Genetic Landscape of Masticatory Muscle Tendon–Aponeurosis Hyperplasia
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Pig Samples
2.3. Whole Genome Sequencing (WGS) and Variant Calling Pipelines
2.4. Variant Prioritization Pipeline
2.5. Transcriptome Sequencing (RNA-seq) of Pig Tissue Samples
3. Results
3.1. Variant Detection by WGS Analysis
3.2. Integration of the WGS Analysis of Patients with the RNA-seq Analysis of Pig Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schiffman, E.; Ohrbach, R.; Truelove, E.; Look, J.; Anderson, G.; Goulet, J.-P.; List, T.; Svensson, P.; Gonzalez, Y.; Lobbezoo, F.; et al. Diagnostic Criteria for Temporomandibular Disorders (DC/TMD) for Clinical and Research Applications: Recommendations of the International RDC/TMD Consortium Network and Orofacial Pain Special Interest Group. J. Oral Facial Pain Headache 2014, 28, 6–27. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.-H.; Lv, K.; Yang, R.-T.; Li, Z.; Yang, X.-W.; Li, Z.-B. Clinical, retrospective case-control study on the mechanics of obstacle in mouth opening and malocclusion in patients with maxillofacial fractures. Sci. Rep. 2018, 8, 7724. [Google Scholar] [CrossRef]
- Smith, M.J.; Myall, R.W. Tetanus: Review of the literature and report of a case. Oral Surg. Oral Med. Oral Pathol. 1976, 41, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Aliko, A.; Ciancaglini, R.; Alushi, A.; Tafaj, A.; Ruci, D. Temporomandibular joint involvement in rheumatoid arthritis, systemic lupus erythematosus and systemic sclerosis. Int. J. Oral Maxillofac. Surg. 2011, 40, 704–709. [Google Scholar] [CrossRef]
- Jager-Wittenaar, H.; Dijkstra, P.U.; Vissink, A.; van Oort, R.P.; Roodenburg, J.L. Variation in repeated mouth-opening measurements in head and neck cancer patients with and without trismus. Int. J. Oral Maxillofac. Surg. 2009, 38, 26–30. [Google Scholar] [CrossRef]
- Çorumlu, U.; Kopuz, C.; Demir, M.T.; Pirzirenli, M.E. Bilateral elongated mandibular coronoid process in an Anatolian skull. Anat. Cell Biol. 2016, 49, 217–220. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Khandelwal, P.; Dhupar, V.; Akkara, F.; Louis, A. Zygomatico-coronoid ankylosis as sequel of inadequate treatment. Ann. Maxillofac. Surg. 2018, 8, 158–161. [Google Scholar] [CrossRef] [PubMed]
- Arika, T.; Kakudo, K. Hyperplasia of tendon and aponeurosis of masticatory muscles (HyTAM)—Clinical appearance. J. Jpn. Soc. Temporomandibular Jt. 2009, 21, 31–34. [Google Scholar]
- Kakudo, K.; Yoda, T. New concept of limited mouth opening associated with square mandible: Diagnosis and treatment for masticatory muscle tendon-aponeurosis hyperplasia. J. Jpn. Soc. Temporomandibular Jt. 2009, 21, 28–30. [Google Scholar]
- Hayashi, N.; Yoda, T.; Tomoda, T.; Yumoto, M.; Kitamura, T.; Okubo, M.; Iwasaki, Y.; Enoki, Y.; Sato, T. Longitudinal epidemiological survey on suspected masticatory muscle tendon-aponeurosis hyperplasia in adolescents. J. Jpn. Soc. Temporomandibular Jt. 2018, 30, 187–194. [Google Scholar] [CrossRef]
- Worni, A.; Mericske-Stern, R.; Iizuka, T.; Büttner, M. Limited mouth opening—What now? Swiss Dent. J. 2014, 124, 935–944. [Google Scholar] [PubMed]
- Yoda, T.; Sato, T.; Abe, T.; Sakamoto, I.; Tomaru, Y.; Omura, K.; Hatano, N.; Takato, T.; Ishii, Y. Long-term results of surgical therapy for masticatory muscle tendon-aponeurosis hyperplasia accompanied by limited mouth opening. Int. J. Oral Maxillofac. Surg. 2009, 38, 1143–1147. [Google Scholar] [CrossRef] [PubMed]
- Ajila, V.; Hegde, S.; Gopakumar, R.; Babu, G.S. Imaging and Histopathological Features of Jacob’s Disease: A Case Study. Head Neck Pathol. 2012, 6, 51–53. [Google Scholar] [CrossRef][Green Version]
- Kobayashi, K.; Shimoda, S.; Yoda, T.; Kakudo, K. Current status of MR imaging for the masticatory muscle tendon-aponeurosis hyperplasia. J. Jpn. Soc. Temporomandibular Jt. 2009, 21, 35–39. [Google Scholar]
- Hayashi, N.; Sato, T.; Fukushima, Y.; Takano, A.; Sakamoto, I.; Yoda, T. A two-year follow-up of surgical and non-surgical treatments in patients with masticatory muscle tendon-aponeurosis hyperplasia. Int. J. Oral Maxillofac. Surg. 2018, 47, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Hori, N.; Nakamoto, N.; Akita, M.; Yoda, T. Masticatory muscle tendon-aponeurosis hyperplasia exhibits heterotopic calcification in tendons. Oral Dis. 2014, 20, 404–408. [Google Scholar] [CrossRef] [PubMed]
- Nakamoto, A.; Sato, T.; Hirosawa, N.; Nakamoto, N.; Enoki, Y.; Chida, D.; Usui, M.; Takeda, S.; Nagai, T.; Sasaki, A.; et al. Proteomics-based identification of novel proteins in temporal tendons of patients with masticatory muscle tendon–aponeurosis hyperplasia. Int. J. Oral Maxillofac. Surg. 2014, 43, 113–119. [Google Scholar] [CrossRef]
- Hayashi, N.; Sato, T.; Kokabu, S.; Usui, M.; Yumoto, M.; Ikami, E.; Sakamoto, Y.; Nifuji, A.; Hayata, T.; Noda, M.; et al. Possible association of oestrogen and Cryba4 with masticatory muscle tendon-aponeurosis hyperplasia. Oral Dis. 2019, 25, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, N.; Sato, T.; Yumoto, M.; Kokabu, S.; Fukushima, Y.; Kawata, Y.; Kajihara, T.; Mizuno, Y.; Mizuno, Y.; Kawakami, T.; et al. Cyclic stretch induces decorin expression via yes-associated protein in tenocytes: A possible mechanism for hyperplasia in masticatory muscle tendon-aponeurosis hyperplasia. J. Oral Maxillofac. Surg. Med. Pathol. 2019, 31, 175–179. [Google Scholar] [CrossRef]
- Ito, K.; Go, Y.; Tatsumoto, S.; Usui, C.; Mizuno, Y.; Ikami, E.; Isozaki, Y.; Usui, M.; Kajihara, T.; Yoda, T.; et al. Gene expression profiling of the masticatory muscle tendons and Achilles tendons under tensile strain in the Japanese macaque Macaca fuscata. PLoS ONE 2023, 18, e0280649. [Google Scholar] [CrossRef] [PubMed]
- Yumoto, M.; Mizuno, Y.; Isozaki, Y.; Ito, K.O.; Yoda, T.; Sato, T. Analysis of Masticatory Muscle Tendon-aponeurosis Hyperplasia by Using Next-generation Sequencing. In Vivo 2022, 36, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Austin-Tse, C.A.; Jobanputra, V.; Perry, D.L.; Bick, D.; Taft, R.J.; Venner, E.; Gibbs, R.A.; Young, T.; Barnett, S.; Belmont, J.W.; et al. Best practices for the interpretation and reporting of clinical whole genome sequencing. NPJ Genom. Med. 2022, 7, 27. [Google Scholar] [CrossRef] [PubMed]
- Marshall, C.R.; Chowdhury, S.; Taft, R.J.; Lebo, M.S.; Buchan, J.G.; Harrison, S.M.; Rowsey, R.; Klee, E.W.; Liu, P.; Worthey, E.A.; et al. Best practices for the analytical validation of clinical whole-genome sequencing intended for the diagnosis of germline disease. NPJ Genom. Med. 2020, 5, 47. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows—Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ Data to high-confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.11–11.10.33. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Kishita, Y.; Shimura, M.; Kohda, M.; Akita, M.; Imai-Okazaki, A.; Yatsuka, Y.; Nakajima, Y.; Ito, T.; Ohtake, A.; Murayama, K.; et al. A novel homozygous variant in MICOS13/QIL1 causes hepato-encephalopathy with mitochondrial DNA depletion syndrome. Mol. Genet. Genom. Med. 2020, 8, e1427. [Google Scholar] [CrossRef] [PubMed]
- Sherry, S.T.; Ward, M.-H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef]
- Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; Abecasis, G.R.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alfoldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef]
- Tadaka, S.; Hishinuma, E.; Komaki, S.; Motoike, I.N.; Kawashima, J.; Saigusa, D.; Inoue, J.; Takayama, J.; Okamura, Y.; Aoki, Y.; et al. jMorp updates in 2020: Large enhancement of multi-omics data resources on the general Japanese population. Nucleic Acids Res. 2021, 49, D536–D544. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. feature Counts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Caruso, N.; Herberth, B.; Bartoli, M.; Puppo, F.; Dumonceaux, J.; Zimmermann, A.; Denadai, S.; Lebossé, M.; Roche, S.; Geng, L.; et al. Deregulation of the protocadherin gene FAT1 alters muscle shapes: Implications for the pathogenesis of facioscapulohumeral dystrophy. PLoS Genet. 2013, 9, e1003550. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.G.; Van Hateren, N.; Tickle, C.; Wilson, S.A. The expression of Fat-1 cadherin during chick limb development. Int. J. Dev. Biol. 2007, 51, 173–176. [Google Scholar] [CrossRef]
- Tissir, F.; Qu, Y.; Montcouquiol, M.; Zhou, L.; Komatsu, K.; Shi, D.; Fujimori, T.; Labeau, J.; Tyteca, D.; Courtoy, P.; et al. Lack of cadherins Celsr2 and Celsr3 impairs ependymal ciliogenesis, leading to fatal hydrocephalus. Nat. Neurosci. 2010, 13, 700–707. [Google Scholar] [CrossRef] [PubMed]
- Seeger-Nukpezah, T.; Golemis, E.A. The extracellular matrix and ciliary signaling. Curr. Opin. Cell Biol. 2012, 24, 652–661. [Google Scholar] [CrossRef] [PubMed]
- Sbardella, D.; Tundo, G.R.; Fasciglione, G.F.; Gioia, M.; Bisicchia, S.; Gasbarra, E.; Ippolito, E.; Tarantino, U.; Coletta, M.; Marini, S. Role of metalloproteinases in tendon pathophysiology. Mini-Rev. Med. Chem. 2014, 14, 978–987. [Google Scholar] [CrossRef]
- Shi, Y.; Hu, G.; Su, J.; Li, W.; Chen, Q.; Shou, P.; Xu, C.; Chen, X.; Huang, Y.; Zhu, Z.; et al. Mesenchymal stem cells: A new strategy for immunosuppression and tissue repair. Cell Res. 2010, 20, 510–518. [Google Scholar] [CrossRef]
- Sadeghi Shaker, M.; Rokni, M.; Mahmoudi, M.; Farhadi, E. Ras family signaling pathway in immunopathogenesis of inflammatory rheumatic diseases. Front. Immunol. 2023, 14, 1151246. [Google Scholar] [CrossRef]
- Ayres, J.A.; Shum, L.; Akarsu, A.; Dashner, R.; Takahashi, K.; Ikura, T.; Slavkin, H.C.; Nuckolls, G.H. DACH: Genomic Characterization, evaluation as a candidate for postaxial polydactyly type A2, and developmental expression pattern of the mouse homologue. Genomics 2001, 77, 18–26. [Google Scholar] [CrossRef]
- Li, X.; Ohgi, K.A.; Rosenfeld, M.G.; Zhang, J.; Krones, A.; Bush, K.T.; Glass, C.K.; Nigam, S.K.; Aggarwal, A.K.; Maas, R.L.; et al. Eya protein phosphatase activity regulates Six1–Dach–Eya transcriptional effects in mammalian organogenesis. Nature 2003, 426, 247–254. [Google Scholar] [CrossRef]
- Maire, P.; Dos Santos, M.; Madani, R.; Sakakibara, I.; Viaut, C.; Wurmser, M. Myogenesis control by SIX transcriptional complexes. Semin. Cell Dev. Biol. 2020, 104, 51–64. [Google Scholar] [CrossRef]
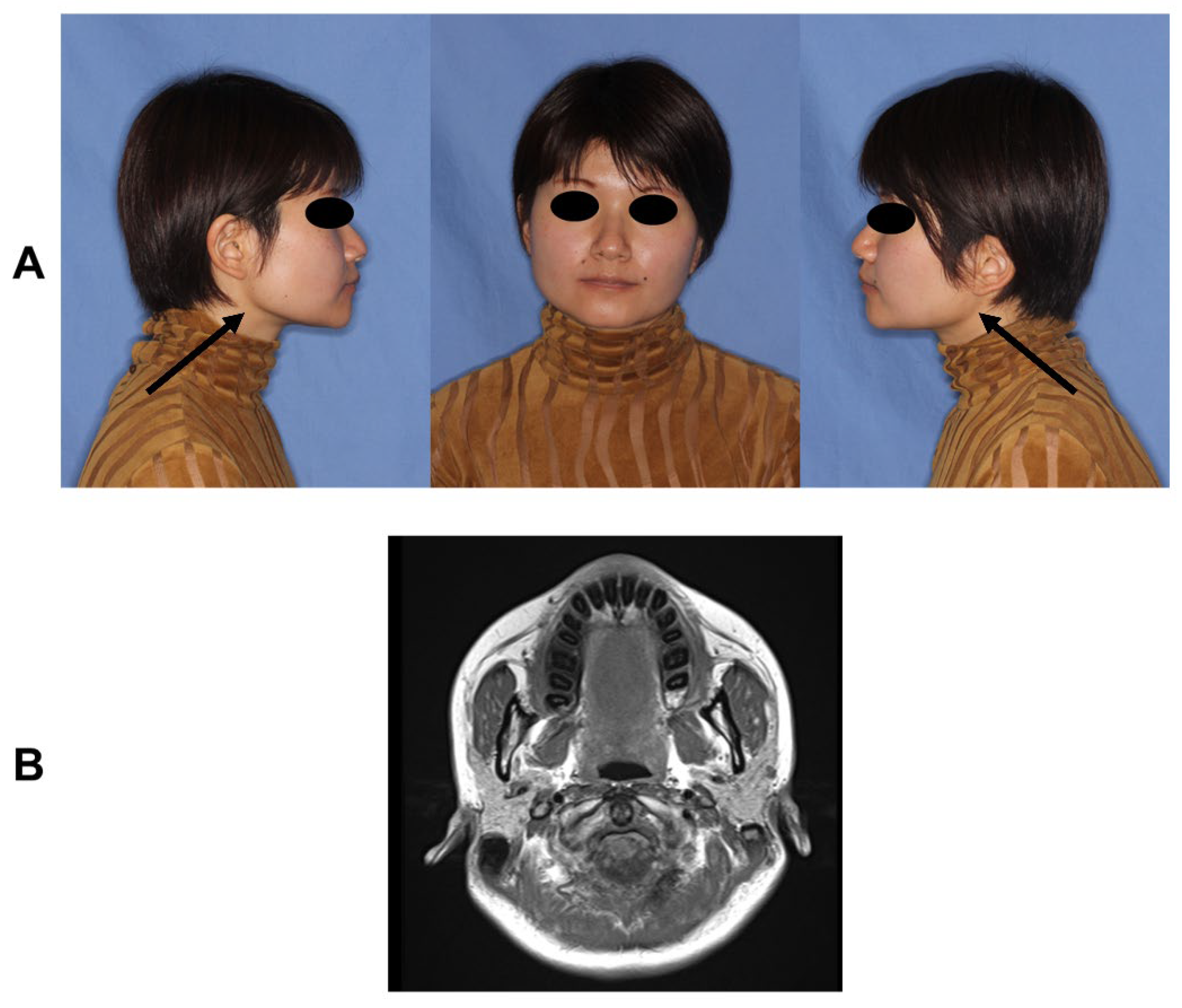
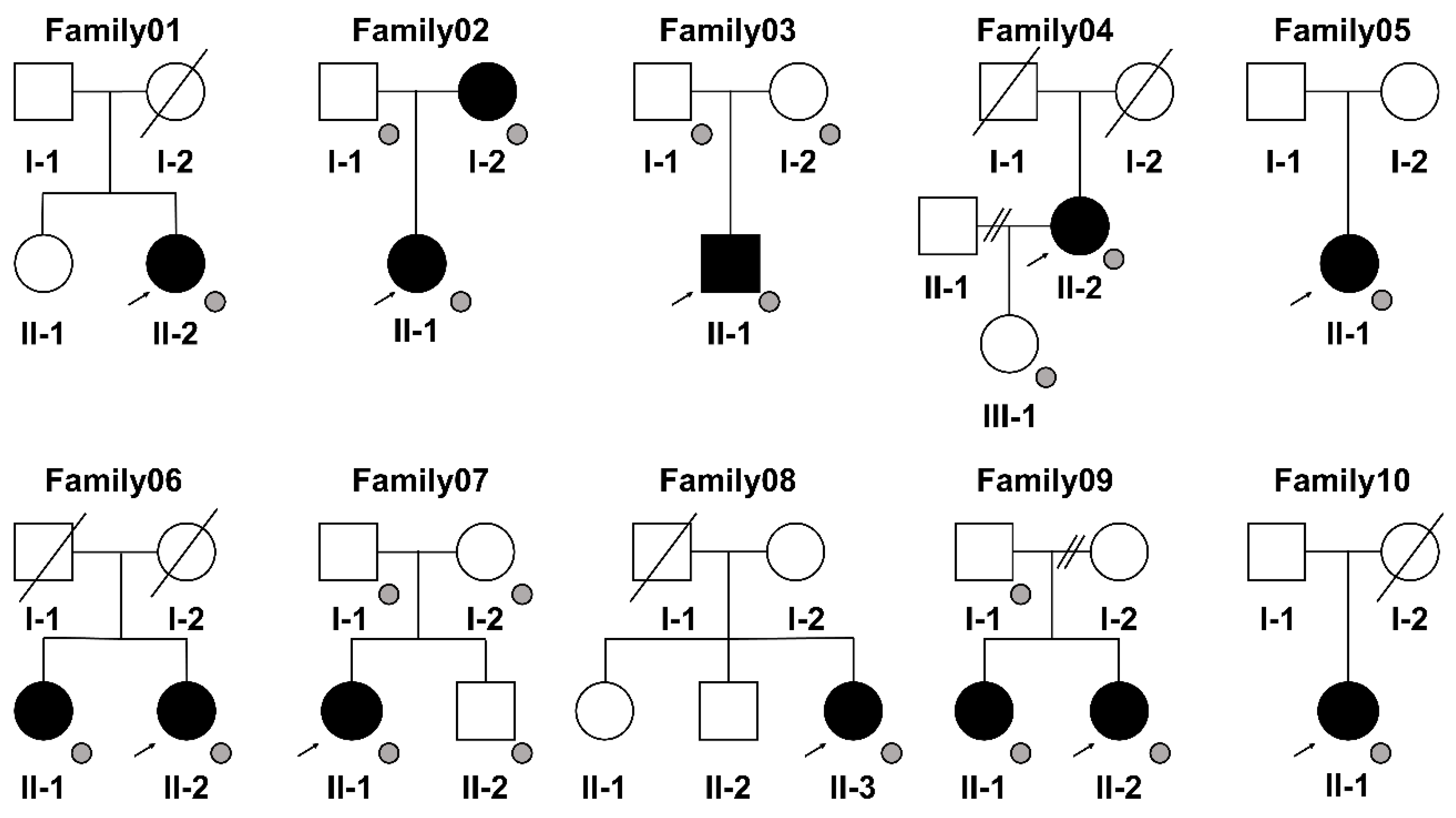
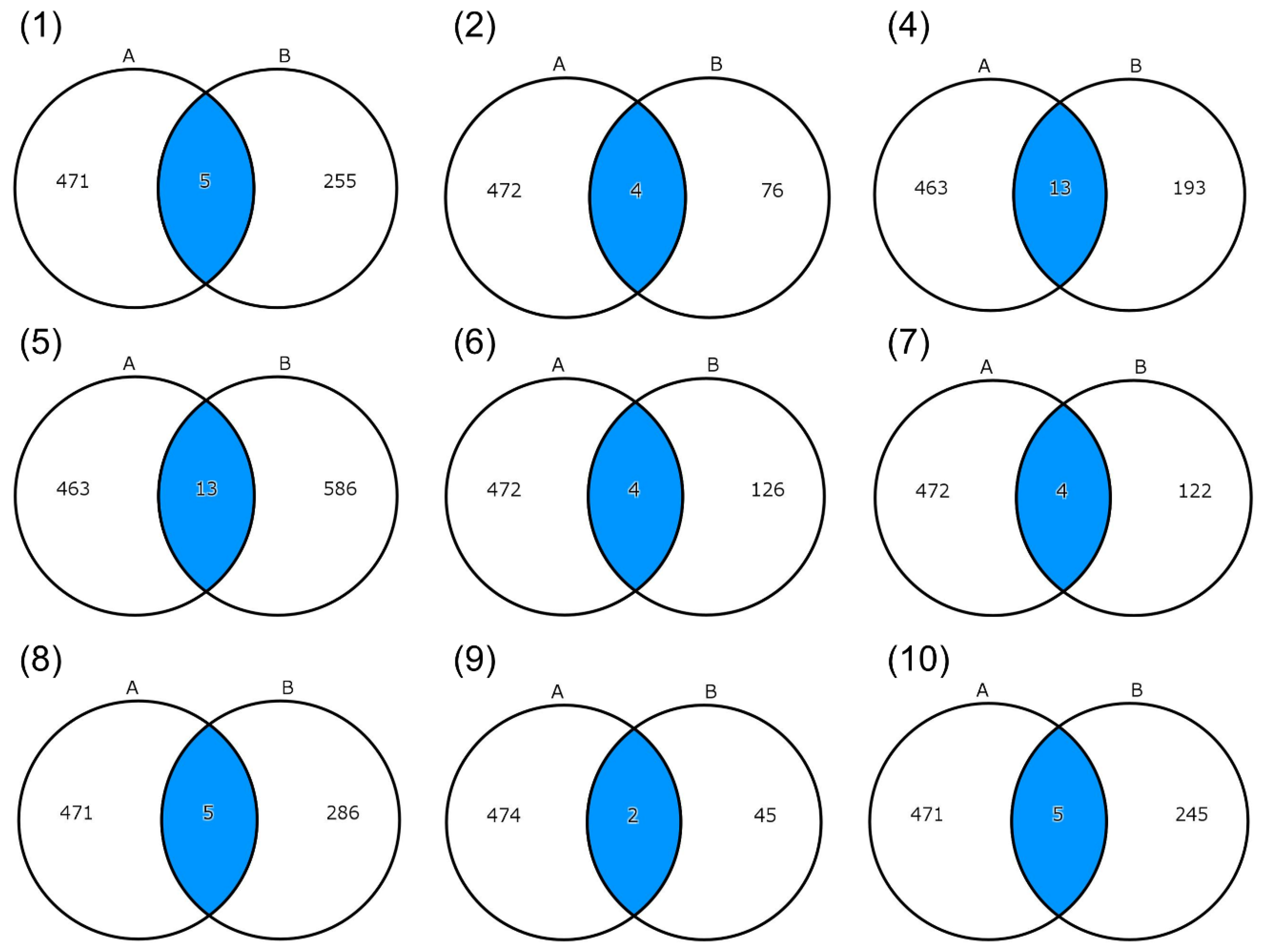
| Patient (Family) | Sex | Age at the Time of Sample Collection (Years) | Maximum Mouth Opening (mm) |
|---|---|---|---|
| Patient01 (II-2 of Family01) | Female | 48 | 28 |
| Patient02 (II-1 of Family02) | Female | 37 | 25 |
| Patient03 (II-1 of Family03) | Male | 43 | 22 |
| Patient04 (II-2 of Family04) | Female | 79 | 22 |
| Patient05 (II-1 of Family05) | Female | 39 | 22 |
| Patient06 (II-2 of Family06) | Female | 60 | 14 |
| Patient07 (II-1 of Family07) | Female | 44 | 23 |
| Patient08 (II-3 of Family08) | Female | 46 | 20 |
| Patient09 (II-2 of Family09) | Female | 47 | 25 |
| Patient10 (II-1 of Family10) | Female | 44 | 23 |
| ID/Gene | Family01 | Family02 | Family03 | Family04 | Family05 | Family06 | Family07 | Family08 | Family09 | Family10 |
|---|---|---|---|---|---|---|---|---|---|---|
| ALPL | DYSF | BCO2 | ARHGAP27 | CAT | DHTKD1 | FUT1 | SOX18 | BAIAP3 | ||
| DACH1 | ESAM | CELSR2 | CD300LG | CCDC68 | EDNRB | HIP1R | SURF2 | CASKIN1 | ||
| EFCAB5 | NCR1 | ENPEP | CTNNBIP1 | GIPR | NEURL1B | MCAT | CYP27A1 | |||
| MMP15 | RASGRP3 | EPN3 | DHTKD1 | NBEA | STOML3 | TDRD10 | LLGL2 | |||
| NECTIN2 | FHIP1B | DYSF | ADAP2 | SULT2B1 | ||||||
| NT5DC3 | ETFB | |||||||||
| PCDH1 | FBLIM1 | |||||||||
| RASIP1 | GPR4 | |||||||||
| TBC1D30 | HPDL | |||||||||
| TMEM140 | PCDH1 | |||||||||
| TMEM266 | SCARB1 | |||||||||
| TRAF3IP3 | ST6GALNAC2 | |||||||||
| BAIAP3 | WIPF3 |
| Gene Symbols | Descriptions | Family_1 | Family_2 |
|---|---|---|---|
| DACH1 | DACHSHUND FAMILY TRANSCRIPTION FACTOR 1; DACH1 | Family01 | |
| EFCAB5 | DIHYDROPYRIMIDINASE-LIKE 2; DPYSL2 | Family01 | |
| MMP15 | MATRIX METALLOPROTEINASE 15; MMP15 | Family01 | |
| NECTIN2 | NECTIN CELL ADHESION MOLECULE 2; NECTIN2 | Family01 | |
| NCR1 | NATURAL CYTOTOXICITY TRIGGERING RECEPTOR 1; NCR1 | Family02 * | |
| RASGRP3 | RAS GUANYL NUCLEOTIDE-RELEASING PROTEIN 3; RASGRP3 | Family02 * | |
| BCO2 | BETA-CAROTENE OXYGENASE 2; BCO2 | Family04 * | |
| CELSR2 | CADHERIN EGF LAG SEVEN-PASS G-TYPE RECEPTOR 2; CELSR2 | Family04 * | |
| ENPEP | GLUTAMYL AMINOPEPTIDASE; ENPEP | Family04 * | |
| EPN3 | EPSIN 3; EPN3 | Family04 * | |
| FHIP1B | FHF COMPLEX SUBUNIT HOOK-INTERACTING PROTEIN 1B; FHIP1B | Family04 * | |
| NT5DC3 | 5-PRIME-NUCLEOTIDASE DOMAIN-CONTAINING PROTEIN 3; NT5DC3 | Family04 * | |
| PCDH1 | PROTOCADHERIN 1; PCDH1 | Family04 * | Family05 |
| RASIP1 | RAS-INTERACTING PROTEIN 1; RASIP1 | Family04 * | |
| TBC1D30 | TBC1 DOMAIN FAMILY, MEMBER 30; TBC1D30 | Family04 * | |
| TMEM140 | TRANSMEMBRANE PROTEIN 140; TMEM140 | Family04 * | |
| TMEM266 | TRANSMEMBRANE PROTEIN 266; TMEM266 | Family04 * | |
| TRAF3IP3 | TRAF3-INTERACTING PROTEIN 3; TRAF3IP3 | Family04 * | |
| ARHGAP27 | RHO GTPase-ACTIVATING PROTEIN 27; ARHGAP27 | Family05 | |
| CD300LG | CD300 ANTIGEN-LIKE FAMILY, MEMBER G; CD300LG | Family05 | |
| CTNNBIP1 | CATENIN, BETA-INTERACTING PROTEIN 1; CTNNBIP1 | Family05 | |
| FBLIM1 | FILAMIN-BINDING LIM PROTEIN 1; FBLIM1 | Family05 | |
| GPR4 | G PROTEIN-COUPLED RECEPTOR 4; GPR4 | Family05 | |
| ST6GALNAC2 | ST6 ALPHA-N-ACETYL-NEURAMINYL-2,3-BETA-GALACTOSYL-1,3-N-ACETYLGALACTOSAMINIDE ALPHA-2,6-SIALYLTRANSFERASE 2; ST6GALNAC2 | Family05 | |
| WIPF3 | WAS/WASL-INTERACTING PROTEIN FAMILY, MEMBER 3; WIPF3 | Family05 | |
| CCDC68 | COILED-COIL DOMAIN-CONTAINING PROTEIN 68; CCDC68 | Family06 * | |
| GIPR | GASTRIC INHIBITORY POLYPEPTIDE RECEPTOR; GIPR | Family06 * | |
| NEURL1B | NEURALIZED E3 UBIQUITIN PROTEIN LIGASE 1B; NEURL1B | Family07 * | |
| STOML3 | STOMATIN-LIKE PROTEIN 3; STOML3 | Family07 * | |
| HIP1R | HUNTINGTIN-INTERACTING PROTEIN 1-RELATED PROTEIN; HIP1R | Family08 | |
| MCAT | MALONYL CoA:ACP ACYLTRANSFERASE, MITOCHONDRIAL; MCAT | Family08 | |
| TDRD10 | TUDOR DOMAIN CONTAINING 10 | Family08 | |
| SURF2 | SURFEIT 2; SURF2 | Family09 * | |
| BAIAP3 | BAI1-ASSOCIATED PROTEIN 3; BAIAP3 | Family04 * | Family10 |
| CASKIN1 | CASK-INTERACTING PROTEIN 1; CASKIN1 | Family10 | |
| LLGL2 | LLGL SCRIBBLE CELL POLARITY COMPLEX COMPONENT 2; LLGL2 | Family10 | |
| ADAP2 | ARFGAP WITH DUAL PLECKSTRIN HOMOLOGY DOMAINS 2; ADAP2 | Family8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tajima, R.; Okazaki, A.; Sato, T.; Ozaki, K.; Motooka, D.; Okazaki, Y.; Yoda, T. Genetic Landscape of Masticatory Muscle Tendon–Aponeurosis Hyperplasia. Genes 2023, 14, 1718. https://doi.org/10.3390/genes14091718
Tajima R, Okazaki A, Sato T, Ozaki K, Motooka D, Okazaki Y, Yoda T. Genetic Landscape of Masticatory Muscle Tendon–Aponeurosis Hyperplasia. Genes. 2023; 14(9):1718. https://doi.org/10.3390/genes14091718
Chicago/Turabian StyleTajima, Rina, Atsuko Okazaki, Tsuyoshi Sato, Kokoro Ozaki, Daisuke Motooka, Yasushi Okazaki, and Tetsuya Yoda. 2023. "Genetic Landscape of Masticatory Muscle Tendon–Aponeurosis Hyperplasia" Genes 14, no. 9: 1718. https://doi.org/10.3390/genes14091718
APA StyleTajima, R., Okazaki, A., Sato, T., Ozaki, K., Motooka, D., Okazaki, Y., & Yoda, T. (2023). Genetic Landscape of Masticatory Muscle Tendon–Aponeurosis Hyperplasia. Genes, 14(9), 1718. https://doi.org/10.3390/genes14091718






