Impact of Long-Term Cannabidiol (CBD) Treatment on Mouse Kidney Transcriptome
Highlights
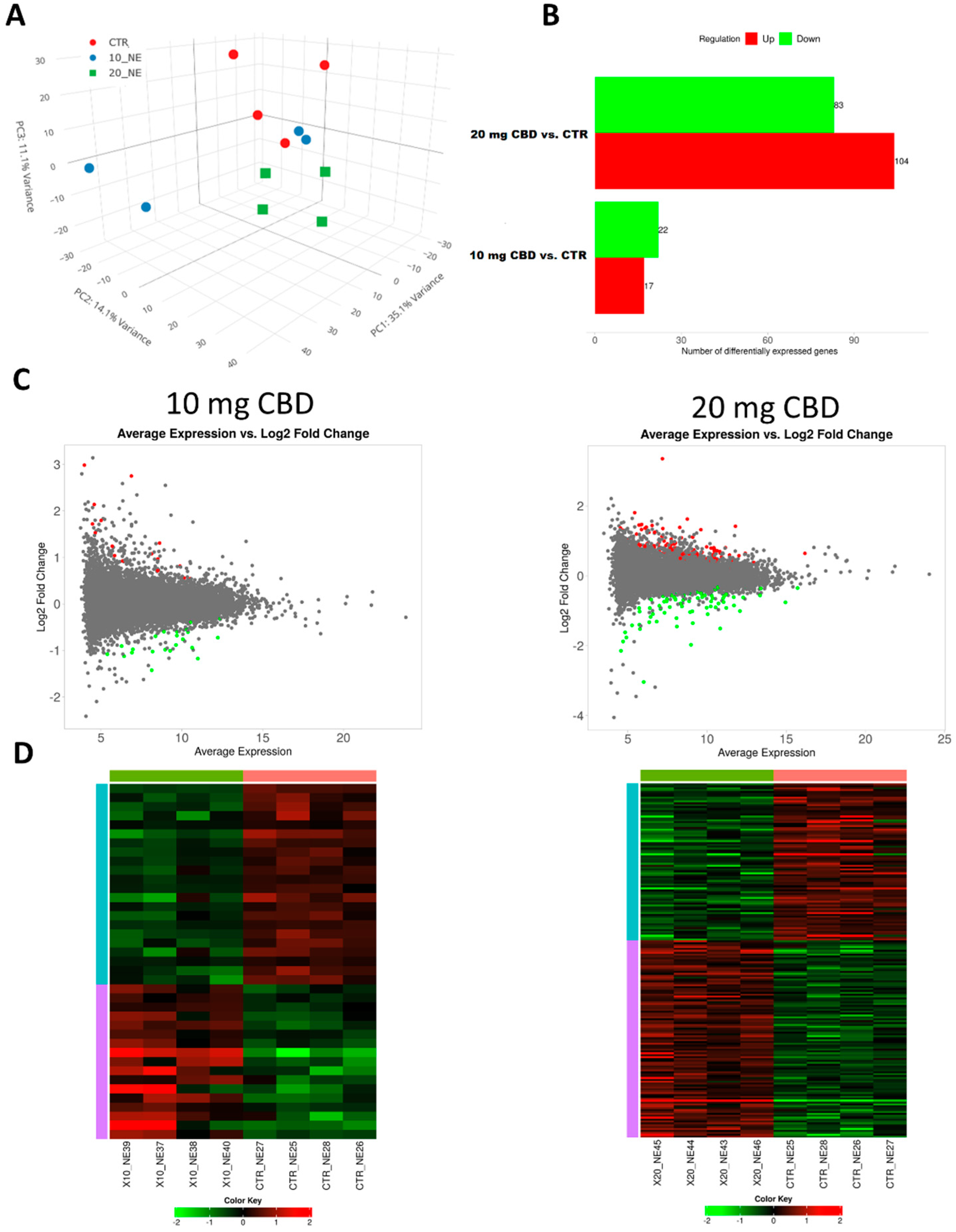
- Chronic CBD treatment significantly impacts the renal transcriptome;
- CBD modulates the expression of genes related to circadian rhythm, glucocorticoid receptor function, inflammatory responses, and other pathways;
- Higher CBD doses influence a larger number of genes implicated in similar biological processes.
- CBD shows potential for treating specific renal conditions; however, this requires validation in future clinical trials;
- This study sheds light on the molecular mechanisms through which CBD may affect kidney function, crucial for assessing its therapeutic applications;
- The findings emphasize the necessity of further research on the safety and long-term effects of CBD use, especially regarding renal health;
- The identification of specific genes and pathways altered by CBD provides a robust foundation for future investigations into its role in kidney-related therapies.
Abstract
1. Introduction
2. Material and Methods
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Burstein, S. Cannabidiol (CBD) and its analogs: A review of their effects on inflammation. Bioorg. Med. Chem. 2015, 23, 1377–1385. [Google Scholar] [CrossRef]
- Radwan, M.; Chandra, S.; Gul, S.; El Sholy, M. Cannabinoids, phenolics, terpenes, and alkaloids of Cannabis. Molecules 2021, 26, 2774. [Google Scholar] [CrossRef] [PubMed]
- Atalay, S.; Jarocka-Karpowicz, I.; Skrzydlewska, E. Antioxidative and anti-inflammatory properties of cannabidiol. Antioxidants 2019, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Raïch, I.; Lillo, J.; Rivas-Santisteban, R.; Rebassa, J.B.; Capó, T.; Santandreu, M.; Cubeles-Juberias, E.; Reyes-Resina, I.; Navarro, G. Potential of CBD acting on cannabinoid receptors CB1 and CB2 in ischemic stroke. Int. J. Mol. Sci. 2024, 25, 6708. [Google Scholar] [CrossRef] [PubMed]
- Page, D.A.; Ruben, P.C. Cannabidiol potentiates hyperpolarization-activated cyclic nucleotide-gated (HCN4) channels. J. Gen. Physiol. 2024, 156, e202313505. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, E.C.; Chamberland, S.; Bazelot, M.; Nebet, E.R.; Wang, X.; McKenzie, S.; Jain, S.; Greenhill, S.; Wilson, M.; Marley, N.; et al. Cannabidiol modulates excitatory-inhibitory ratio to counter hippocampal hyperactivity. Neuron 2023, 111, 1282–1300.e8. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Aguirre, C.; Carmona-Cruz, F.; Velasco, A.L.; Velasco, F.; Aguado-Carrillo, G.; Cuéllar-Herrera, M.; Rocha, L. Cannabidiol acts at 5-HT1A receptors in the human brain: Relevance for treating temporal lobe epilepsy. Front. Behav. Neurosci. 2020, 14, 611278. [Google Scholar] [CrossRef] [PubMed]
- Etemad, L.; Karimi, G.; Alavi, M.S.; Roohbakhsh, A. Pharmacological effects of cannabidiol by transient receptor potential channels. Life Sci. 2022, 300, 120582. [Google Scholar] [CrossRef]
- Worth, H.; O’Hara, D.; Agarwal, N.; Collister, D.; Brennan, F.; Smyth, B. Cannabinoids for symptom management in patients with kidney failure: A narrative review. Clin. J. Am. Soc. Nephrol. 2022, 17, 911–921. [Google Scholar] [CrossRef]
- Zhao, Z.; Yan, Q.; Xie, J.; Liu, Z.; Liu, F.; Liu, Y.; Zhou, S.; Pan, S.; Liu, D.; Duan, J.; et al. The intervention of cannabinoid receptor in chronic and acute kidney disease animal models: A systematic review and meta-analysis. Diabetol. Metab. Syndr. 2024, 16, 45. [Google Scholar] [CrossRef] [PubMed]
- Soliman, N.A.; Dahmy SI, E.; Shalaby, A.A.; Mohammed, K.A. Prospective affirmative therapeutics of cannabidiol oil mitigates doxorubicin-induced abnormalities in kidney function, inflammation, and renal tissue changes. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2024, 397, 3897–3906. [Google Scholar] [CrossRef]
- Naya, N.; Kelly, J.; Corna, G.; Golino, M.; Abbate, A.; Toldo, S. Molecular and cellular mechanisms of action of cannabidiol. Molecules 2023, 28, 5980. [Google Scholar] [CrossRef]
- Dodt, M.; Roehr, J.T.; Ahmed, R.; Dieterich, C. FLEXBAR—Flexible barcode and adapter processing for next-generation sequencing platforms. Biology 2012, 1, 895–905. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Ge, X. iDEP Web application for RNA-Seq data analysis. In RNA Bioinformatics; Methods in Molecular Biology; Humana: New York, NY, USA, 2021. [Google Scholar] [CrossRef]
- Ge, S.X.; Son, E.W.; Yao, R. iDEP: An integrated web application for differential expression and pathway analysis of RNA-seq data. BMC Bioinform. 2018, 19, 534. [Google Scholar] [CrossRef]
- Muñoz, J.J.; Anauate, A.C.; Amaral, A.G.; Ferreira, F.M.; Watanabe, E.H.; Meca, R.; Ormanji, M.S.; Boim, M.A.; Onuchic, L.F.; Heilberg, I.P. Ppia is the most stable housekeeping gene for qRT-PCR normalization in kidneys of three Pkd1-deficient mouse models. Sci. Rep. 2021, 11, 19798. [Google Scholar] [CrossRef] [PubMed]
- Secio-Silva, A.; Emrich, F.; Evangelista-Silva, P.H.; Prates, R.P.; Hijo, A.H.; Figueira-Costa, T.N.; Schaeffer, M.; Goulart-Silva, F.; Peliciari-Garcia, R.A.; Bargi-Souza, P. Which housekeeping gene is the best choice for RT-qPCR analysis in mice fed with a high-fat diet? Studies in the liver, kidney, pancreas, and intestines. Gene Rep. 2023, 31, 101756. [Google Scholar] [CrossRef]
- Moreno, J.A.; Hamza, E.; Guerrero-Hue, M.; Rayego-Mateos, S.; García-Caballero, C.; Vallejo-Mudarra, M.; Metzinger, L.; Metzinger-Le Meuth, V. Non-Coding RNAs in Kidney Diseases: The Long and Short of Them. Int. J. Mol. Sci. 2021, 22, 6077. [Google Scholar] [CrossRef]
- Costello, H.M.; Johnston, J.G.; Juffre, A.; Crislip, G.R.; Gumz, M.L. Circadian clocks of the kidney: Function, mechanism, and regulation. Physiol. Rev. 2022, 102, 1669–1701. [Google Scholar] [CrossRef] [PubMed]
- Stow, L.R.; Gumz, M.L. The circadian clock in the kidney. J. Am. Soc. Nephrol. JASN 2011, 22, 598–604. [Google Scholar] [CrossRef]
- Goldman, R. Studies in diurnal variation of water and electrolyte excretion; nocturnal diuresis of water and sodium in congestive cardiac failure and cirrhosis of the liver. J. Clin. Investig. 1951, 30, 1191–1199. [Google Scholar] [CrossRef]
- Dyer, A.R.; Martin, G.J.; Burton, W.N.; Levin, M.; Stamler, J. Blood pressure and diurnal variation in sodium, potassium, and water excretion. J. Hum. Hypertens. 1998, 12, 363–371. [Google Scholar] [CrossRef]
- Mohandas, R.; Douma, L.G.; Scindia, Y.; Gumz, M.L. Circadian rhythms and renal pathophysiology. J. Clin. Investig. 2022, 132, e148277. [Google Scholar] [CrossRef] [PubMed]
- Firsov, D.; Bonny, O. Circadian rhythms and the kidney. Nat. Rev. Nephrol. 2018, 14, 626–635. [Google Scholar] [CrossRef]
- Lafaye, G.; Desterke, C.; Marulaz, L.; Benyamina, A. Cannabidiol affects circadian clock core complex and its regulation in microglia cells. Addict. Biol. 2019, 24, 921–934. [Google Scholar] [CrossRef] [PubMed]
- Acuña-Goycolea, C.; Obrietan, K.; van den Pol, A.N. Cannabinoids excite circadian clock neurons. J. Neurosci. 2010, 30, 10061–10066. [Google Scholar] [CrossRef]
- Noh, S.G.; Jung, H.J.; Kim, S.; Arulkumar, R.; Kim, D.H.; Park, D.; Chung, H.Y. Regulation of Circadian Genes Nr1d1 and Nr1d2 in Sex-Different Manners during Liver Aging. Int. J. Mol. Sci. 2022, 23, 10032. [Google Scholar] [CrossRef]
- Dumas, B.; Harding, H.P.; Choi, H.S.; Lehmann, K.A.; Chung, M.; Lazar, M.A.; Moore, D.D. A new orphan member of the nuclear hormone receptor superfamily closely related to Rev-Erb. Mol. Endocrinol. 1994, 8, 996–1005. [Google Scholar] [CrossRef]
- Burris, T.P. Nuclear hormone receptors for heme: REV-ERBα and REV-ERBβ are ligand-regulated components of the mammalian clock. Mol. Endocrinol. 2008, 22, 1509–1520. [Google Scholar] [CrossRef]
- Wang, J.; Harris, C. Glucocorticoid Signaling: From Molecules to Mice to Man; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar] [CrossRef]
- Vandewalle, J.; Luypaert, A.; De Bosscher, K.; Libert, C. Therapeutic mechanisms of glucocorticoids. Trends Endocrinol. Metab. 2018, 29, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Mangos, G.; Whitworth, J.; Williamson, P.; Kelly, J. Glucocorticoids and the kidney. Nephrology 2003, 8, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Zannas, A.S.; Wiechmann, T.; Gassen, N.C.; Binder, E.B. Gene–stress–epigenetic regulation of FKBP5: Clinical and translational implications. Neuropsychopharmacol. Rev. 2016, 41, 261–274. [Google Scholar] [CrossRef] [PubMed]
- Wochnik, G.M.; Ruegg, J.; Abel, G.A.; Schmidt, U.; Holsboer, F.; Rein, T. FK506-binding proteins 51 and 52 differentially regulate dynein interaction and nuclear translocation of the glucocorticoid receptor in mammalian cells. J. Biol. Chem. 2005, 280, 4609–4616. [Google Scholar] [CrossRef] [PubMed]
- Chourpiliadis, C.; Aeddula, N.R. Physiology, Glucocorticoids. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2024. Available online: https://www.ncbi.nlm.nih.gov/books/NBK560897/ (accessed on 17 July 2023).
| 10 mg CBD vs. Control | 20 mg CBD vs. Control | |||||
|---|---|---|---|---|---|---|
| Symbol | Ensembl ID | Description | log2FC * | FDR ** | log2FC | FDR ** |
| Dbp | ENSMUSG00000059824 | D site albumin promoter binding protein | 2.75 | 9.29 × 10−34 | 3.36 | 4.79 × 10−52 |
| Ppard | ENSMUSG00000002250 | peroxisome proliferator activator receptor delta | −0.98 | 3.11 × 10−10 | −1.03 | 1.30 × 10−11 |
| Per3 | ENSMUSG00000028957 | period circadian clock 3 | 1.08 | 1.93 × 10−7 | 1.39 | 1.09 × 10−13 |
| Npas2 | ENSMUSG00000026077 | neuronal PAS domain protein 2 | −1.42 | 1.67 × 10−5 | −1.45 | 4.00 × 10−6 |
| Ppm1h | ENSMUSG00000034613 | protein phosphatase 1H (PP2C domain containing) | −0.55 | 8.22 × 10−5 | −0.56 | 1.63 × 10−5 |
| Ypel2 | ENSMUSG00000018427 | yippee like 2 | −1.01 | 1.00 × 10−4 | −1.35 | 8.63 × 10−10 |
| Acmsd | ENSMUSG00000026348 | amino carboxymuconate semialdehyde decarboxylase | −0.76 | 1.05 × 10−3 | −0.91 | 3.50 × 10−6 |
| Rorc | ENSMUSG00000028150 | RAR-related orphan receptor γ | −0.68 | 1.05 × 10−3 | −0.57 | 7.06 × 10−3 |
| Nfil3 | ENSMUSG00000056749 | nuclear factor. interleukin 3, regulated | −0.98 | 2.06 × 10−3 | −1.31 | 6.30 × 10−7 |
| Cyp2a5 | ENSMUSG00000005547 | cytochrome P450, family 2, subfamily a, polypeptide 5 | 1.42 | 4.59 × 10−8 | 1.42 | 4.59 × 10−8 |
| Bhlhe41 | ENSMUSG00000030256 | basic helix-loop-helix family, member e41 | 1.46 | 1.45 × 10−6 | 1.46 | 1.45 × 10−6 |
| ENSMUSG00000120875 | novel transcript | 1.25 | 1.75 × 10−6 | 1.25 | 1.75 × 10−6 | |
| Biological Process | CBD Dose (mg/kg b.w.) | Gene Regulation | FDR * | Enrichment ** |
|---|---|---|---|---|
| Response to stimulus | 10 | Up | 0.049 | 72 |
| Circadian rhythm | 10 | Up | 0.049 | 23.1 |
| Circadian behavior | 10 | Up | 0.049 | 76.2 |
| Nutrient response | 10 | Down | 0.025 | 17.5 |
| Response to redox states | 10 | Down | 0.025 | 157.8 |
| Negative regulation of the glucocorticoid receptor signaling pathway | 10 | Down | 0.031 | 210.4 |
| Diurnal regulation of gene expression | 10 | Down | 0.031 | 32.7 |
| Intracellular receptor signaling pathway | 10 | Down | 0.031 | 12.4 |
| Glucocorticoid receptor signaling pathway | 10 | Down | 0.031 | 114.8 |
| Corticosteroid receptor signaling pathway | 10 | Down | 0.031 | 78.9 |
| Physiological rhythm | 20 | Up | 0.026 | 6.4 |
| Response to the stimulus | 20 | Up | 0.025 | 19.3 |
| Circadian rhythm | 20 | Up | 0.025 | 20.4 |
| Metabolism process of arachidonic acid | 20 | Up | 0.047 | 17.8 |
| Cellular response to a corticosteroid stimulus | 20 | Up | 0.015 | 10.8 |
| Process of qualitative biological regulation | 20 | Up | 0.010 | 1.8 |
| Diurnal regulation of gene expression | 20 | Up | 0.011 | 12.8 |
| Cellular response to peptides | 20 | Up | 0.010 | 4.7 |
| Metabolism process of unsaturated fatty acids | 20 | Up | 0.010 | 5.1 |
| Cellular response to potassium ions | 20 | Up | 0.015 | 95.2 |
| Organic acid biosynthesis process | 20 | Down | 0.031 | 5.9 |
| Monosaccharide biosynthesis process | 20 | Down | 0.031 | 4.3 |
| The biosynthetic process of carboxylic acids | 20 | Down | 0.031 | 5.9 |
| Organic acid metabolism process | 20 | Down | 0.036 | 3.2 |
| The metabolism process of monocarboxylic acids | 20 | Down | 0.010 | 3.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rokicki, M.; Żurowski, J.; Sawicki, S.; Ocłoń, E.; Szmatoła, T.; Jasielczuk, I.; Mizera-Szpilka, K.; Semik-Gurgul, E.; Gurgul, A. Impact of Long-Term Cannabidiol (CBD) Treatment on Mouse Kidney Transcriptome. Genes 2024, 15, 1640. https://doi.org/10.3390/genes15121640
Rokicki M, Żurowski J, Sawicki S, Ocłoń E, Szmatoła T, Jasielczuk I, Mizera-Szpilka K, Semik-Gurgul E, Gurgul A. Impact of Long-Term Cannabidiol (CBD) Treatment on Mouse Kidney Transcriptome. Genes. 2024; 15(12):1640. https://doi.org/10.3390/genes15121640
Chicago/Turabian StyleRokicki, Mikołaj, Jakub Żurowski, Sebastian Sawicki, Ewa Ocłoń, Tomasz Szmatoła, Igor Jasielczuk, Karolina Mizera-Szpilka, Ewelina Semik-Gurgul, and Artur Gurgul. 2024. "Impact of Long-Term Cannabidiol (CBD) Treatment on Mouse Kidney Transcriptome" Genes 15, no. 12: 1640. https://doi.org/10.3390/genes15121640
APA StyleRokicki, M., Żurowski, J., Sawicki, S., Ocłoń, E., Szmatoła, T., Jasielczuk, I., Mizera-Szpilka, K., Semik-Gurgul, E., & Gurgul, A. (2024). Impact of Long-Term Cannabidiol (CBD) Treatment on Mouse Kidney Transcriptome. Genes, 15(12), 1640. https://doi.org/10.3390/genes15121640






