Complete Mitochondrial Genomes of Pluvialis fulva and Charadrius dubius with Phylogenetic Analysis of Charadriiformes
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimen Collection and DNA Extraction
2.2. Library Preparation and Sequencing
2.3. Mitogenome Assembly and Annotation
2.4. Comparative Mitogenomic Analyses
2.5. Mitogenomic Phylogenetic Analyses
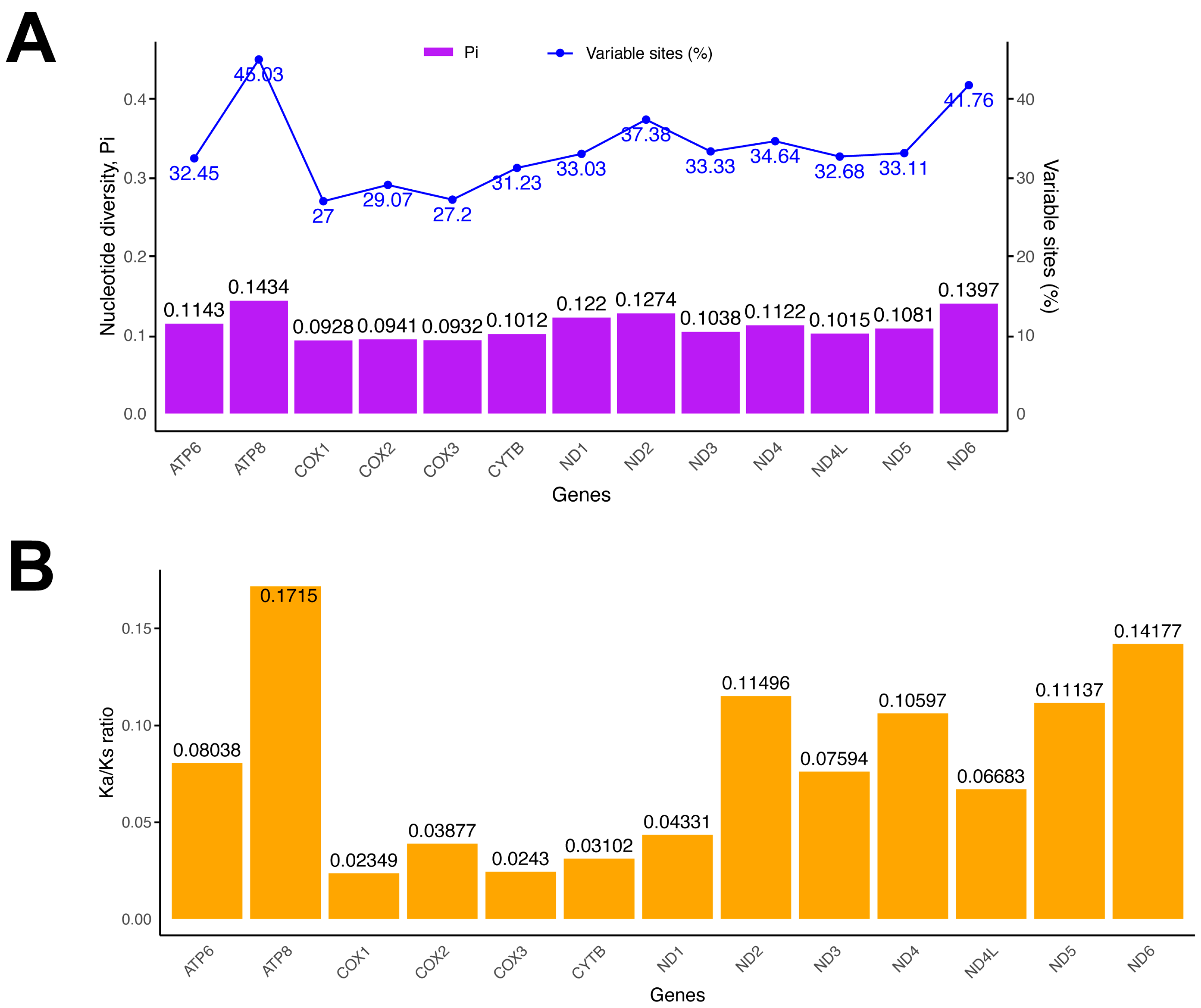
3. Results and Discussion
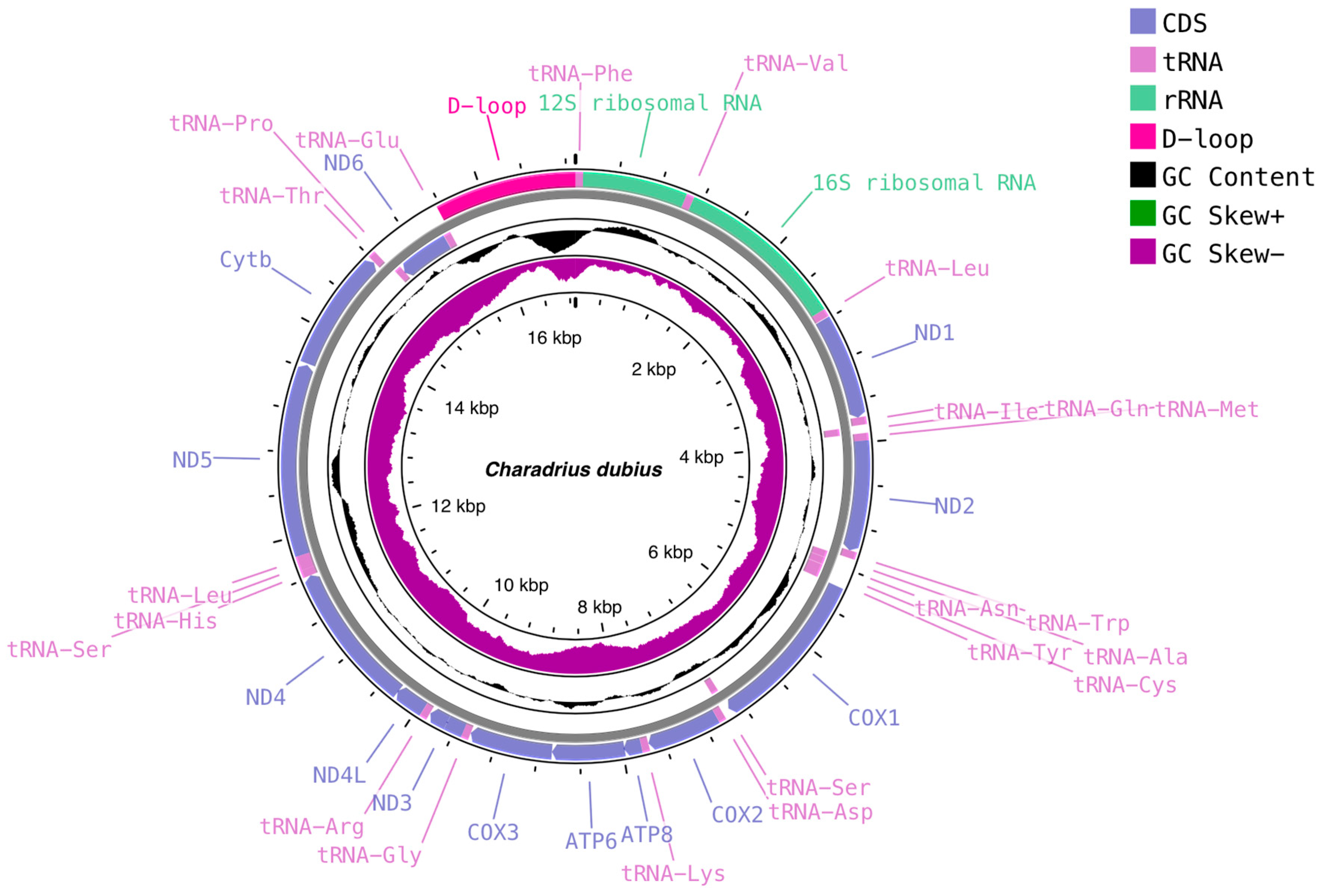
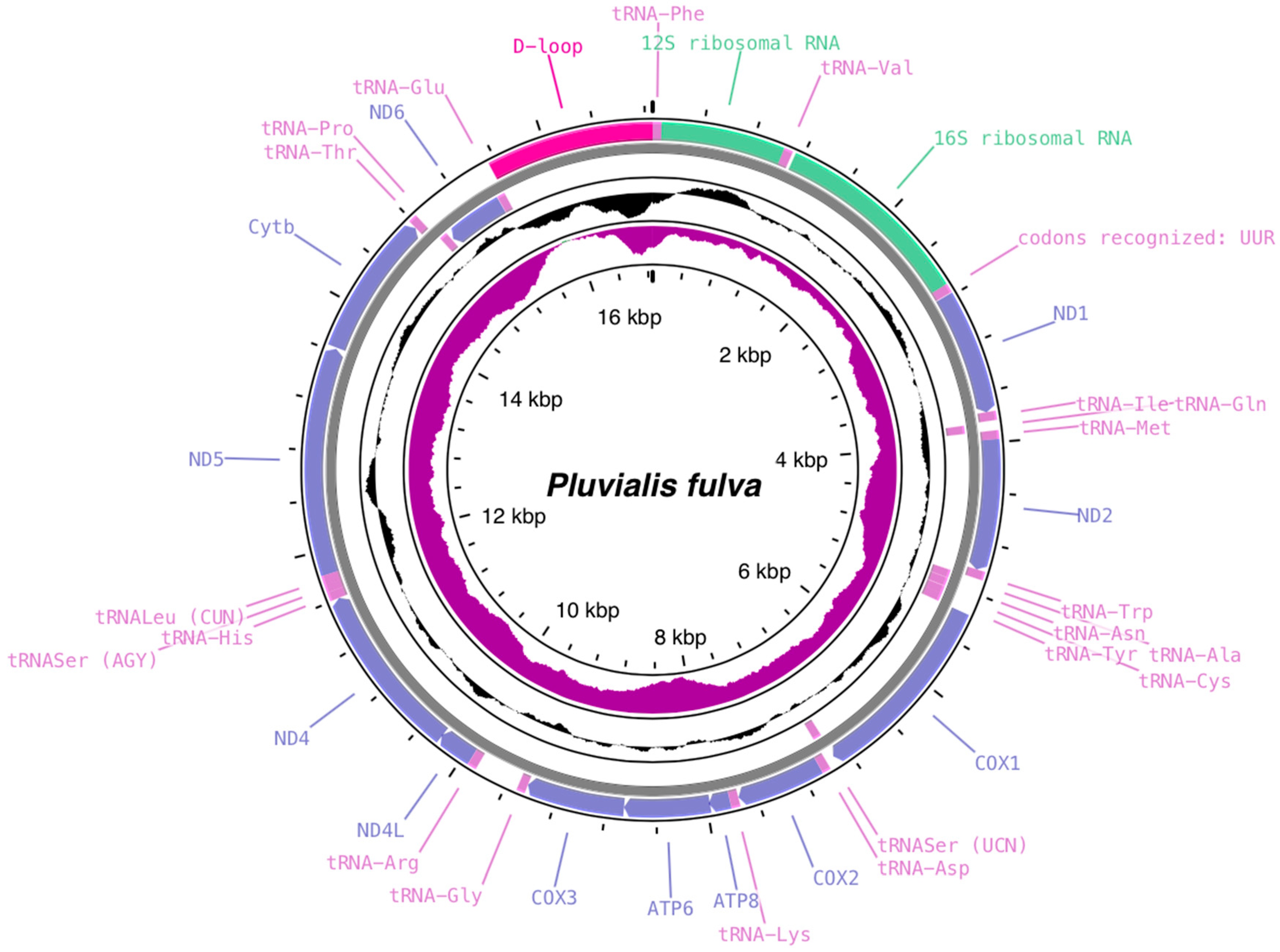
3.1. C. dubius and P. fulva Mitogenome Structure and Organization
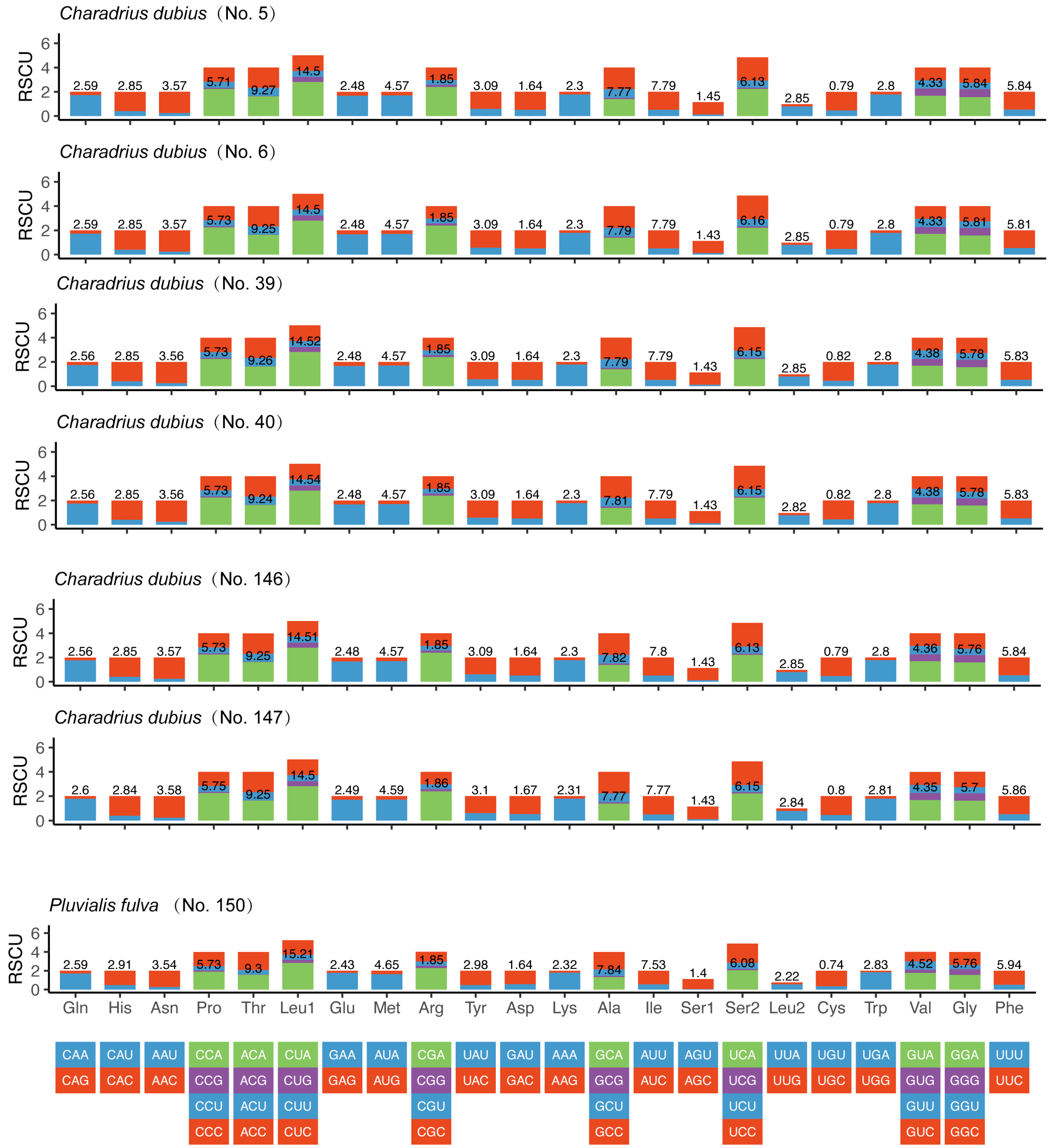
3.2. Codon Usage
3.3. Phylogenetic Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sullivan, B.L.; Wood, C.L.; Iliff, M.J.; Bonney, R.E.; Fink, D.; Kelling, S. eBird: A Citizen-Based Bird Observation Network in the Biological Sciences. Biol. Conserv. 2009, 142, 2282–2292. [Google Scholar] [CrossRef]
- Černý, D.; Natale, R. Comprehensive Taxon Sampling and Vetted Fossils Help Clarify the Time Tree of Shorebirds (Aves, Charadriiformes). Mol. Phylogenet. Evol. 2022, 177, 107620. [Google Scholar] [CrossRef] [PubMed]
- Winkler, D.W.; Billerman, S.M.; Lovette, I.J. Plovers and Lapwings (Charadriidae); Version 1.0; Birds of the World; Billerman, S.M., Keeney, B.K., Rodewald, P.G., Schulenberg, T.S., Eds.; Cornell Lab of Ornithology: Ithaca, NY, USA, 2020. [Google Scholar]
- Dos Remedios, N.; Lee, P.L.M.; Burke, T.; Székely, T.; Küpper, C. North or South? Phylogenetic and Biogeographic Origins of a Globally Distributed Avian Clade. Mol. Phylogenet. Evol. 2015, 89, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Barth, J.M.I.; Matschiner, M.; Robertson, B.C. Phylogenetic Position and Subspecies Divergence of the Endangered New Zealand Dotterel (Charadrius obscurus). PLoS ONE 2013, 8, e78068. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhang, C.; Pan, T.; Liu, W.; Li, K.; Hu, C.; Chang, Q. The Mitochondrial Genome of the Kentish Plover Charadrius alexandrinus (Charadriiformes: Charadriidae) and Phylogenetic Analysis of Charadrii. Genes Genom. 2018, 40, 955–963. [Google Scholar] [CrossRef]
- Baker, A.J.; Yatsenko, Y.; Tavares, E.S. Eight Independent Nuclear Genes Support Monophyly of the Plovers: The Role of Mutational Variance in Gene Trees. Mol. Phylogenet. Evol. 2012, 65, 631–641. [Google Scholar] [CrossRef] [PubMed]
- Fain, M.G.; Krajewski, C.; Houde, P. Phylogeny of “Core Gruiformes” (Aves: Grues) and Resolution of the Limpkin–Sungrebe Problem. Mol. Phylogenet. Evol. 2007, 43, 515–529. [Google Scholar] [CrossRef] [PubMed]
- Baker, A.J.; Pereira, S.L.; Paton, T.A. Phylogenetic Relationships and Divergence Times of Charadriiformes Genera: Multigene Evidence for the Cretaceous Origin of at Least 14 Clades of Shorebirds. Biol. Lett. 2007, 3, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Ericson, P.G.; Envall, I.; Irestedt, M.; Norman, J.A. Inter-Familial Relationships of the Shorebirds (Aves: Charadriiformes) Based on Nuclear DNA Sequence Data. BMC Evol. Biol. 2003, 3, 16. [Google Scholar] [CrossRef]
- Ballard, J.W.O.; Whitlock, M.C. The Incomplete Natural History of Mitochondria. Mol. Ecol. 2004, 13, 729–744. [Google Scholar] [CrossRef]
- James, J.E.; Piganeau, G.; Eyre-Walker, A. The Rate of Adaptive Evolution in Animal Mitochondria. Mol. Ecol. 2016, 25, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Zardoya, R. Recent Advances in Understanding Mitochondrial Genome Diversity. F1000Research 2020, 9, 270. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Peng, X.; Jing, S.; Liu, B.; Zhu, L.; He, G. Intraspecific and Interspecific Variations in the Mitochondrial Genomes of Nilaparvata (Hemiptera: Delphacidae). J. Econ. Entomol. 2015, 108, 2021–2029. [Google Scholar] [CrossRef] [PubMed]
- Morón-López, J.; Vergara, K.; Sato, M.; Gajardo, G.; Ueki, S. Intraspecies Variation of the Mitochondrial Genome: An Evaluation for Phylogenetic Approaches Based on the Conventional Choices of Genes and Segments on Mitogenome. PLoS ONE 2022, 17, e0273330. [Google Scholar] [CrossRef] [PubMed]
- Burleigh, J.G.; Kimball, R.T.; Braun, E.L. Building the Avian Tree of Life Using a Large-Scale, Sparse Supermatrix. Mol. Phylogenet. Evol. 2015, 84, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Colwell, M.A.; Haig, S.M. An Overview of the World’s Plovers*. In The Population Ecology and Conservation of Charadrius Plovers; CRC Press: Boca Raton, FL, USA, 2019; ISBN 978-1-315-15288-2. [Google Scholar]
- Ding, J.; Liu, W.; Zhang, Y.; Chang, Q.; Hu, C. The Complete Mitochondrial Genome of Pacific Golden Plover Pluvialis fulva (Charadriiformes, Charadriidae). Mitochondrial DNA B Resour. 2016, 1, 701–702. [Google Scholar] [CrossRef]
- Johnson, O.; Fielding, L.; Fox, J.; Gold, R.; Goodwill, R.; Johnson, P. Tracking the Migrations of Pacific Golden-Plovers (Pluvialis fulva) between Hawaii and Alaska: New Insight on Flight Performance, Breeding Ground Destinations, and Nesting from Birds Carrying Light Level Geolocators. Wader Study Group Bull. 2011, 118, 26–31. [Google Scholar]
- Connors, P.G. Taxonomy, Distribution, and Evolution of Golden Plovers (Pluvialis dominica and Pluvialis fulva). Auk 1983, 100, 607–620. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de Novo Metazoan Mitochondrial Genome Annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Ding, J.; Qian, R.; Tai, D.; Yao, W.; Hu, C.; Chang, Q. The Complete Mitochondrial Genome of Grey Plover Pluvialis squatarola (Charadriiformes, Charadriidae). Mitochondrial DNA Part B 2020, 5, 2738–2739. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An Integrated and Scalable Desktop Platform for Streamlined Molecular Sequence Data Management and Evolutionary Phylogenetics Studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Charif, D.; Lobry, J.R. SeqinR 1.0-2: A Contributed Package to the R Project for Statistical Computing Devoted to Biological Sequences Retrieval and Analysis. In Structural Approaches to Sequence Evolution: Molecules, Networks, Populations; Bastolla, U., Porto, M., Roman, H.E., Vendruscolo, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 207–232. ISBN 978-3-540-35306-5. [Google Scholar]
- Paradis, E. Pegas: An R Package for Population Genetics with an Integrated-Modular Approach. Bioinformatics 2010, 26, 419–420. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, E.D.; Mirarab, S.; Aberer, A.J.; Li, B.; Houde, P.; Li, C.; Ho, S.Y.W.; Faircloth, B.C.; Nabholz, B.; Howard, J.T.; et al. Whole-Genome Analyses Resolve Early Branches in the Tree of Life of Modern Birds. Science 2014, 346, 1320–1331. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.-Y. Ggtree: An r Package for Visualization and Annotation of Phylogenetic Trees with Their Covariates and Other Associated Data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Mindell, D.P.; Sorenson, M.D.; Dimcheff, D.E. Multiple Independent Origins of Mitochondrial Gene Order in Birds. Proc. Natl. Acad. Sci. USA 1998, 95, 10693–10697. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.Y.; Roh, S.J.; Kim, S.H.; Jung, T.W.; Lee, D.J.; Kim, H.K.; Jung, J.H.; Cho, S.-Y.; Kim, Y.J.; Kook, J.W.; et al. Complete Mitochondrial Genome of Little Ringed Plover Charadrius dubius (Charadriiformes, Charadriidae). Mitochondrial DNA Part B 2022, 7, 1896–1898. [Google Scholar] [CrossRef]
- Rousselle, M.; Laverré, A.; Figuet, E.; Nabholz, B.; Galtier, N. Influence of Recombination and GC-Biased Gene Conversion on the Adaptive and Nonadaptive Substitution Rate in Mammals versus Birds. Mol. Biol. Evol. 2019, 36, 458–471. [Google Scholar] [CrossRef]
- Rand, D.M. The Units of Selection on Mitochondrial DNA. Annu. Rev. Ecol. Syst. 2001, 32, 415–448. [Google Scholar] [CrossRef]
- Li, X.; Huang, Y.; Lei, F. Comparative Mitochondrial Genomics and Phylogenetic Relationships of the Crossoptilon Species (Phasianidae, Galliformes). BMC Genom. 2015, 16, 42. [Google Scholar] [CrossRef]
- Lavrov, D.V.; Pett, W. Animal Mitochondrial DNA as We Do Not Know It: Mt-Genome Organization and Evolution in Nonbilaterian Lineages. Genome Biol. Evol. 2016, 8, 2896–2913. [Google Scholar] [CrossRef] [PubMed]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA Punctuation Model of RNA Processing in Human Mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: Oxford, UK, 2000. [Google Scholar]
- Zhou, X.; Lin, Q.; Fang, W.; Chen, X. The Complete Mitochondrial Genomes of Sixteen Ardeid Birds Revealing the Evolutionary Process of the Gene Rearrangements. BMC Genom. 2014, 15, 573. [Google Scholar] [CrossRef]
- Lan, G.; Yu, J.; Liu, J.; Zhang, Y.; Ma, R.; Zhou, Y.; Zhu, B.; Wei, W.; Liu, J.; Qi, G. Complete Mitochondrial Genome and Phylogenetic Analysis of Tarsiger indicus (Aves: Passeriformes: Muscicapidae). Genes 2024, 15, 90. [Google Scholar] [CrossRef] [PubMed]
- Pons, J.-M.; Hassanin, A.; Crochet, P.-A. Phylogenetic Relationships within the Laridae (Charadriiformes: Aves) Inferred from Mitochondrial Markers. Mol. Phylogenet. Evol. 2005, 37, 686–699. [Google Scholar] [CrossRef] [PubMed]
- Paton, T.A.; Baker, A.J.; Groth, J.G.; Barrowclough, G.F. RAG-1 Sequences Resolve Phylogenetic Relationships within Charadriiform Birds. Mol. Phylogenet. Evol. 2003, 29, 268–278. [Google Scholar] [CrossRef]


| Species | Sample ID | Collection Date | Gender | Collection Site |
|---|---|---|---|---|
| C. dubius | 5 | 4 June 2018 | * | Hongjiannao Lake, Shenmu, Shaanxi, China |
| C. dubius | 6 | 4 June 2018 | * | Hongjiannao Lake, Shenmu, Shaanxi, China |
| C. dubius | 39 | 5 July 2018 | Female | Tangyu, Lantian, Shaanxi, China |
| C. dubius | 40 | 5 July 2018 | Male | Tangyu, Lantian, Shaanxi, China |
| C. dubius | 146 | 11 June 2020 | * | Yuyang River, Yulin, Shaanxi, China |
| C. dubius | 147 | 11 June 2020 | * | Yuyang River, Yulin, Shaanxi, China |
| P. fulva | 150 | 2 March 2020 | Male | Xi’an Xianyang International Airport, Xi’an, Shaanxi, China |
| Speices | Sample ID | Length (bp) | GC Content (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mitogenome | Of Protein-Coding Genes | rRNA | tRNA | Mitogenome | Of Protein-Coding Genes | rRNA | tRNA | ||
| C. dubius | 5 | 16,866 | 11,418 | 2567 | 1553 | 44.6 | 45.3 | 45.6 | 41.7 |
| 6 | 16,888 | 11,418 | 2538 | 1553 | 44.6 | 45.4 | 45.6 | 41.6 | |
| 39 | 16,911 | 11,400 | 2566 | 1556 | 44.6 | 45.3 | 45.7 | 41.5 | |
| 40 | 16,833 | 11,397 | 2568 | 1552 | 44.6 | 45.3 | 45.7 | 41.6 | |
| 146 | 16,926 | 11,400 | 2544 | 1553 | 44.5 | 45.3 | 45.5 | 41.6 | |
| 147 | 16,911 | 11,397 | 2544 | 1553 | 44.6 | 45.3 | 45.5 | 41.7 | |
| P. fulva | 150 | 16,859 | 11,391 | 2539 | 1551 | 45.2 | 46.2 | 44.8 | 41.3 |
| Species | AT Skew | GC Skew | AT% | GC% |
|---|---|---|---|---|
| Charadrius alexandrinus | 0.14 | −0.39 | 55.24 | 44.76 |
| Charadrius dubius | 0.14 | −0.39 | 54.99 | 45.01 |
| Charadrius dubius (No. 146) | 0.14 | −0.38 | 55.48 | 44.52 |
| Charadrius dubius (No. 147) | 0.14 | −0.39 | 55.41 | 44.59 |
| Charadrius dubius (No. 39) | 0.14 | −0.38 | 55.38 | 44.62 |
| Charadrius dubius (No. 40) | 0.14 | −0.38 | 55.37 | 44.63 |
| Charadrius dubius (No. 5) | 0.14 | −0.38 | 55.43 | 44.57 |
| Charadrius dubius (No. 6) | 0.14 | −0.38 | 55.34 | 44.66 |
| Charadrius leschenaultii | 0.14 | −0.39 | 55.47 | 44.53 |
| Charadrius mongolus | 0.13 | −0.39 | 55.38 | 44.62 |
| Charadrius vociferus | 0.14 | −0.40 | 55.58 | 44.42 |
| Pluvialis apricaria | 0.15 | −0.40 | 54.37 | 45.63 |
| Pluvialis fulva | 0.15 | −0.39 | 54.87 | 45.13 |
| Pluvialis fulva (No. 150) | 0.14 | −0.39 | 54.76 | 45.24 |
| Pluvialis squatarola | 0.14 | −0.38 | 54.27 | 45.73 |
| Vanellus cinereus | 0.15 | −0.39 | 55.15 | 44.85 |
| Vanellus vanellus | 0.13 | −0.38 | 55.47 | 44.53 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, K.; Wang, Q.; Bian, K.; Li, F.; Tang, J.; Suo, L.; Hou, X.; Yang, C. Complete Mitochondrial Genomes of Pluvialis fulva and Charadrius dubius with Phylogenetic Analysis of Charadriiformes. Genes 2024, 15, 1642. https://doi.org/10.3390/genes15121642
Sun K, Wang Q, Bian K, Li F, Tang J, Suo L, Hou X, Yang C. Complete Mitochondrial Genomes of Pluvialis fulva and Charadrius dubius with Phylogenetic Analysis of Charadriiformes. Genes. 2024; 15(12):1642. https://doi.org/10.3390/genes15121642
Chicago/Turabian StyleSun, Kuo, Qingxiong Wang, Kun Bian, Feiran Li, Jie Tang, Lijuan Suo, Xiang Hou, and Chao Yang. 2024. "Complete Mitochondrial Genomes of Pluvialis fulva and Charadrius dubius with Phylogenetic Analysis of Charadriiformes" Genes 15, no. 12: 1642. https://doi.org/10.3390/genes15121642
APA StyleSun, K., Wang, Q., Bian, K., Li, F., Tang, J., Suo, L., Hou, X., & Yang, C. (2024). Complete Mitochondrial Genomes of Pluvialis fulva and Charadrius dubius with Phylogenetic Analysis of Charadriiformes. Genes, 15(12), 1642. https://doi.org/10.3390/genes15121642






