The Stem Cell Expression Profile of Odontogenic Tumors and Cysts: A Systematic Review and Meta-Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Research Question and Study Protocol
2.2. Search Strategy
2.3. Eligibility Screening and Study Selection
2.4. Data Extraction and Synthesis
2.5. Risk of Bias Assessment
2.6. Meta-Analysis
3. Results
3.1. Study Cohort
3.2. Study Population Characteristics
3.3. Study Comparator Characteristics
3.4. Outcome of Stem Cell Markers’ Expression
3.4.1. Benign Odontogenic Tumors
3.4.2. Malignant Odontogenic Tumors
3.4.3. Developmental Odontogenic Cysts
3.4.4. Inflammatory Odontogenic Cysts
3.5. RoB Assessment
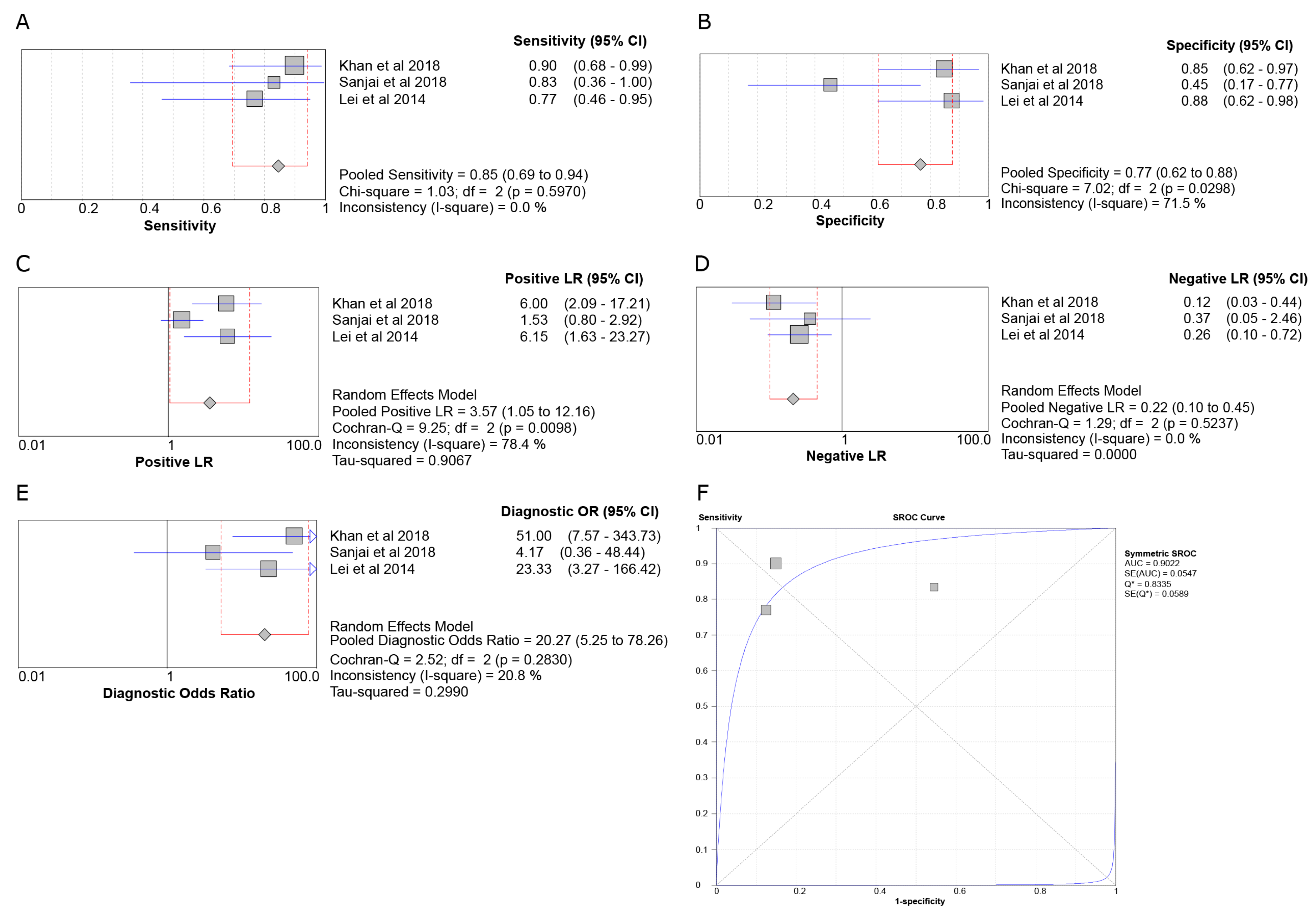
3.6. Meta-Analysis
3.6.1. SOX2 in ACA vs. AMBL
3.6.2. SOX2 in OKC vs. AMBL
3.6.3. SOX2 in AMBL vs. DF
3.6.4. OCT4 in OKC vs. AMBL
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- El-Naggar, A.K.; Chan, J.K.C.; Grandis, J.R.; Takata, T.; Slootweg, P.J. Odontogenic and maxillofacial bone tumors. In WHO Classification of Head and Neck Tumours, 4th ed.; IARC: Lyon, France, 2017; pp. 215–236. [Google Scholar]
- Patil, S.; Halgatti, V.; Khandelwal, S.; Santosh, B.S.; Maheshwari, S. Prevalence of cysts and tumors around the retained and unerupted third molars in the Indian population. J. Oral Biol. Craniofac. Res. 2014, 4, 82–87. [Google Scholar] [CrossRef]
- Guimaraes, L.M.; Coura, B.P.; Gomez, R.S.; Gomes, C.C. The Molecular Pathology of Odontogenic Tumors: Expanding the Spectrum of MAPK Pathway Driven Tumors. Front. Oral Health 2021, 2, 740788. [Google Scholar] [CrossRef] [PubMed]
- Tamiolakis, P.; Thermos, G.; Tosios, K.I.; Sklavounou-Andrikopoulou, A. Demographic and Clinical Characteristics of 5294 Jaw Cysts: A Retrospective Study of 38 Years. Head Neck Pathol. 2019, 13, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Rojo, L.; Granchi, Z.; Graf, D.; Mitsiadis, T.A. Stem Cell Fate Determination during Development and Regeneration of Ectodermal Organs. Front. Physiol. 2012, 3, 107. [Google Scholar] [CrossRef] [PubMed]
- Juuri, E.; Isaksson, S.; Jussila, M.; Heikinheimo, K.; Thesleff, I. Expression of the stem cell marker, SOX2, in ameloblastoma and dental epithelium. Eur. J. Oral Sci. 2013, 121, 509–516. [Google Scholar] [CrossRef]
- Xiong, J.; Mrozik, K.; Gronthos, S.; Bartold, P.M. Epithelial cell rests of Malassez contain unique stem cell populations capable of undergoing epithelial-mesenchymal transition. Stem Cells Dev. 2012, 21, 2012–2025. [Google Scholar] [CrossRef]
- Klonisch, T.; Wiechec, E.; Hombach-Klonisch, S.; Ande, S.R.; Wesselborg, S.; Schulze-Osthoff, K.; Los, M. Cancer stem cell markers in common cancers—Therapeutic implications. Trends Mol. Med. 2008, 14, 450–460. [Google Scholar] [CrossRef]
- Park, C.Y.; Tseng, D.; Weissman, I.L. Cancer stem cell-directed therapies: Recent data from the laboratory and clinic. Mol. Ther. 2009, 17, 219–230. [Google Scholar] [CrossRef]
- Chacham, M.; Almoznino, G.; Zlotogorski-Hurvitz, A.; Buchner, A.; Vered, M. Expression of stem cell markers in stroma of odontogenic cysts and tumors. J. Oral Pathol. Med. Off. Publ. Int. Assoc. Oral Pathol. Am. Acad. Oral Pathol. 2020, 49, 1068–1077. [Google Scholar] [CrossRef]
- Estrela, C.; Carmo Souza, P.O.; Barbosa, M.G.; Aburad de Carvalhosa, A.; Batista, A.C.; Pinto Junior, D.D.S.; Yamamoto-Silva, F.P.; de Freitas Silva, B.S. Mesenchymal Stem Cell Marker Expression in Periapical Abscess. J. Endod. 2019, 45, 716–723. [Google Scholar] [CrossRef]
- Khan, W.; Augustine, D.; Rao, R.S.; Sowmya, S.V.; Haragannavar, V.C.; Nambiar, S. Stem Cell Markers SOX-2 and OCT-4 Enable to Resolve the Diagnostic Dilemma between Ameloblastic Carcinoma and Aggressive Solid Multicystic Ameloblastoma. Adv. Biomed. Res. 2018, 7, 149. [Google Scholar] [CrossRef] [PubMed]
- Kumamoto, H.; Ohki, K.; Kumamoto, H.; Ohki, K. Detection of CD133, Bmi-1, and ABCG2 in ameloblastic tumors. J. Oral Pathol. Med. 2010, 39, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Martins Balbinot, K.; Almeida Loureiro, F.J.; Chemelo, G.P.; Alves Mesquita, R.; Cruz Ramos, A.M.P.; Ramos, R.T.J.; da Costa da Silva, A.L.; de Menezes, S.A.F.; da Silva Kataoka, M.S.; Alves Junior, S.M.; et al. Immunoexpression of stem cell markers SOX-2, NANOG AND OCT4 in ameloblastoma. PeerJ 2023, 11, e14349. [Google Scholar] [CrossRef] [PubMed]
- Mack, D.L.; Guan, X.; Wagoner, A.; Walker, S.J.; Childers, M.K. Disease-in-a-dish: The contribution of patient-specific induced pluripotent stem cell technology to regenerative rehabilitation. Am. J. Phys. Med. Rehabil. 2014, 93, S155–S168. [Google Scholar] [CrossRef]
- Chen, K.; Huang, Y.H.; Chen, J.L. Understanding and targeting cancer stem cells: Therapeutic implications and challenges. Acta Pharmacol. Sin. 2013, 34, 732–740. [Google Scholar] [CrossRef]
- Spagnuolo, G.; Codispoti, B.; Marrelli, M.; Rengo, C.; Rengo, S.; Tatullo, M. Commitment of Oral-Derived Stem Cells in Dental and Maxillofacial Applications. Dent. J. 2018, 6, 72. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Booth, A.; Clarke, M.; Dooley, G.; Ghersi, D.; Moher, D.; Petticrew, M.; Stewart, L. The nuts and bolts of PROSPERO: An international prospective register of systematic reviews. Syst. Rev. 2012, 1, 2. [Google Scholar] [CrossRef]
- Calloni, R.; Cordero, E.A.; Henriques, J.A.; Bonatto, D. Reviewing and updating the major molecular markers for stem cells. Stem Cells Dev. 2013, 22, 1455–1476. [Google Scholar] [CrossRef]
- Hadjimichael, C.; Chanoumidou, K.; Papadopoulou, N.; Arampatzi, P.; Papamatheakis, J.; Kretsovali, A. Common stemness regulators of embryonic and cancer stem cells. World. J. Stem Cells 2015, 7, 1150–1184. [Google Scholar] [CrossRef]
- Neradil, J.; Veselska, R. Nestin as a marker of cancer stem cells. Cancer Sci. 2015, 106, 803–811. [Google Scholar] [CrossRef] [PubMed]
- Seal, R.L.; Braschi, B.; Gray, K.; Jones, T.E.M.; Tweedie, S.; Haim-Vilmovsky, L.; Bruford, E.A. Genenames.org: The HGNC resources in 2023. Nucleic Acids Res. 2023, 51, D1003–D1009. [Google Scholar] [CrossRef] [PubMed]
- Munn, Z.; Barker, T.H.; Moola, S.; Tufanaru, C.; Stern, C.; McArthur, A.; Stephenson, M.; Aromataris, E. Methodological quality of case series studies: An introduction to the JBI critical appraisal tool. JBI Evid. Synth. 2020, 18, 2127–2133. [Google Scholar] [CrossRef] [PubMed]
- Kalogirou, E.M.; Thermos, G.; Zogopoulos, V.; Foutadakis, S.; Michalopoulos, I.; Agelopoulos, M.; Tosios, K.I. The immunohistochemical profile of basal cell nevus syndrome-associated and sporadic odontogenic keratocysts: A systematic review and meta-analysis. Clin. Oral Investig. 2021, 25, 3351–3367. [Google Scholar] [CrossRef] [PubMed]
- Review Manager (RevMan) [Computer Program]. Version 5.4, T.C.C. Available online: https://revman.cochrane.org/#/myReviews (accessed on 17 July 2023).
- Zamora, J.; Abraira, V.; Muriel, A.; Khan, K.; Coomarasamy, A. Meta-DiSc: A software for meta-analysis of test accuracy data. BMC Med. Res. Methodol. 2006, 6, 31. [Google Scholar] [CrossRef]
- Bandyopadhyay, A.; Nishat, R.; Behura, S.S.; Panda, A.; Ramachandra, S.; Mohiddin, G. Cancer stem cell markers, SOX 2 and OCT 4 in ameloblastoma and keratocystic odontogenic tumor: An immunohistochemical study. J. Int. Oral Health 2017, 9, 28. [Google Scholar]
- Lei, Y.; Jaradat, J.M.; Owosho, A.A.; Adebiyi, K.E.; Bilodeau, E.A. Identification of SOX2 as a marker for ameloblastic carcinoma. Lab. Investig. 2013, 93, 308A–309A. [Google Scholar]
- Sanjai, K.; Sangappa, S.B.; Shivalingaiah, D.; Kumar, H.M.; Baker, A. Expression of SOX2 and EGFR in ameloblastoma, odontoameloblastoma and ameloblastic carcinoma. J. Clin. Diagn. Res. 2018, 12, ZC48–ZC52. [Google Scholar] [CrossRef]
- Monroy, E.A.C.; de Andrade Santos, P.P.; de Sousa Lopes, M.L.D.; Mosqueda-Taylor, A.; Pinto, L.P.; de Souza, L.B. Oct-4 and CD44 in epithelial stem cells like of benign odontogenic lesions. Histochem. Cell Biol. 2018, 150, 371–377. [Google Scholar] [CrossRef]
- Phattarataratip, E.; Panitkul, T.; Khodkaew, W.; Anupuntanun, P.; Jaroonvechatam, J.; Pitarangsikul, S. Expression of SOX2 and OCT4 in odontogenic cysts and tumors. Head Face Med. 2021, 17, 29. [Google Scholar] [CrossRef]
- Silva, B.S.; Silva, L.R.; Lima, K.L.; Dos Santos, A.C.; Oliveira, A.C.; Dezzen-Gomide, A.C.; Batista, A.C.; Yamamoto-Silva, F.P. SOX2 and BCL-2 Expressions in Odontogenic Keratocyst and Ameloblastoma. Med. Oral Patol. Oral Cir. Bucal 2020, 25, e283–e290. [Google Scholar] [CrossRef] [PubMed]
- da Trindade, G.A.; da Silva, L.P.; de Andrade Santos, P.P.; Pinto, L.P.; de Souza, L.B. Expression of a Tumor Stem Cell Marker (Aldehyde Dehydrogenase 1-ALDH1) in Benign Epithelial Odontogenic Lesions. Head Neck Pathol. 2022, 16, 785–791. [Google Scholar] [CrossRef] [PubMed]
- Farias, Z.; Silva, L.P.D.; de Arruda, J.A.A.; Cavalcante, J.S.; Almeida, H.C.R.; Oliveira, M.C.V.; Souza, L.B.; Sobral, A.P.V. ALDH1 expression and potential clinical implications in chronic inflammatory periapical lesions. Braz. Oral Res. 2022, 36, e019. [Google Scholar] [CrossRef]
- Júnior, J.F.; de França, G.M.; da Silva Barros, C.C.; Felix, F.A.; da Silva, W.R.; de Lucena, H.F.; Oliveira, C.N.; Galvão, H.C. Biomarkers involved in the proliferation of the odontogenic keratocyst, glandular odontogenic cyst and botryoid odontogenic cyst. Oral Maxillofac. Surg. 2022, 26, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Kalogirou, E.M.; Foutadakis, S.; Koutsi, M.A.; Vatsellas, G.; Vlachodimitropoulos, D.; Petsinis, V.; Sklavounou, A.; Agelopoulos, M.; Tosios, K.I. Decoding a gene expression program that accompanies the phenotype of sporadic and basal cell nevus syndrome-associated odontogenic keratocyst. J. Oral Pathol. Med. Off. Publ. Int. Assoc. Oral Pathol. Am. Acad. Oral Pathol. 2022, 51, 649–658. [Google Scholar] [CrossRef]
- Tseng, C.H.; Lu, P.H.; Wang, Y.P.; Chang, J.Y.F. Enrichment of SOX2-Positive Cells in BRAF V600E Mutated and Recurrent Ameloblastoma. J. Pers. Med. 2022, 12, 77. [Google Scholar] [CrossRef]
- Chang, T.H.; Shanti, R.M.; Liang, Y.; Zeng, J.; Shi, S.; Alawi, F.; Carrasco, L.; Zhang, Q.; Le, A.D. LGR5(+) epithelial tumor stem-like cells generate a 3D-organoid model for ameloblastoma. Cell Death Dis. 2020, 11, 338. [Google Scholar] [CrossRef]
- Fraser, G.J.; Hamed, S.S.; Martin, K.J.; Hunter, K.D. Shark tooth regeneration reveals common stem cell characters in both human rested lamina and ameloblastoma. Sci. Rep. 2019, 9, 15956. [Google Scholar] [CrossRef]
- Andisheh-Tadbir, A.; Gorgizadeh, A. CD166 expression in dentigerous cyst, keratocystic odontogenic tumor and ameloblastoma. J. Clin. Exp. Dent. 2016, 8, e236–e240. [Google Scholar] [CrossRef]
- Heikinheimo, K.; Kurppa, K.J.; Laiho, A.; Peltonen, S.; Berdal, A.; Bouattour, A.; Ruhin, B.; Catón, J.; Thesleff, I.; Leivo, I.; et al. Early dental epithelial transcription factors distinguish ameloblastoma from keratocystic odontogenic tumor. J. Dent. Res. 2015, 94, 101–111. [Google Scholar] [CrossRef]
- Marrelli, M.; Paduano, F.; Tatullo, M. Human periapical cyst-mesenchymal stem cells differentiate into neuronal cells. J. Dent. Res. 2015, 94, 843–852. [Google Scholar] [CrossRef]
- Srinath, S.; Iyengar, A.; Mysorekar, V. CD 44 expression in dentigerous cyst, radicular cyst and ameloblastoma, by immunohistochemical analysis. IOSR J. Dent. Med. Sci. 2014, 13, 80–83. [Google Scholar] [CrossRef]
- Moosvi, Z.; Rekha, K. c-Myc oncogene expression in selected odontogenic cysts and tumors: An immunohistochemical study. J. Oral Maxillofac. Pathol. JOMFP 2013, 17, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Salehinejad, J.; Saghafi, S.; Sharifi, N.; Zare-Mahmoodabadi, R.; Saghravanian, N.; Ghazi, N.; Shakeri, M.T. Evaluation of osteopontin and CD44v6 expression in odontogenic cystic lesions by immunohistochemistry. Pathol. Res. Pract. 2012, 208, 410–414. [Google Scholar] [CrossRef] [PubMed]
- Song, J.S.; Stefanik, D.; Damek-Poprawa, M.; Alawi, F.; Akintoye, S.O. Differentiation and regenerative capacities of human odontoma-derived mesenchymal cells. Differ. Res. Biol. Divers. 2009, 77, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.P.; Liu, B.Y. High expression of osteopontin and CD44v6 in odontogenic keratocysts. J. Formos. Med. Assoc. 2009, 108, 286–292. [Google Scholar] [CrossRef][Green Version]
- Fujita, S.; Hideshima, K.; Ikeda, T. Nestin expression in odontoblasts and odontogenic ectomesenchymal tissue of odontogenic tumours. J. Clin. Pathol. 2006, 59, 240–245. [Google Scholar] [CrossRef]
- Leonardi, R.; Lanteri, E.; Stivala, F.; Travali, S. Immunolocalization of CD44 adhesion molecules in human periradicular lesions. Oral Surg Oral Med. Oral Pathol. Oral Radiol. Endod. 2000, 89, 480–485. [Google Scholar] [CrossRef]
- Barnes, L.; Eveson, J.W.; Reichart, P.; Sidransky, D. Odontogenic Tumours. In WHO Classification of Tumors: Pathology and Genetics of Head and Neck Tumours, 3rd ed.; IARC: Lyon, France, 2005. [Google Scholar]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Balaji, S.; Li, H.; Steen, E.; Keswani, S.G. Considerations for Immunohistochemistry. In Success in Academic Surgery: Basic Science; Springer International Publishing: Cham, Germany, 2019; pp. 105–144. [Google Scholar]
- Vandromme, M.; Gauthier-Rouviere, C.; Lamb, N.; Fernandez, A. Regulation of transcription factor localization: Fine-tuning of gene expression. Trends Biochem. Sci. 1996, 21, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.; Hochedlinger, K. The sox family of transcription factors: Versatile regulators of stem and progenitor cell fate. Cell Stem Cell 2013, 12, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Weina, K.; Utikal, J. SOX2 and cancer: Current research and its implications in the clinic. Clin. Transl. Med. 2014, 3, 19. [Google Scholar] [CrossRef] [PubMed]
- da Cunha, J.M.; da Costa-Neves, A.; Kerkis, I.; da Silva, M.C. Pluripotent stem cell transcription factors during human odontogenesis. Cell Tissue Res. 2013, 353, 435–441. [Google Scholar] [CrossRef]
- Li, T.J. The odontogenic keratocyst: A cyst, or a cystic neoplasm? J. Dent. Res. 2011, 90, 133–142. [Google Scholar] [CrossRef]
- Shi, G.; Jin, Y. Role of Oct4 in maintaining and regaining stem cell pluripotency. Stem Cell Res. Ther. 2010, 1, 39. [Google Scholar] [CrossRef]
- Avilion, A.A.; Nicolis, S.K.; Pevny, L.H.; Perez, L.; Vivian, N.; Lovell-Badge, R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003, 17, 126–140. [Google Scholar] [CrossRef]
- Villodre, E.S.; Kipper, F.C.; Pereira, M.B.; Lenz, G. Roles of OCT4 in tumorigenesis, cancer therapy resistance and prognosis. Cancer Treat. Rev. 2016, 51, 1–9. [Google Scholar] [CrossRef]
- Cai, J.; He, B.; Li, X.; Sun, M.; Lam, A.K.; Qiao, B.; Qiu, W. Regulation of tumorigenesis in oral epithelial cells by defined reprogramming factors Oct4 and Sox2. Oncol. Rep. 2016, 36, 651–658. [Google Scholar] [CrossRef]
- Ghaleb, A.M.; Yang, V.W. Kruppel-like factor 4 (KLF4): What we currently know. Gene 2017, 611, 27–37. [Google Scholar] [CrossRef]
- Tiwari, A.; Loughner, C.L.; Swamynathan, S.; Swamynathan, S.K. KLF4 Plays an Essential Role in Corneal Epithelial Homeostasis by Promoting Epithelial Cell Fate and Suppressing Epithelial-Mesenchymal Transition. Invest. Ophthalmol. Vis. Sci. 2017, 58, 2785–2795. [Google Scholar] [CrossRef] [PubMed]
- Pelengaris, S.; Khan, M.; Evan, G. c-MYC: More than just a matter of life and death. Nat. Rev. Cancer 2002, 2, 764–776. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, J.L.; Feldman, R.A. Tinkering with transcription factors uncovers plasticity of somatic cells. Genes. Cancer 2010, 1, 1089–1099. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liebau, S.; Mahaddalkar, P.U.; Kestler, H.A.; Illing, A.; Seufferlein, T.; Kleger, A. A hierarchy in reprogramming capacity in different tissue microenvironments: What we know and what we need to know. Stem Cells Dev. 2013, 22, 695–706. [Google Scholar] [CrossRef]
- Naor, D.; Sionov, R.V.; Ish-Shalom, D. CD44: Structure, function, and association with the malignant process. Adv. Cancer Res. 1997, 71, 241–319. [Google Scholar] [CrossRef]
- Compagnone, M.; Gatti, V.; Presutti, D.; Ruberti, G.; Fierro, C.; Markert, E.K.; Vousden, K.H.; Zhou, H.; Mauriello, A.; Anemone, L.; et al. DeltaNp63-mediated regulation of hyaluronic acid metabolism and signaling supports HNSCC tumorigenesis. Proc. Natl Acad. Sci. USA 2017, 114, 13254–13259. [Google Scholar] [CrossRef]
- Paduano, F.; Marrelli, M.; Palmieri, F.; Tatullo, M. CD146 Expression Influences Periapical Cyst Mesenchymal Stem Cell Properties. Stem Cell Rev. Rep. 2016, 12, 592–603. [Google Scholar] [CrossRef]
- Shear, M. The aggressive nature of the odontogenic keratocyst: Is it a benign cystic neoplasm? Part 2. Proliferation and genetic studies. Oral Oncol. 2002, 38, 323–331. [Google Scholar] [CrossRef]

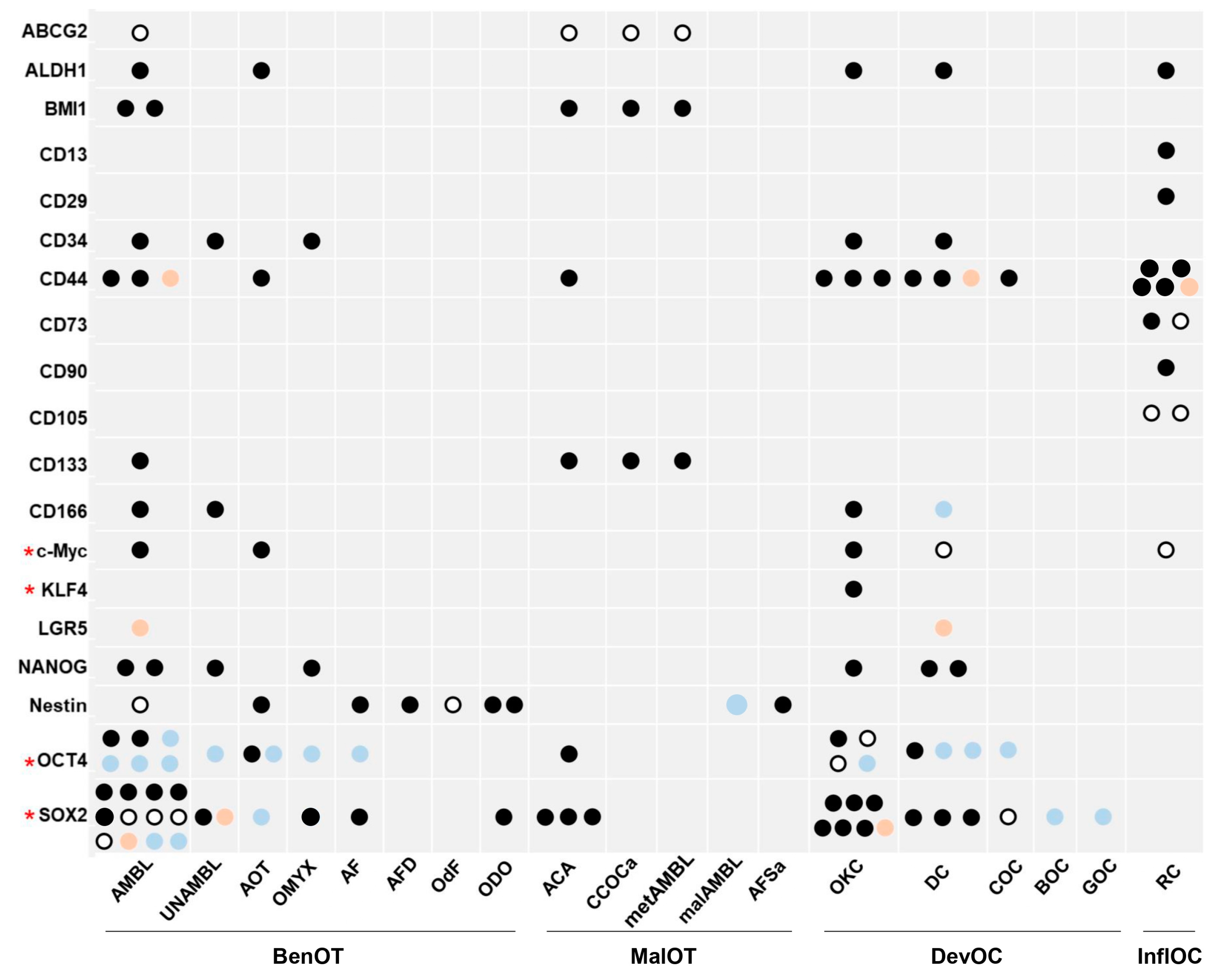


| Reference | Odontogenic Lesions; Controls (N) | Method | Marker * | Outcome of Comparison |
|---|---|---|---|---|
| Martins Balbinot et al., 2023 [14] | AMBL (23), DC (10); DF (10) | IHC | SOX2, OCT4, NANOG | Significantly higher scores of SOX2, OCT4, and NANOG were found in AMBL than DC (p < 0.001) and DF (p < 0.001). |
| da Trindade et al., 2022 [34] | AMBL (20), AOT (20), OKC (20), DC (20) | IHC | ALDH1 | DC/OKC showed a significantly higher ALDH1 score than AMBL/AOT (p < 0.0001). No differences were found between the two cyst groups or between the two tumor groups. |
| Farias et al., 2022 [35] | RC (26); PG (25) | IHC | ALDH1 | A lower ALDH1 immunoexpression score was found in RC than PG. |
| Júnior et al., 2022 [36] | OKC (20), BOC (10), GOC (10) | IHC | SOX2 | SOX2 was expressed in OKC, while not in BOC or GOC. |
| Kalogirou et al., 2022 [37] | OKC (12); DF (6) | RNA-seq, IHC | SOX2, KLF4 | SOX2 and KLF4 were upregulated in OKC compared to DF (p < 0.000). Higher immunoscores of SOX2 and KLF4 were found in OKC compared with DF. |
| Tseng et al., 2022 [38] | AMBL (49), UNAMBL (25); DF (6) | IHC | SOX2 | DF showed a higher SOX2 score than all AMBL types. |
| Phattarataratip et al., 2021 [32] | AMBL (15), AOT (5), AF (5), OKC (15), DC (10), COC (5); DF (2) | IHC | SOX2, OCT4 | A significantly higher SOX2 score was found in OKC compared with DC and AMBL (p < 0.001), and in AF compared with COC and AOT (p < 0.001). No difference was found in OCT4 score. |
| Chacham et al., 2020 [10] | AMBL (10), UNAMBL (10), OMYX (10), OKC (27), DC (10) | IHC | NANOG, SOX2, CD34, OCT4 | A significantly higher NANOG immunoexpression score was found in DC/pOKC/UNAMBL than the other groups (except for pOKC vs. sOKC) (p < 0.05). A significantly higher SOX2 immunoscore was found in odontogenic tumors than in odontogenic cysts (except for UNAMBL vs. DC) (p < 0.05). A significantly higher CD34 immunoscore was found in rOKC than DC, and in pOKC/rOKC than UNAMBL/OMYX (p < 0.05). OCT4 was only expressed in a limited number of pOKC cells. |
| Silva et al., 2020 [33] | AMBL (20), OKC (20) | IHC | SOX2 | A significantly higher SOX2 score was found in OKC than AMBL (p < 0.001). |
| Chang et al., 2020 [39] | AMBL (15), DC (6) | IF | LGR5 | A significantly higher staining score of LGR5 was found in AMBL than DC (p < 0.0001). |
| Estrela et al., 2019 [11] | RC (10); PG (10), PAbs (10), APap (10) | IHC | CD44, CD73, CD105 | Significantly higher expressions of CD44, CD73, and CD105 were found in the PAbs than RC/PG (p < 0.05). |
| Fraser et al., 2019 [40] | AMBL (5); DF (5) | IF | SOX2, BMI1 | No difference was found. |
| Khan et al., 2018 [12] | AMBL (20), ACA (20); OSCC (5), DF (5) | IHC | SOX2, OCT4, CD44 | Significantly higher SOX2 and OCT4 expressions were found in ACA than AMBL (p < 0.001). |
| Monroy et al., 2018 [31] | AMBL (20), AOT (20), OKC (20) | IHC | OCT4, CD44 | A significantly higher CD44 score was found in OKC compared with AMBL and AOT (p = 0.034). No difference was found in OCT4 score. |
| Sanjai et al., 2018 [30] | AMBL (11), ODAMBL (2), ACA (6) | IHC | SOX2 | No difference was found. |
| Bandyopadhyay et al., 2017 [28] | AMBL (15), OKC (15) | IHC | SOX2, OCT4 | SOX2 was expressed only in OKC. No difference in OCT4 score. |
| Andisheh-Tadbir & Gorgizadeh, 2016 [41] | AMBL (17), UNAMBL (15), OKC (18), DC (19) | IHC | CD166 | A significantly higher CD166 expression was found in AMBL/UNAMBL/OKC than DC (p < 0.005). |
| Heikinheimo et al., 2015 [42] | AMBL (15), OKC (12); NOM (4) | microarrays, real-time RT-PCR, IF | SOX2 | SOX2 was upregulated in OKC compared to AMBL (p < 0.000). SOX2 immunoexpression was found only in OKC. |
| Marrelli et al., 2015 [43] | HRCMCs (4); DPSCs (4) | flow cytometry, real-time RT-PCR | CD13, CD29, CD44, CD73, CD90, CD105 | Lower CD105 expression was found in HRCMSs than DPSCs. No other differences were found. |
| Lei et al., 2014 [29] | AMBL (16), IGaAMBL (6), ACA (13) | IHC | SOX2 | A significantly higher SOX2 score was found in ACA than AMBL/IGaAMBL (p = 0.0021). |
| Srinath et al., 2014 [44] | AMBL (10), DC (5) RC (5) | IHC | CD44 | No difference was found. |
| Juuri et al., 2013 [6] | AMBL (5); HPMDE (NA) | IHC | SOX2 | SOX2 was expressed in AMBL and dental lamina. |
| Moosvi & Rekha, 2013 [45] | AMBL (10), AOT (10), OKC (10), DC (10), RC (10) | IHC | c-Myc | A significantly higher intensity score was found in OKC than DC (p = 0.0257) and RC (p = 0.0452); in AMBL than DC (p = 0.0312); and in AOT than DC (p = 0.0257) and RC (p = 0.0452). No differences between OKC, AMBL, and AOT. |
| Salehinejad et al., 2012 [46] | OKC (14), DC (14), RC (14), COC (14) | IHC | CD44v6 | A significantly higher CD44v6 score was found in OKC compared with DC (p < 0.001), RC (p < 0.001), and COC (p = 0.024), and in COC compared with DC (p < 0.001) and RC (p < 0.001). |
| Kumamoto et al., 2010 [13] | AMBL (47), metAMBL (2), ACA (2), CCOCa (2); TG (12) | IHC, RT-PCR | CD133, BMI1, ABCG2 | A significantly higher ABCG2 score was found in AMBL/metAMBL/ACA than TG (p < 0.05), and a significantly higher CD133 score was found in malAMBL/ACA than AMBL (p < 0.001) and TG (p < 0.001). No difference was found in BMI1 score. |
| Song et al., 2009 [47] | HODCs; DPSCs, BMSCs | IF, real-time RT-PCR | Nestin, SOX2 | A significantly higher expression level of Nestin mRNA was found in HODCs compared with DPSCs/BMSCs (p < 0.04). SOX2 was expressed only in HODCs. |
| Wang & Liu, 2009 [48] | OKC (20), DC (8), RC (10) | IHC | CD44v6 | No difference was found. |
| Fujita et al., 2006 [49] | AMBL (44), malAMBL (3), ODO (62), AF (2), AFD (7), AFSa (2), AOT (6), OdF (3) | IHC | Nestin | Nestin was expressed in odontogenic mixed tumors, mainly in sites of odontogenic ectomesenchyme. |
| Leonardi et al., 2000 [50] | RC (4); PG (16) | IHC | CD44 | No difference was found. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalogirou, E.-M.; Lekakis, G.; Petroulias, A.; Chavdoulas, K.; Zogopoulos, V.L.; Michalopoulos, I.; Tosios, K.I. The Stem Cell Expression Profile of Odontogenic Tumors and Cysts: A Systematic Review and Meta-Analysis. Genes 2023, 14, 1735. https://doi.org/10.3390/genes14091735
Kalogirou E-M, Lekakis G, Petroulias A, Chavdoulas K, Zogopoulos VL, Michalopoulos I, Tosios KI. The Stem Cell Expression Profile of Odontogenic Tumors and Cysts: A Systematic Review and Meta-Analysis. Genes. 2023; 14(9):1735. https://doi.org/10.3390/genes14091735
Chicago/Turabian StyleKalogirou, Eleni-Marina, Georgios Lekakis, Aristodimos Petroulias, Konstantinos Chavdoulas, Vasileios L. Zogopoulos, Ioannis Michalopoulos, and Konstantinos I. Tosios. 2023. "The Stem Cell Expression Profile of Odontogenic Tumors and Cysts: A Systematic Review and Meta-Analysis" Genes 14, no. 9: 1735. https://doi.org/10.3390/genes14091735
APA StyleKalogirou, E.-M., Lekakis, G., Petroulias, A., Chavdoulas, K., Zogopoulos, V. L., Michalopoulos, I., & Tosios, K. I. (2023). The Stem Cell Expression Profile of Odontogenic Tumors and Cysts: A Systematic Review and Meta-Analysis. Genes, 14(9), 1735. https://doi.org/10.3390/genes14091735








