Scholar] [CrossRef] [PubMed]
- Prakash, A.; Natarajan, A.; Marky, L.A.; Ouellette, M.M.; Borgstahl, G.E. Identification of the DNA-Binding Domains of Human Replication Protein A That Recognize G-Quadruplex DNA. J. Nucleic Acids 2011, 2011, 896947. [Google Scholar] [CrossRef]
- Lysetska, M.; Knoll, A.; Boehringer, D.; Hey, T.; Krauss, G.; Krausch, G. UV light-damaged DNA and its interaction with human replication protein A: An atomic force microscopy study. Nucleic Acids Res. 2002, 30, 2686–2691. [Google Scholar] [CrossRef] [PubMed]
- You, J.S.; Wang, M.; Lee, S.H. Functional characterization of zinc-finger motif in redox regulation of RPA-ssDNA interaction. Biochemistry 2000, 39, 12953–12958. [Google Scholar] [CrossRef]
- Prakash, A.; Kieken, F.; Marky, L.A.; Borgstahl, G.E. Stabilization of a G-Quadruplex from Unfolding by Replication Protein A Using Potassium and the Porphyrin TMPyP4. J. Nucleic Acids 2011, 2011, 529828. [Google Scholar] [CrossRef]
- Ding, J.; Li, X.; Shen, J.; Zhao, Y.; Zhong, S.; Lai, L.; Niu, H.; Qi, Z. ssDNA accessibility of Rad51 is regulated by orchestrating multiple RPA dynamics. Nat. Commun. 2023, 14, 3864. [Google Scholar] [CrossRef]
- Kang, Y.; Han, Y.G.; Khim, K.W.; Choi, W.G.; Ju, M.K.; Park, K.; Shin, K.J.; Chae, Y.C.; Choi, J.H.; Kim, H.; et al. Alteration of replication protein A binding mode on single-stranded DNA by NSMF potentiates RPA phosphorylation by ATR kinase. Nucleic Acids Res. 2023, 51, 7936–7950. [Google Scholar] [CrossRef]
- Braun, K.A.; Lao, Y.; He, Z.; Ingles, C.J.; Wold, M.S. Role of protein-protein interactions in the function of replication protein A (RPA): RPA modulates the activity of DNA polymerase Alpha by multiple mechanisms. Biochemistry 1997, 36, 8443–8454. [Google Scholar] [CrossRef]
- Golub, E.I.; Gupta, R.C.; Haaf, T.; Wold, M.S.; Radding, C.M. Interaction of human rad51 recombination protein with single-stranded DNA binding protein, RPA. Nucleic Acids Res. 1998, 26, 5388–5393. [Google Scholar] [CrossRef]
- Riva, F.; Zuco, V.; Vink, A.A.; Supino, R.; Prosperi, E. UV-induced DNA incision and proliferating cell nuclear antigen recruitment to repair sites occur independently of p53-replication protein A interaction in p53 wild type and mutant ovarian carcinoma cells. Carcinogenesis 2001, 22, 1971–1978. [Google Scholar] [CrossRef]
- Wang, M.; Mahrenholz, A.; Lee, S.H. RPA stabilizes the XPA-damaged DNA complex through protein-protein interaction. Biochemistry 2000, 39, 6433–6439. [Google Scholar] [CrossRef]
- Yoo, E.; Kim, B.U.; Lee, S.Y.; Cho, C.H.; Chung, J.H.; Lee, C.H. 53BP1 is associated with replication protein A and is required for RPA2 hyperphosphorylation following DNA damage. Oncogene 2005, 24, 5423–5430. [Google Scholar] [CrossRef]
- Chaudhuri, J.; Khuong, C.; Alt, F.W. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature 2004, 430, 992–998. [Google Scholar] [CrossRef] [PubMed]
- Kalan, S.; Matveyenko, A.; Loayza, D. LIM Protein Ajuba Participates in the Repression of the ATR-Mediated DNA Damage Response. Front. Genet. 2013, 4, 95. [Google Scholar] [CrossRef]
- Fowler, S.; Maguin, P.; Kalan, S.; Loayza, D. LIM Protein Ajuba associates with the RPA complex through direct cell cycle-dependent interaction with the RPA70 subunit. Sci. Rep. 2018, 8, 9536. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Elledge, S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003, 300, 1542–1548. [Google Scholar] [CrossRef] [PubMed]
- Ball, H.L.; Myers, J.S.; Cortez, D. ATRIP binding to replication protein A-single-stranded DNA promotes ATR-ATRIP localization but is dispensable for Chk1 phosphorylation. Mol. Biol. Cell 2005, 16, 2372–2381. [Google Scholar] [CrossRef]
- Namiki, Y.; Zou, L. ATRIP associates with replication protein A-coated ssDNA through multiple interactions. Proc. Natl. Acad. Sci. USA 2006, 103, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Vaithiyalingam, S.; Shi, Q.; Chazin, W.J.; Zinkel, S.S. BID binds to replication protein A and stimulates ATR function following replicative stress. Mol. Cell Biol. 2011, 31, 4298–4309. [Google Scholar] [CrossRef] [PubMed]
- Brosh, R.M., Jr.; Li, J.L.; Kenny, M.K.; Karow, J.K.; Cooper, M.P.; Kureekattil, R.P.; Hickson, I.D.; Bohr, V.A. Replication protein A physically interacts with the Bloom’s syndrome protein and stimulates its helicase activity. J. Biol. Chem. 2000, 275, 23500–23508. [Google Scholar] [CrossRef] [PubMed]
- Doherty, K.M.; Sommers, J.A.; Gray, M.D.; Lee, J.W.; von Kobbe, C.; Thoma, N.H.; Kureekattil, R.P.; Kenny, M.K.; Brosh, R.M., Jr. Physical and functional mapping of the replication protein a interaction domain of the werner and bloom syndrome helicases. J. Biol. Chem. 2005, 280, 29494–29505. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, L.J.; Borowiec, J.A.; Mastrangelo, I.A. Single-stranded-DNA binding alters human replication protein A structure and facilitates interaction with DNA-dependent protein kinase. Mol. Cell Biol. 1996, 16, 4798–4807. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Zhao, Y.; Xu, Y.; Ning, S.; Huo, W.; Hou, M.; Gao, G.; Ji, J.; Guo, R.; Xu, D. Ewing Tumor-associated Antigen 1 Interacts with Replication Protein A to Promote Restart of Stalled Replication Forks. J. Biol. Chem. 2016, 291, 21956–21962. [Google Scholar] [CrossRef]
- Haahr, P.; Hoffmann, S.; Tollenaere, M.A.; Ho, T.; Toledo, L.I.; Mann, M.; Bekker-Jensen, S.; Raschle, M.; Mailand, N. Activation of the ATR kinase by the RPA-binding protein ETAA1. Nat. Cell Biol. 2016, 18, 1196–1207. [Google Scholar] [CrossRef]
- Bass, T.E.; Luzwick, J.W.; Kavanaugh, G.; Carroll, C.; Dungrawala, H.; Glick, G.G.; Feldkamp, M.D.; Putney, R.; Chazin, W.J.; Cortez, D. ETAA1 acts at stalled replication forks to maintain genome integrity. Nat. Cell Biol. 2016, 18, 1185–1195. [Google Scholar] [CrossRef]
- Lee, Y.C.; Zhou, Q.; Chen, J.; Yuan, J. RPA-Binding Protein ETAA1 Is an ATR Activator Involved in DNA Replication Stress Response. Curr. Biol. 2016, 26, 3257–3268. [Google Scholar] [CrossRef]
- Zhao, W.; Vaithiyalingam, S.; San Filippo, J.; Maranon, D.G.; Jimenez-Sainz, J.; Fontenay, G.V.; Kwon, Y.; Leung, S.G.; Lu, L.; Jensen, R.B.; et al. Promotion of BRCA2-Dependent Homologous Recombination by DSS1 via RPA Targeting and DNA Mimicry. Mol. Cell 2015, 59, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, M.; Takaki, E.; Takii, R.; Tan, K.; Prakasam, R.; Hayashida, N.; Iemura, S.; Natsume, T.; Nakai, A. RPA assists HSF1 access to nucleosomal DNA by recruiting histone chaperone FACT. Mol. Cell 2012, 48, 182–194. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Sharma, S.; Sommers, J.A.; Kenny, M.K.; Cantor, S.B.; Brosh, R.M., Jr. FANCJ (BACH1) helicase forms DNA damage inducible foci with replication protein A and interacts physically and functionally with the single-stranded DNA-binding protein. Blood 2007, 110, 2390–2398. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Zhu, M.; Wu, W.; Rokutanda, N.; Togashi, Y.; Liang, W.; Ohta, T. HERC2 regulates RPA2 by mediating ATR-induced Ser33 phosphorylation and ubiquitin-dependent degradation. Sci. Rep. 2019, 9, 14257. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Rokutanda, N.; Takeuchi, J.; Lai, Y.; Maruyama, R.; Togashi, Y.; Nishikawa, H.; Arai, N.; Miyoshi, Y.; Suzuki, N.; et al. HERC2 Facilitates BLM and WRN Helicase Complex Interaction with RPA to Suppress G-Quadruplex DNA. Cancer Res. 2018, 78, 6371–6385. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Gan, H.; Wang, Z.; Lee, J.H.; Zhou, H.; Ordog, T.; Wold, M.S.; Ljungman, M.; Zhang, Z. RPA Interacts with HIRA and Regulates H3.3 Deposition at Gene Regulatory Elements in Mammalian Cells. Mol. Cell 2017, 65, 272–284. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Xu, Z.; Leng, H.; Zheng, P.; Yang, J.; Chen, K.; Feng, J.; Li, Q. RPA binds histone H3-H4 and functions in DNA replication-coupled nucleosome assembly. Science 2017, 355, 415–420. [Google Scholar] [CrossRef]
- Dueva, R.; Iliakis, G. Replication protein A: A multifunctional protein with roles in DNA replication, repair and beyond. NAR Cancer 2020, 2, zcaa022. [Google Scholar] [CrossRef]
- Sukhodolets, K.E.; Hickman, A.B.; Agarwal, S.K.; Sukhodolets, M.V.; Obungu, V.H.; Novotny, E.A.; Crabtree, J.S.; Chandrasekharappa, S.C.; Collins, F.S.; Spiegel, A.M.; et al. The 32-kilodalton subunit of replication protein A interacts with menin, the product of the MEN1 tumor suppressor gene. Mol. Cell Biol. 2003, 23, 493–509. [Google Scholar] [CrossRef]
- Chen, C.C.; Juan, C.W.; Chen, K.Y.; Chang, Y.C.; Lee, J.C.; Chang, M.C. Upregulation of RPA2 promotes NF-kappaB activation in breast cancer by relieving the antagonistic function of menin on NF-kappaB-regulated transcription. Carcinogenesis 2017, 38, 196–206. [Google Scholar] [CrossRef][Green Version]
- Robison, J.G.; Elliott, J.; Dixon, K.; Oakley, G.G. Replication protein A and the Mre11.Rad50.Nbs1 complex co-localize and interact at sites of stalled replication forks. J. Biol. Chem. 2004, 279, 34802–34810. [Google Scholar] [CrossRef]
- Oakley, G.G.; Tillison, K.; Opiyo, S.A.; Glanzer, J.G.; Horn, J.M.; Patrick, S.M. Physical interaction between replication protein A (RPA) and MRN: Involvement of RPA2 phosphorylation and the N-terminus of RPA1. Biochemistry 2009, 48, 7473–7481. [Google Scholar] [CrossRef]
- Daniely, Y.; Borowiec, J.A. Formation of a complex between nucleolin and replication protein A after cell stress prevents initiation of DNA replication. J. Cell Biol. 2000, 149, 799–810. [Google Scholar] [CrossRef]
- Kim, K.; Dimitrova, D.D.; Carta, K.M.; Saxena, A.; Daras, M.; Borowiec, J.A. Novel checkpoint response to genotoxic stress mediated by nucleolin-replication protein a complex formation. Mol. Cell Biol. 2005, 25, 2463–2474. [Google Scholar] [CrossRef]
- Wang, Y.; Guan, J.; Wang, H.; Wang, Y.; Leeper, D.; Iliakis, G. Regulation of DNA replication after heat shock by replication protein a-nucleolin interactions. J. Biol. Chem. 2001, 276, 20579–20588. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Botchan, M.R. The acidic transcriptional activation domains of VP16 and p53 bind the cellular replication protein A and stimulate in vitro BPV-1 DNA replication. Cell 1993, 73, 1207–1221. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Brinton, B.T.; Greenblatt, J.; Hassell, J.A.; Ingles, C.J. The transactivator proteins VP16 and GAL4 bind replication factor A. Cell 1993, 73, 1223–1232. [Google Scholar] [CrossRef] [PubMed]
- Dutta, A.; Ruppert, J.M.; Aster, J.C.; Winchester, E. Inhibition of DNA replication factor RPA by p53. Nature 1993, 365, 79–82. [Google Scholar] [CrossRef]
- Romanova, L.Y.; Willers, H.; Blagosklonny, M.V.; Powell, S.N. The interaction of p53 with replication protein A mediates suppression of homologous recombination. Oncogene 2004, 23, 9025–9033. [Google Scholar] [CrossRef] [PubMed]
- Serrano, M.A.; Li, Z.; Dangeti, M.; Musich, P.R.; Patrick, S.; Roginskaya, M.; Cartwright, B.; Zou, Y. DNA-PK, ATM and ATR collaboratively regulate p53-RPA interaction to facilitate homologous recombination DNA repair. Oncogene 2013, 32, 2452–2462. [Google Scholar] [CrossRef] [PubMed]
- Murphy, A.K.; Fitzgerald, M.; Ro, T.; Kim, J.H.; Rabinowitsch, A.I.; Chowdhury, D.; Schildkraut, C.L.; Borowiec, J.A. Phosphorylated RPA recruits PALB2 to stalled DNA replication forks to facilitate fork recovery. J. Cell Biol. 2014, 206, 493–507. [Google Scholar] [CrossRef]
- Wan, L.; Lou, J.; Xia, Y.; Su, B.; Liu, T.; Cui, J.; Sun, Y.; Lou, H.; Huang, J. hPrimpol1/CCDC111 is a human DNA primase-polymerase required for the maintenance of genome integrity. EMBO Rep. 2013, 14, 1104–1112. [Google Scholar] [CrossRef]
- Guilliam, T.A.; Jozwiakowski, S.K.; Ehlinger, A.; Barnes, R.P.; Rudd, S.G.; Bailey, L.J.; Skehel, J.M.; Eckert, K.A.; Chazin, W.J.; Doherty, A.J. Human PrimPol is a highly error-prone polymerase regulated by single-stranded DNA binding proteins. Nucleic Acids Res. 2015, 43, 1056–1068. [Google Scholar] [CrossRef]
- Marechal, A.; Li, J.M.; Ji, X.Y.; Wu, C.S.; Yazinski, S.A.; Nguyen, H.D.; Liu, S.; Jimenez, A.E.; Jin, J.; Zou, L. PRP19 transforms into a sensor of RPA-ssDNA after DNA damage and drives ATR activation via a ubiquitin-mediated circuitry. Mol. Cell 2014, 53, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, Y.; Wang, P.; Liang, H.; Cui, M.; Zhu, M.; Guo, L.; Su, Q.; Sun, Y.; McNutt, M.A.; et al. PTEN regulates RPA1 and protects DNA replication forks. Cell Res. 2015, 25, 1189–1204. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Shell, S.M.; Zou, Y. Interaction and colocalization of Rad9/Rad1/Hus1 checkpoint complex with replication protein A in human cells. Oncogene 2005, 24, 4728–4735. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Liu, D.; Elledge, S.J. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc. Natl. Acad. Sci. USA 2003, 100, 13827–13832. [Google Scholar] [CrossRef]
- Hedglin, M.; Aitha, M.; Pedley, A.; Benkovic, S.J. Replication protein A dynamically regulates monoubiquitination of proliferating cell nuclear antigen. J. Biol. Chem. 2019, 294, 5157–5168. [Google Scholar] [CrossRef]
- Wu, X.; Yang, Z.; Liu, Y.; Zou, Y. Preferential localization of hyperphosphorylated replication protein A to double-strand break repair and checkpoint complexes upon DNA damage. Biochem. J. 2005, 391, 473–480. [Google Scholar] [CrossRef]
- Ma, C.J.; Kwon, Y.; Sung, P.; Greene, E.C. Human RAD52 interactions with replication protein A and the RAD51 presynaptic complex. J. Biol. Chem. 2017, 292, 11702–11713. [Google Scholar] [CrossRef]
- Stauffer, M.E.; Chazin, W.J. Physical interaction between replication protein A and Rad51 promotes exchange on single-stranded DNA. J. Biol. Chem. 2004, 279, 25638–25645. [Google Scholar] [CrossRef]
- Park, M.S.; Ludwig, D.L.; Stigger, E.; Lee, S.H. Physical interaction between human RAD52 and RPA is required for homologous recombination in mammalian cells. J. Biol. Chem. 1996, 271, 18996–19000. [Google Scholar] [CrossRef]
- Shinohara, A.; Shinohara, M.; Ohta, T.; Matsuda, S.; Ogawa, T. Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes. Cells 1998, 3, 145–156. [Google Scholar] [CrossRef]
- Sugiyama, T.; Kowalczykowski, S.C. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J. Biol. Chem. 2002, 277, 31663–31672. [Google Scholar] [CrossRef]
- Plate, I.; Hallwyl, S.C.; Shi, I.; Krejci, L.; Muller, C.; Albertsen, L.; Sung, P.; Mortensen, U.H. Interaction with RPA is necessary for Rad52 repair center formation and for its mediator activity. J. Biol. Chem. 2008, 283, 29077–29085. [Google Scholar] [CrossRef]
- Seong, C.; Sehorn, M.G.; Plate, I.; Shi, I.; Song, B.; Chi, P.; Mortensen, U.; Sung, P.; Krejci, L. Molecular anatomy of the recombination mediator function of Saccharomyces cerevisiae Rad52. J. Biol. Chem. 2008, 283, 12166–12174. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Chu, J.; Yucer, N.; Leng, M.; Wang, S.Y.; Chen, B.P.; Hittelman, W.N.; Wang, Y. RING finger and WD repeat domain 3 (RFWD3) associates with replication protein A (RPA) and facilitates RPA-mediated DNA damage response. J. Biol. Chem. 2011, 286, 22314–22322. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Chen, J. E3 ligase RFWD3 participates in replication checkpoint control. J. Biol. Chem. 2011, 286, 22308–22313. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.D.; Yadav, T.; Giri, S.; Saez, B.; Graubert, T.A.; Zou, L. Functions of Replication Protein A as a Sensor of R Loops and a Regulator of RNaseH1. Mol. Cell 2017, 65, 832–847.e834. [Google Scholar] [CrossRef] [PubMed]
- Yusufzai, T.; Kong, X.; Yokomori, K.; Kadonaga, J.T. The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes. Dev. 2009, 23, 2400–2404. [Google Scholar] [CrossRef] [PubMed]
- Ciccia, A.; Bredemeyer, A.L.; Sowa, M.E.; Terret, M.E.; Jallepalli, P.V.; Harper, J.W.; Elledge, S.J. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes. Dev. 2009, 23, 2415–2425. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Ghosal, G.; Chen, J. The annealing helicase HARP protects stalled replication forks. Genes. Dev. 2009, 23, 2394–2399. [Google Scholar] [CrossRef] [PubMed]
- Postow, L.; Woo, E.M.; Chait, B.T.; Funabiki, H. Identification of SMARCAL1 as a component of the DNA damage response. J. Biol. Chem. 2009, 284, 35951–35961. [Google Scholar] [CrossRef] [PubMed]
- Kemp, M.G.; Akan, Z.; Yilmaz, S.; Grillo, M.; Smith-Roe, S.L.; Kang, T.H.; Cordeiro-Stone, M.; Kaufmann, W.K.; Abraham, R.T.; Sancar, A.; et al. Tipin-replication protein A interaction mediates Chk1 phosphorylation by ATR in response to genotoxic stress. J. Biol. Chem. 2010, 285, 16562–16571. [Google Scholar] [CrossRef] [PubMed]
- Unsal-Kacmaz, K.; Chastain, P.D.; Qu, P.P.; Minoo, P.; Cordeiro-Stone, M.; Sancar, A.; Kaufmann, W.K. The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement. Mol. Cell Biol. 2007, 27, 3131–3142. [Google Scholar] [CrossRef] [PubMed]
- Nagelhus, T.A.; Haug, T.; Singh, K.K.; Keshav, K.F.; Skorpen, F.; Otterlei, M.; Bharati, S.; Lindmo, T.; Benichou, S.; Benarous, R.; et al. A sequence in the N-terminal region of human uracil-DNA glycosylase with homology to XPA interacts with the C-terminal part of the 34-kDa subunit of replication protein A. J. Biol. Chem. 1997, 272, 6561–6566. [Google Scholar] [CrossRef] [PubMed]
- Machwe, A.; Lozada, E.; Wold, M.S.; Li, G.M.; Orren, D.K. Molecular cooperation between the Werner syndrome protein and replication protein A in relation to replication fork blockage. J. Biol. Chem. 2011, 286, 3497–3508. [Google Scholar] [CrossRef]
- Hyun, M.; Park, S.; Kim, E.; Kim, D.H.; Lee, S.J.; Koo, H.S.; Seo, Y.S.; Ahn, B. Physical and functional interactions of Caenorhabditis elegans WRN-1 helicase with RPA-1. Biochemistry 2012, 51, 1336–1345. [Google Scholar] [CrossRef]
- Brosh, R.M., Jr.; Orren, D.K.; Nehlin, J.O.; Ravn, P.H.; Kenny, M.K.; Machwe, A.; Bohr, V.A. Functional and physical interaction between WRN helicase and human replication protein A. J. Biol. Chem. 1999, 274, 18341–18350. [Google Scholar] [CrossRef]
- Shen, J.C.; Lao, Y.; Kamath-Loeb, A.; Wold, M.S.; Loeb, L.A. The N-terminal domain of the large subunit of human replication protein A binds to Werner syndrome protein and stimulates helicase activity. Mech. Ageing Dev. 2003, 124, 921–930. [Google Scholar] [CrossRef]
- Li, L.; Lu, X.; Peterson, C.A.; Legerski, R.J. An interaction between the DNA repair factor XPA and replication protein A appears essential for nucleotide excision repair. Mol. Cell Biol. 1995, 15, 5396–5402. [Google Scholar] [CrossRef]
- Saijo, M.; Kuraoka, I.; Masutani, C.; Hanaoka, F.; Tanaka, K. Sequential binding of DNA repair proteins RPA and ERCC1 to XPA in vitro. Nucleic Acids Res. 1996, 24, 4719–4724. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, T.; Saijo, M.; Kuraoka, I.; Kobayashi, T.; Nakatsu, Y.; Nagai, A.; Enjoji, T.; Masutani, C.; Sugasawa, K.; Hanaoka, F.; et al. DNA repair protein XPA binds replication protein A (RPA). J. Biol. Chem. 1995, 270, 4152–4157. [Google Scholar] [CrossRef]
- de Laat, W.L.; Appeldoorn, E.; Sugasawa, K.; Weterings, E.; Jaspers, N.G.; Hoeijmakers, J.H. DNA-binding polarity of human replication protein A positions nucleases in nucleotide excision repair. Genes. Dev. 1998, 12, 2598–2609. [Google Scholar] [CrossRef]
- Matsunaga, T.; Park, C.H.; Bessho, T.; Mu, D.; Sancar, A. Replication protein A confers structure-specific endonuclease activities to the XPF-ERCC1 and XPG subunits of human DNA repair excision nuclease. J. Biol. Chem. 1996, 271, 11047–11050. [Google Scholar] [CrossRef] [PubMed]
- Bessho, T.; Sancar, A.; Thompson, L.H.; Thelen, M.P. Reconstitution of human excision nuclease with recombinant XPF-ERCC1 complex. J. Biol. Chem. 1997, 272, 3833–3837. [Google Scholar] [CrossRef]
- Fisher, L.A.; Bessho, M.; Wakasugi, M.; Matsunaga, T.; Bessho, T. Role of interaction of XPF with RPA in nucleotide excision repair. J. Mol. Biol. 2011, 413, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Ellison, V.; Stillman, B. Biochemical characterization of DNA damage checkpoint complexes: Clamp loader and clamp complexes with specificity for 5’ recessed DNA. PLoS Biol. 2003, 1, E33. [Google Scholar] [CrossRef] [PubMed]
- Guler, G.D.; Liu, H.; Vaithiyalingam, S.; Arnett, D.R.; Kremmer, E.; Chazin, W.J.; Fanning, E. Human DNA helicase B (HDHB) binds to replication protein A and facilitates cellular recovery from replication stress. J. Biol. Chem. 2012, 287, 6469–6481. [Google Scholar] [CrossRef]
- Tkac, J.; Xu, G.; Adhikary, H.; Young, J.T.F.; Gallo, D.; Escribano-Diaz, C.; Krietsch, J.; Orthwein, A.; Munro, M.; Sol, W.; et al. HELB Is a Feedback Inhibitor of DNA End Resection. Mol. Cell 2016, 61, 405–418. [Google Scholar] [CrossRef]
- Sparks, J.L.; Kumar, R.; Singh, M.; Wold, M.S.; Pandita, T.K.; Burgers, P.M. Human exonuclease 5 is a novel sliding exonuclease required for genome stability. J. Biol. Chem. 2012, 287, 42773–42783. [Google Scholar] [CrossRef] [PubMed]
- Loo, Y.M.; Melendy, T. Recruitment of replication protein A by the papillomavirus E1 protein and modulation by single-stranded DNA. J. Virol. 2004, 78, 1605–1615. [Google Scholar] [CrossRef]
- Han, Y.; Loo, Y.M.; Militello, K.T.; Melendy, T. Interactions of the papovavirus DNA replication initiator proteins, bovine papillomavirus type 1 E1 and simian virus 40 large T antigen, with human replication protein A. J. Virol. 1999, 73, 4899–4907. [Google Scholar] [CrossRef]
- Weisshart, K.; Taneja, P.; Fanning, E. The replication protein A binding site in simian virus 40 (SV40) T antigen and its role in the initial steps of SV40 DNA replication. J. Virol. 1998, 72, 9771–9781. [Google Scholar] [CrossRef] [PubMed]
- Arunkumar, A.I.; Klimovich, V.; Jiang, X.; Ott, R.D.; Mizoue, L.; Fanning, E.; Chazin, W.J. Insights into hRPA32 C-terminal domain--mediated assembly of the simian virus 40 replisome. Nat. Struct. Mol. Biol. 2005, 12, 332–339. [Google Scholar] [CrossRef]
- Park, C.J.; Lee, J.H.; Choi, B.S. Solution structure of the DNA-binding domain of RPA from Saccharomyces cerevisiae and its interaction with single-stranded DNA and SV40 T antigen. Nucleic Acids Res. 2005, 33, 4172–4181. [Google Scholar] [CrossRef] [PubMed]
- Christensen, J.; Tattersall, P. Parvovirus initiator protein NS1 and RPA coordinate replication fork progression in a reconstituted DNA replication system. J. Virol. 2002, 76, 6518–6531. [Google Scholar] [CrossRef]
- Yuzhakov, A.; Kelman, Z.; Hurwitz, J.; O’Donnell, M. Multiple competition reactions for RPA order the assembly of the DNA polymerase delta holoenzyme. EMBO J. 1999, 18, 6189–6199. [Google Scholar] [CrossRef]
- Kim, H.S.; Brill, S.J. Rfc4 interacts with Rpa1 and is required for both DNA replication and DNA damage checkpoints in Saccharomyces cerevisiae. Mol. Cell Biol. 2001, 21, 3725–3737. [Google Scholar] [CrossRef]
- Otterlei, M.; Warbrick, E.; Nagelhus, T.A.; Haug, T.; Slupphaug, G.; Akbari, M.; Aas, P.A.; Steinsbekk, K.; Bakke, O.; Krokan, H.E. Post-replicative base excision repair in replication foci. EMBO J. 1999, 18, 3834–3844. [Google Scholar] [CrossRef]
- Loor, G.; Zhang, S.J.; Zhang, P.; Toomey, N.L.; Lee, M.Y. Identification of DNA replication and cell cycle proteins that interact with PCNA. Nucleic Acids Res. 1997, 25, 5041–5046. [Google Scholar] [CrossRef] [PubMed]
- Dianov, G.L.; Jensen, B.R.; Kenny, M.K.; Bohr, V.A. Replication protein A stimulates proliferating cell nuclear antigen-dependent repair of abasic sites in DNA by human cell extracts. Biochemistry 1999, 38, 11021–11025. [Google Scholar] [CrossRef] [PubMed]
- Farina, A.; Shin, J.H.; Kim, D.H.; Bermudez, V.P.; Kelman, Z.; Seo, Y.S.; Hurwitz, J. Studies with the human cohesin establishment factor, ChlR1. Association of ChlR1 with Ctf18-RFC and Fen1. J. Biol. Chem. 2008, 283, 20925–20936. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.T.; Rossi, M.; Cermak, L.; Saraf, A.; Florens, L.; Washburn, M.P.; Sung, P.; Schildkraut, C.L.; Pagano, M. FBH1 promotes DNA double-strand breakage and apoptosis in response to DNA replication stress. J. Cell Biol. 2013, 200, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Elia, A.E.; Wang, D.C.; Willis, N.A.; Boardman, A.P.; Hajdu, I.; Adeyemi, R.O.; Lowry, E.; Gygi, S.P.; Scully, R.; Elledge, S.J. RFWD3-Dependent Ubiquitination of RPA Regulates Repair at Stalled Replication Forks. Mol. Cell 2015, 60, 280–293. [Google Scholar] [CrossRef] [PubMed]
- MacKay, C.; Toth, R.; Rouse, J. Biochemical characterisation of the SWI/SNF family member HLTF. Biochem. Biophys. Res. Commun. 2009, 390, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Sommers, J.A.; Banerjee, T.; Hinds, T.; Wan, B.; Wold, M.S.; Lei, M.; Brosh, R.M., Jr. Novel function of the Fanconi anemia group J or RECQ1 helicase to disrupt protein-DNA complexes in a replication protein A-stimulated manner. J. Biol. Chem. 2014, 289, 19928–19941. [Google Scholar] [CrossRef]
- Cui, S.; Arosio, D.; Doherty, K.M.; Brosh, R.M., Jr.; Falaschi, A.; Vindigni, A. Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A. Nucleic Acids Res. 2004, 32, 2158–2170. [Google Scholar] [CrossRef]
- Cui, S.; Klima, R.; Ochem, A.; Arosio, D.; Falaschi, A.; Vindigni, A. Characterization of the DNA-unwinding activity of human RECQ1, a helicase specifically stimulated by human replication protein A. J. Biol. Chem. 2003, 278, 1424–1432. [Google Scholar] [CrossRef]
- Garcia, P.L.; Liu, Y.; Jiricny, J.; West, S.C.; Janscak, P. Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004, 23, 2882–2891. [Google Scholar] [CrossRef]
- Hu, Y.; Raynard, S.; Sehorn, M.G.; Lu, X.; Bussen, W.; Zheng, L.; Stark, J.M.; Barnes, E.L.; Chi, P.; Janscak, P.; et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes. Dev. 2007, 21, 3073–3084. [Google Scholar] [CrossRef]
- Galanty, Y.; Belotserkovskaya, R.; Coates, J.; Jackson, S.P. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes. Dev. 2012, 26, 1179–1195. [Google Scholar] [CrossRef]
- Dornreiter, I.; Erdile, L.F.; Gilbert, I.U.; von Winkler, D.; Kelly, T.J.; Fanning, E. Interaction of DNA polymerase Alpha-primase with cellular replication protein A and SV40 T antigen. EMBO J. 1992, 11, 769–776. [Google Scholar] [CrossRef]
- Feng, J.; Wakeman, T.; Yong, S.; Wu, X.; Kornbluth, S.; Wang, X.F. Protein phosphatase 2A-dependent dephosphorylation of replication protein A is required for the repair of DNA breaks induced by replication stress. Mol. Cell Biol. 2009, 29, 5696–5709. [Google Scholar] [CrossRef]
- Majka, J.; Binz, S.K.; Wold, M.S.; Burgers, P.M. Replication protein A directs loading of the DNA damage checkpoint clamp to 5’-DNA junctions. J. Biol. Chem. 2006, 281, 27855–27861. [Google Scholar] [CrossRef]
- Davis, A.P.; Symington, L.S. The Rad52-Rad59 complex interacts with Rad51 and replication protein A. DNA Repair 2003, 2, 1127–1134. [Google Scholar] [CrossRef]
- Grimme, J.M.; Honda, M.; Wright, R.; Okuno, Y.; Rothenberg, E.; Mazin, A.V.; Ha, T.; Spies, M. Human Rad52 binds and wraps single-stranded DNA and mediates annealing via two hRad52-ssDNA complexes. Nucleic Acids Res. 2010, 38, 2917–2930. [Google Scholar] [CrossRef] [PubMed]
- Bansbach, C.E.; Betous, R.; Lovejoy, C.A.; Glick, G.G.; Cortez, D. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes. Dev. 2009, 23, 2405–2414. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.G.; Liu, Y.; Mao, L.Y.; Zhang, J.T.; Zou, Y. Dimerization of human XPA and formation of XPA2-RPA protein complex. Biochemistry 2002, 41, 13012–13020. [Google Scholar] [CrossRef] [PubMed]
- Stigger, E.; Drissi, R.; Lee, S.H. Functional analysis of human replication protein A in nucleotide excision repair. J. Biol. Chem. 1998, 273, 9337–9343. [Google Scholar] [CrossRef] [PubMed]
- Daughdrill, G.W.; Buchko, G.W.; Botuyan, M.V.; Arrowsmith, C.; Wold, M.S.; Kennedy, M.A.; Lowry, D.F. Chemical shift changes provide evidence for overlapping single-stranded DNA- and XPA-binding sites on the 70 kDa subunit of human replication protein A. Nucleic Acids Res. 2003, 31, 4176–4183. [Google Scholar] [CrossRef] [PubMed]
- Aboussekhra, A.; Biggerstaff, M.; Shivji, M.K.; Vilpo, J.A.; Moncollin, V.; Podust, V.N.; Protic, M.; Hubscher, U.; Egly, J.M.; Wood, R.D. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell 1995, 80, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.M.; Ionescu, D.; Ingles, C.J. Interaction between BRCA2 and replication protein A is compromised by a cancer-predisposing mutation in BRCA2. Oncogene 2003, 22, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Doty, T.; Gibson, B.; Heyer, W.D. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 2010, 17, 1260–1262. [Google Scholar] [CrossRef] [PubMed]
- Yates, L.A.; Tannous, E.A.; Morgan, R.M.; Burgers, P.M.; Zhang, X. A DNA damage-induced phosphorylation circuit enhances Mec1(ATR) Ddc2(ATRIP) recruitment to Replication Protein A. Proc. Natl. Acad. Sci. USA 2023, 120, e2300150120. [Google Scholar] [CrossRef]
- Lee, S.; Heo, J.; Park, C.J. Determinants of replication protein A subunit interactions revealed using a phosphomimetic peptide. J. Biol. Chem. 2020, 295, 18449–18458. [Google Scholar] [CrossRef]
- Stephan, H.; Concannon, C.; Kremmer, E.; Carty, M.P.; Nasheuer, H.P. Ionizing radiation-dependent and independent phosphorylation of the 32-kDa subunit of replication protein A during mitosis. Nucleic Acids Res. 2009, 37, 6028–6041. [Google Scholar] [CrossRef]
- Nuss, J.E.; Patrick, S.M.; Oakley, G.G.; Alter, G.M.; Robison, J.G.; Dixon, K.; Turchi, J.J. DNA damage induced hyperphosphorylation of replication protein A. 1. Identification of novel sites of phosphorylation in response to DNA damage. Biochemistry 2005, 44, 8428–8437. [Google Scholar] [CrossRef]
- Patrick, S.M.; Oakley, G.G.; Dixon, K.; Turchi, J.J. DNA damage induced hyperphosphorylation of replication protein A. 2. Characterization of DNA binding activity, protein interactions, and activity in DNA replication and repair. Biochemistry 2005, 44, 8438–8448. [Google Scholar] [CrossRef]
- Zernik-Kobak, M.; Vasunia, K.; Connelly, M.; Anderson, C.W.; Dixon, K. Sites of UV-induced phosphorylation of the p34 subunit of replication protein A from HeLa cells. J. Biol. Chem. 1997, 272, 23896–23904. [Google Scholar] [CrossRef]
- Gregoire, S.; Irwin, J.; Kwon, I. Techniques for Monitoring Protein Misfolding and Aggregation in Vitro and in Living Cells. Korean J. Chem. Eng. 2012, 29, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.L.; Aria, V.; Baris, Y.; Yeeles, J.T.P. How Pol Alpha-primase is targeted to replisomes to prime eukaryotic DNA replication. Mol. Cell 2023, 83, 2911–2924.e2916. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Georgescu, R.; Yao, N.Y.; Li, H.; O’Donnell, M.E. Cryo-EM structures reveal that RFC recognizes both the 3′- and 5′-DNA ends to load PCNA onto gaps for DNA repair. Elife 2022, 11, e77469. [Google Scholar] [CrossRef] [PubMed]

| Subunits/Domain | Experiment Information | PDB ID | Resolution (Å) | Citation |
|---|---|---|---|---|
| X-ray; MRE11; RPA70(1-121) | 8K00 | 1.4 | [76] | |
| RPA70-F | X-ray; RAD9; RPA70(1-120) | 8JZY | 1.5 | [76] |
| X-ray; ETAA1; RPA70(1-120) | 8JZV | 1.5 | [76] | |
| X-ray; ATRIP; RPA70(1-123) | 7XV4 | 1.6 | [76] | |
| X-ray; HelB; RPA70(1-120) | 7XV1 | 1.8 | [76] | |
| X-ray; BLMp1; RPA70(1-120) | 7XV0 | 1.5 | [76] | |
| X-ray; RMI1; RPA70(1-120) | 7XUV | 1.6 | [76] | |
| X-ray; WRN; RPA70(1-120) | 7XUT | 1.6 | [76] | |
| X-ray; BLMp2; RPA70(1-120) | 7XUW | 1.8 | [76] | |
| X-ray; drug; RPA70(1-123) | 5E7N | 1.21 | [77] | |
| X-ray; Dna2 peptide; RPA70(1-123) | 5EAY | 1.55 | [70] | |
| X-ray; PrimPol(514-525); RPA70(1-123) | 5N85 | 2 | [74] | |
| X-ray; PrimPol(480-560); RPA70(1-123) | 5N8A | 1.28 | [74] | |
| X-ray; drug; RPA70(1-123) | 4R4T | 1.28 | [78] | |
| X-ray; drug; RPA70(1-123) | 4R4Q | 1.35 | [78] | |
| X-ray; drug; RPA70(1-123) | 4R4O | 1.33 | [78] | |
| X-ray; drug; RPA70(1-123) | 4R4I | 1.4 | [78] | |
| X-ray; drug; RPA70(1-123) | 4R4C | 1.4 | [78] | |
| X-ray; drug; RPA70(1-123) | 4O0A | 1.2 | [73] | |
| X-ray; drug; RPA70(1-123) | 4LWC | 1.61 | [73] | |
| X-ray; drug; RPA70(1-123) | 4LW1 | 1.631 | [73] | |
| X-ray; drug; RPA70(1-123) | 4LUZ | 1.9 | [73] | |
| X-ray; drug; RPA70(1-123) | 4LUV | 1.4 | [73] | |
| X-ray; drug; RPA70(1-123) | 4LUO | 1.54 | [73] | |
| X-ray; drug; RPA70(1-123) | 4IPH | 1.94 | [79] | |
| X-ray; drug; RPA70(1-123) | 4IPG | 1.58 | [79] | |
| X-ray; drug; RPA70(1-123) | 4IPD | 1.51 | [79] | |
| X-ray; drug; RPA70(1-123) | 4IPC | 1.22 | [79] | |
| X-ray; drug; RPA70(1-123) | 4IJL | 1.7 | [80] | |
| X-ray; drug; RPA70(1-123) | 4IJH | 1.498 | [80] | |
| X-ray; p53N(33-60)/RPA70(1-123) | 2B3G | 1.6 | [72] | |
| X-ray; RPA70(1-123) | 2B29 | 1.6 | [72] | |
| NMR; RPA70(1-114) | 1EWI | - | [68] | |
| RPA70-A/B | X-ray; RPA70(181-422) | 1FGU | 2.5 | [63] |
| X-ray; ssDNA; RPA70(181-422) | 1JMC | 2.4 | [66] | |
| NMR; XPA-MBD/RPA70(168-326) | 1D4U | - | [20] | |
| RPA32-wHLH | NMR; UNG2(73-88); RPA32(171-270) | 1DPU | - | [69] |
| X-ray; RPA32(197-270) | 4OU0 | 1.4 | [82] | |
| RPA32/RPA14 | X-ray; full length | 2Z6K | 3 | [75] |
| X-ray; full length | 2PQA | 2.5 | [75] | |
| X-ray; full length | 2PI2 | 2 | [75] | |
| X-ray; RPA32(43-172)/RPA14(1-121) | 1QUQ | 2.5 | [81] | |
| RPA70/RPA32/RPA14 | X-ray; Zn2+; RPA70(435-616)/RPA32(43-171)/RPA14(1-121) | 1L1O | 2.8 | [71] |
| Interacting Protein | Interaction Site on RPA | DNA Metabolism Pathway | Citation |
|---|---|---|---|
| AID | RPA32 | Immunoglobulin diversification | [107] |
| * Ajuba | RPA70 | DNA damage response (DDR) | [108,109] |
| ** ATR/ATRIP | RPA70-F | Checkpoint signaling, DNA repair | [110,111,112] |
| BID | RPA70-F | Replication stress response | [113] |
| * BLM | RPA70-A/B | DNA unwinding, resection | [114,115,125] |
| ** BRCA2 | ? | Homologous Recombination (HR) | [105] |
| DDX11 | ? | Chromosome segregation | [193] |
| ** DNA-PKcs | ? | DNA repair | [18,116] |
| ** DSS1 | RPA70 | HR | [121] |
| * ETAA1 | RPA70-F/RPA32 | ATR activation, repair at stalled replication forks | [117,118,119,120] |
| * EXO5 | RPA70-F | Intrastrand crosslink repair | [181] |
| * FANCJ | RPA70 | DNA repair, genome stability | [123] |
| * FBH1 | RPA32 | DNA unwinding, resection | [193] |
| HELB | RPA70-F | Replication stress response | [179,180] |
| HERC2 | RPA70 | Replication | [124,125] |
| HIRA | RPA70-C | Chromatin remodeling | [126] |
| Histones H3 and H4 | RPA70-F | Chromatin remodeling | [127] |
| * HLTF | RPA70 | Genome stability | [195,196] |
| HSF1 | RPA70 | Gene expression | [122] |
| * Menin | RPA32 | Genome stability | [129,130] |
| ** MRE11-RAD50-NBS1 | RPA70-F | HR | [131,132] |
| Nucleolin | RPA14 | Replication (stress) | [133,134,135] |
| NSMF | RPA32 | DDR | [101] |
| ** p53 | RPA70-F | HR | [136,137,138,139,140] |
| * p53BP1 | RPA70/RPA32 | DDR | [106] |
| * PALB2 | RPA32 | Recovery of stalled replication forks | [141] |
| Papillomavirus E1 | RPA70-A | Replication | [182,183] |
| Parvovirus NS1 | RPA70/RPA32 | DNA unwinding, resection | [187] |
| PCNA | RPA70 | Replication | [191] |
| Polδ | RPA70 | Replication | [188] |
| Pol-Prim | RPA70-F/A/B | Replication restart, DNA damage tolerance | [102,203] |
| ** PP2A | RPA32 | DDR | [204] |
| * PRP19/BCAS2 | RPA70-F/C | DNA repair | [144] |
| * PTEN | RPA70 | Genome stability | [145] |
| * RAD9/RAD1/HUS1 (9-1-1) | RPA70/RPA32 | DDR | [146] |
| RAD17 | RPA70-F | DDR, replication stress response | [147,178,205] |
| ** RAD51 | RPA70-A | Recombination | [103,150,151,206] |
| ** RAD52 | RPA70-A/B & RPA32-wHLH | DNA repair | [21,69,152,153,154,155,156,207] |
| RECQL1 | RPA70 | DNA unwinding | [198,199] |
| RECQ5β | ? | DNA unwinding | [200,201] |
| RFC | RPA70 | DNA unwinding | [188,189] |
| * RFWD3 | RPA32-wHLH | DNA repair | [157,158,195] |
| * RNaseH | ? | Transcription, DNA repair | [159] |
| * RNF4 | RPA70 | DNA DSB repair by HR | [202] |
| SENP6 | RPA70 | Unperturbed DNA replication | [38] |
| SMARCAL1/HARP | RPA32-wHLH | Replication fork restart | [160,161,162,208] |
| SV40 Large T antigen | RPA70-A/B & RPA32-wHLH | Replication | [182,183,184,185,186] |
| * Tipin | RPA32-wHLH | DDR | [164,165] |
| * UDG | RPA32-wHLH | Base excision repair | [69,166,190] |
| * UNG2 | RPA32-wHLH | Base excision repair | [69,166] |
| * WRN | RPA70-A/B | DNA unwinding, resection | [115,170] |
| * XPA | RPA70-A & RPA32-wHLH | Nucleotide excision repair (NER) | [69,166,171,209,210,211] |
| * XPF-ERCC1 | RPA70 | NER | [174,175,176] |
| * XPG | ? | NER | [174,176,212] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fousek-Schuller, V.J.; Borgstahl, G.E.O. The Intriguing Mystery of RPA Phosphorylation in DNA Double-Strand Break Repair. Genes 2024, 15, 167. https://doi.org/10.3390/genes15020167
Fousek-Schuller VJ, Borgstahl GEO. The Intriguing Mystery of RPA Phosphorylation in DNA Double-Strand Break Repair. Genes. 2024; 15(2):167. https://doi.org/10.3390/genes15020167
Chicago/Turabian StyleFousek-Schuller, Valerie J., and Gloria E. O. Borgstahl. 2024. "The Intriguing Mystery of RPA Phosphorylation in DNA Double-Strand Break Repair" Genes 15, no. 2: 167. https://doi.org/10.3390/genes15020167
APA StyleFousek-Schuller, V. J., & Borgstahl, G. E. O. (2024). The Intriguing Mystery of RPA Phosphorylation in DNA Double-Strand Break Repair. Genes, 15(2), 167. https://doi.org/10.3390/genes15020167







