Genome-Wide RADseq Reveals Genetic Differentiation of Wild and Cultured Populations of Large Yellow Croaker
Abstract
1. Introduction
2. Materials and Methods
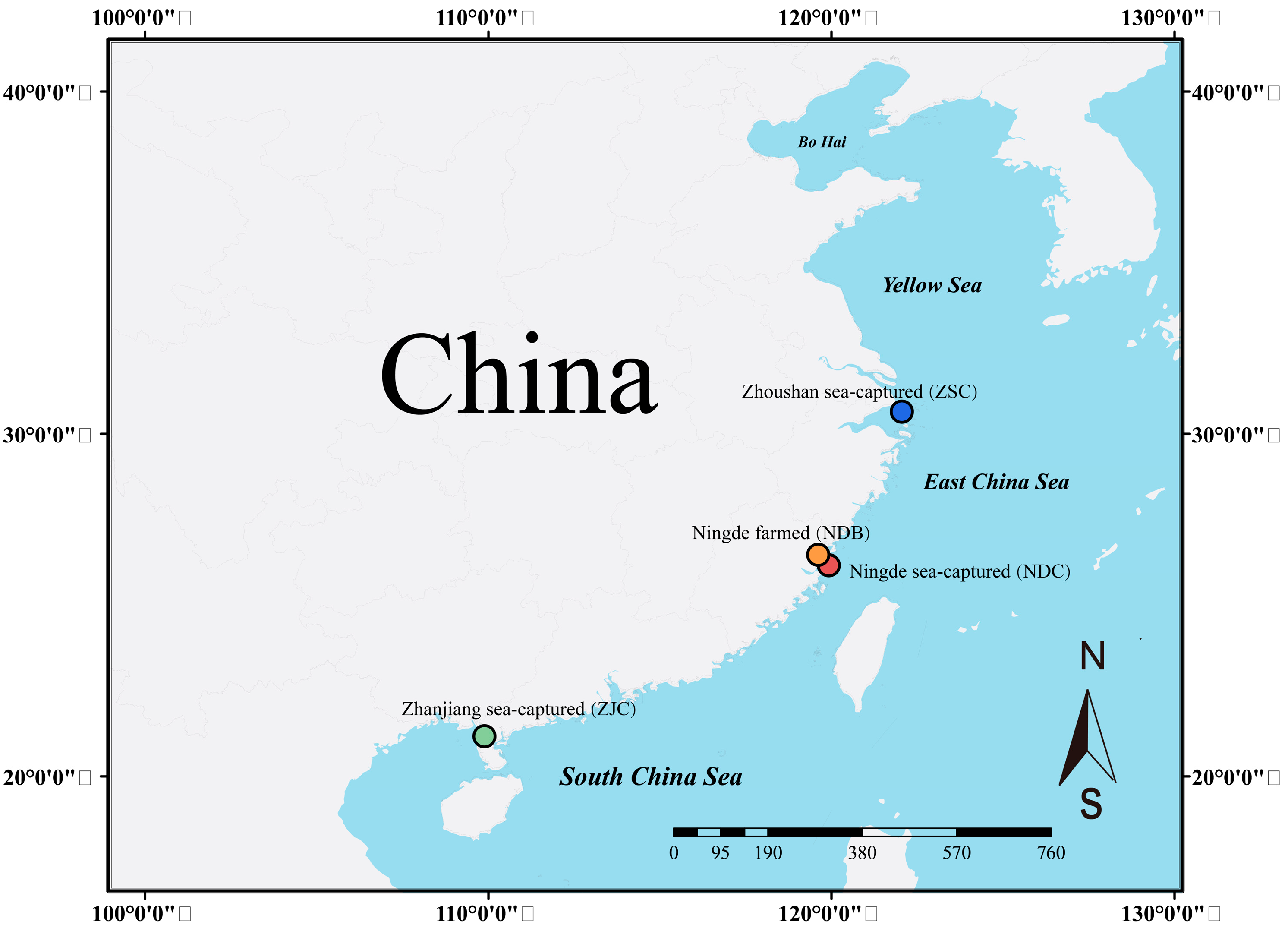
2.1. Sample
2.2. Sequencing
2.3. Variants Calling and Annotation
2.4. Population Diversity and Structure
2.5. Linkage Disequilibrium Decay
2.6. Analysis of Effective Population Sizes
2.7. Signatures of Selection
3. Results
3.1. Sequencing and SNP Discovery
3.2. Genetic Diversity
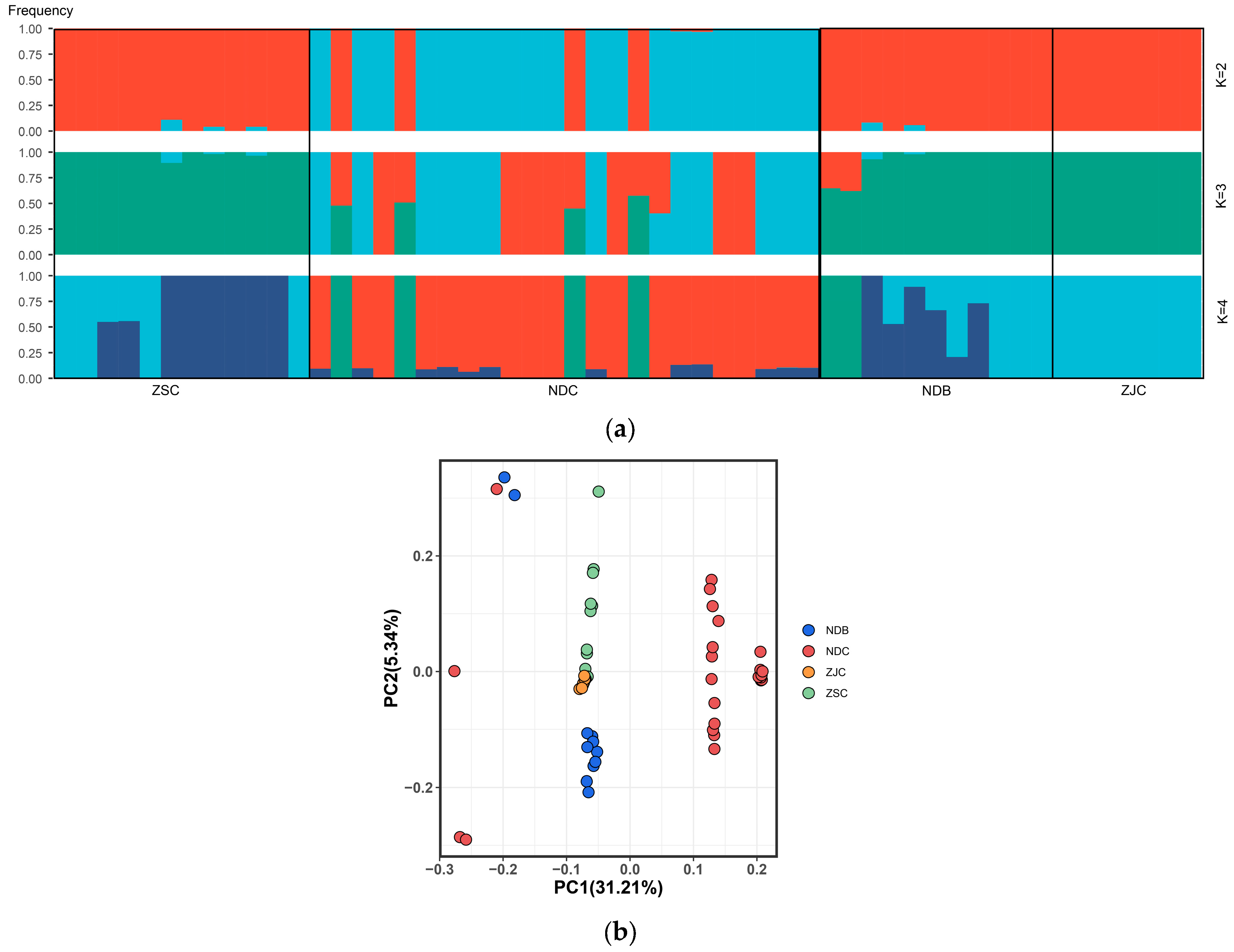
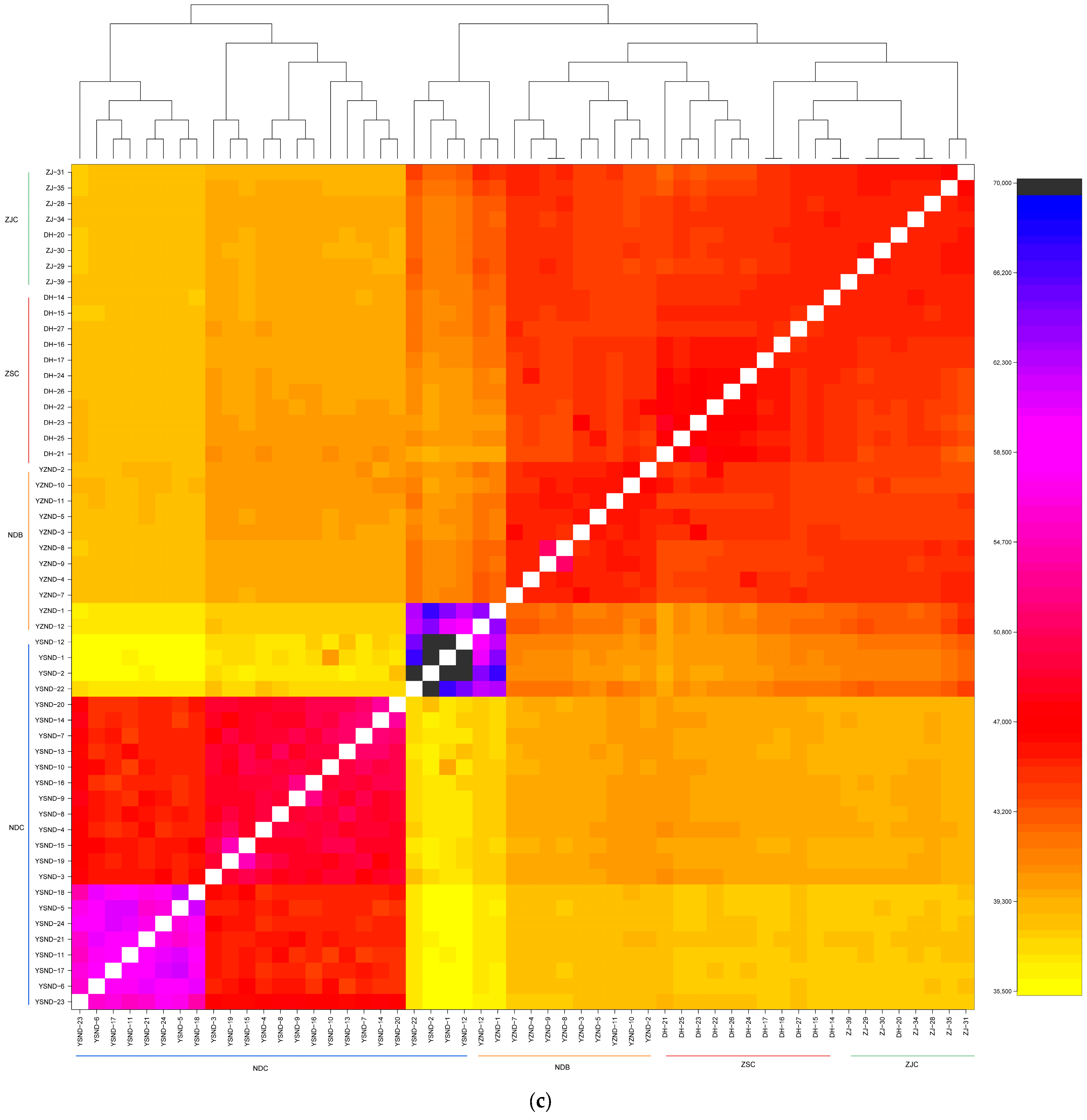
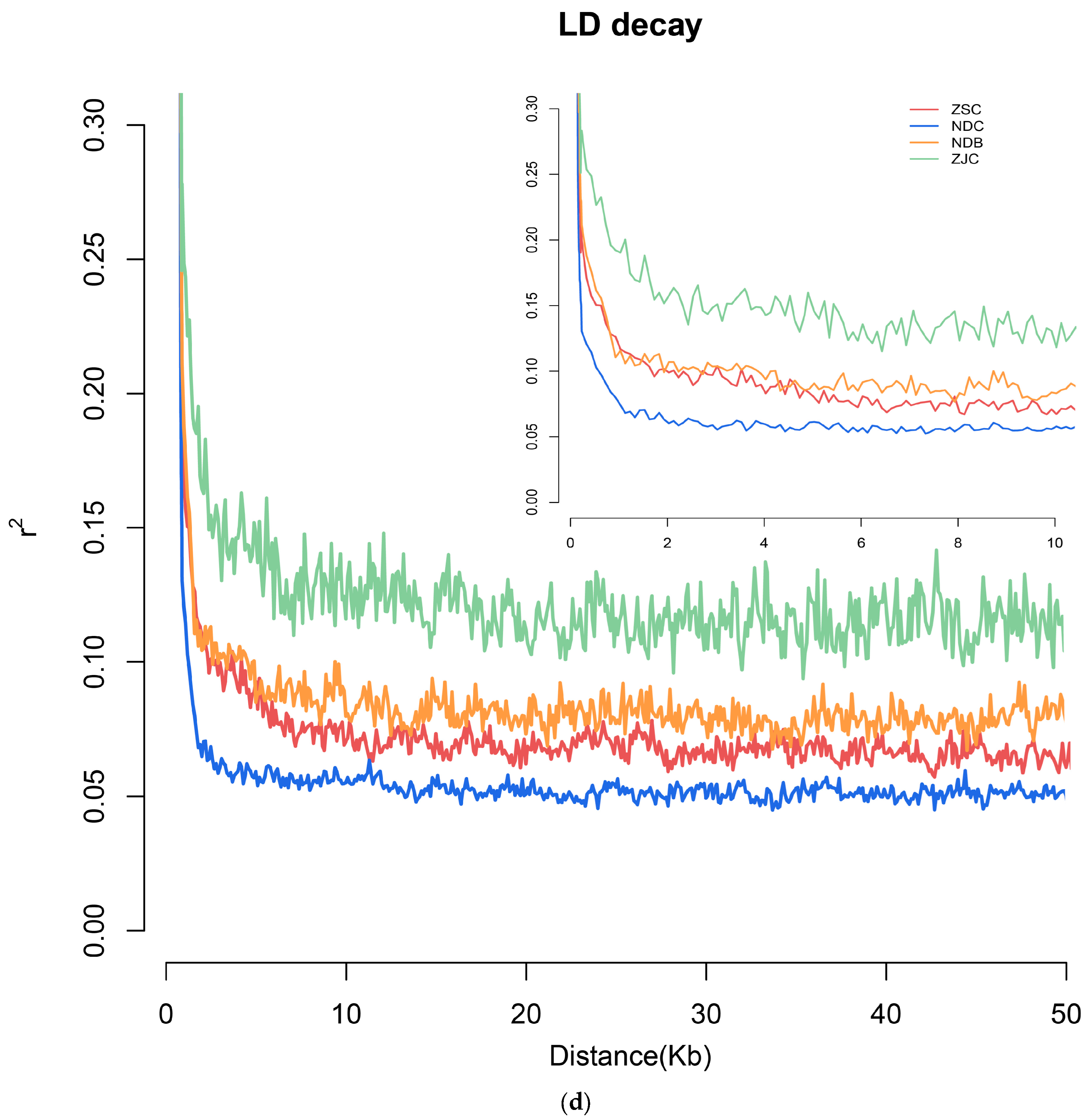
3.3. Population Structure and LD Decay
3.4. Genome-Wide Selection Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Wu, C.; Zhang, D.; Kan, M.; Lv, Z.; Zhu, A.; Su, Y.; Zhou, D.; Zhang, J.; Zhang, Z.; Xu, M.; et al. The draft genome of the large yellow croaker reveals well-developed innate immunity. Nat. Commun. 2014, 5, 5227. [Google Scholar] [CrossRef] [PubMed]
- Ao, J.; Mu, Y.; Xiang, L.X.; Fan, D.; Feng, M.; Zhang, S.; Shi, Q.; Zhu, L.Y.; Li, T.; Ding, Y.; et al. Genome sequencing of the perciform fish Larimichthys crocea provides insights into molecular and genetic mechanisms of stress adaptation. PLoS Genet. 2015, 11, e1005118. [Google Scholar] [CrossRef]
- Quan, C.G.; Wang, J.; Ding, S.X.; Su, Y.Q. Genetic diversity of cultured Pseudosciaena crocea (Richardson) stock by PAGE. J. Xiamen Univ. Nat. Sci. 1999, 38, 584–588. (In Chinese) [Google Scholar] [CrossRef]
- Li, Z.B.; Fang, X.; Chen, J.; Chang, J.B.; Lei, G.G.; Zhang, G.L.; Zhao, B.L.; Wang, Z.L. Loss of the genetic diversity in cultivated populations of Pseudosciaena crocea by AFLP. Oceanol. Limnol. Sin. 2009, 40, 446–450. [Google Scholar] [CrossRef]
- Wang, Y. Germplasm Identification of Wild and Cultured Stock of Large Yellow Croaker (Larimichthys crocea). Master’s Thesis, Shanghai Ocean University, Shanghai, China, 2017. (In Chinese). [Google Scholar]
- Sun, L.Y.; Yang, T.Y.; Meng, W.; Yang, B.Q.; Zhang, T. Analysis of the mitochondrial genome characteristics and phylogenetic relationships of eight sciaenid fishes. Mar. Sci. 2017, 7, 73–77. (In Chinese) [Google Scholar] [CrossRef]
- Baird, N.A.; Etter, P.D.; Atwood, T.S.; Currey, M.C.; Shiver, A.L.; Lewis, Z.A.; Selker, E.U.; Cresko, W.A.; Johnson, E.A. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 2008, 3, e3376. [Google Scholar] [CrossRef]
- Pan, Y.Z.; Wang, X.Q.; Sun, G.L.; Li, F.S.; Gong, X. Application of RAD sequencing for evaluating the genetic diversity of domesticated Panax notoginseng (Araliaceae). PLoS ONE 2016, 11, e0166419. [Google Scholar] [CrossRef] [PubMed]
- Valdisser, P.A.M.; Pappas, G.J.; de Menezes, I.P.; Müller, B.S.; Pereira, W.J.; Narciso, M.G.; Brondani, C.; Souza, T.L.; Borba, T.C.; Vianello, R.P. SNP discovery in common bean by restriction-associated DNA (RAD) sequencing for genetic diversity and population structure analysis. Mol. Genet. Genom. 2016, 291, 1277–1291. [Google Scholar] [CrossRef]
- Qin, M.; Li, C.H.; Li, Z.X.; Chen, W.; Zeng, Y.Q. Genetic Diversities and Differentially Selected Regions Between Shandong Indigenous Pig Breeds and Western Pig Breeds. Front. Genet. 2020, 10, 1351. [Google Scholar] [CrossRef]
- Perrier, C.; Ferchaud, A.-L.; Sirois, P.; Thibault, I.; Bernatchez, L. Do genetic drift and accumulation of deleterious mutations preclude adaptation? Empirical investigation using RADseq in a northern lacustrine fish. Mol. Ecol. 2017, 26, 6317–6335. [Google Scholar] [CrossRef]
- Sherman, K.D.; Paris, J.R.; King, R.A.; Moore, K.A.; Dahlgren, C.P.; Knowles, L.C.; Stump, K.; Tyler, C.R.; Stevens, J.R. RAD-Seq Analysis and in situ Monitoring of Nassau Grouper Reveal Fine-Scale Population Structure and Origins of Aggregating Fish. Front. Mar. Sci. 2020, 7, 157. [Google Scholar] [CrossRef]
- DiBattista, J.D.; Saenz-Agudelo, P.; Piatek, M.J.; Wang, X.; Aranda, M.; Berumen, M.L. Using a butterflyfish genome as a general tool for RAD-Seq studies in specialized reef fish. Mol. Ecol. Resour. 2017, 17, 1330–1341. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.-Y.; Euclide, P.T.; Ludsin, S.A.; Larson, W.A.; Sovic, M.G.; Gibbs, H.L.; Marschall, E.A. RAD-Seq Refines Previous Estimates of Genetic Structure in Lake Erie Walleye. Trans. Am. Fish. Soc. 2020, 149, 159–173. [Google Scholar] [CrossRef]
- Xiao, S.; Wang, P.; Zhang, Y.; Fang, L.; Liu, Y.; Li, J.T.; Wang, Z.Y. Gene map of large yellow croaker (Larimichthys crocea) provides insights into teleost genome evolution and conserved regions associated with growth. Sci. Rep. 2015, 5, 18661. [Google Scholar] [CrossRef]
- Zhou, Z.; Han, K.; Wu, Y.; Bai, H.; Xu, P. Genome-Wide Association Study of Growth and Body-Shape-Related Traits in Large Yellow Croaker (Larimichthys crocea) Using ddRAD Sequencing. Mar. Biotechnol. 2019, 21, 655–670. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [PubMed]
- Malinsky, M.; Trucchi, E.; Lawson, D.J.; Falush, D. RADpainter and fineRADstructure: Population Inference from RADseq Data. Mol. Biol. Evol. 2018, 35, 1284–1290. [Google Scholar] [CrossRef] [PubMed]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.S.; Xu, J.Y.; He, W.M.; Yang, T.L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef]
- Barbato, M.; Orozco-terWengel, P.; Tapio, M.; Bruford, M.W. SNeP: A tool to estimate trends in recent effective population size trajectories using genome-wide SNP data. Front. Genet. 2015, 6, 109. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Liu, Z.; Pan, Q.; Chen, X.; Wang, H.; Guo, H.; Liu, S.; Lu, H.; Tian, S.; Li, R.; et al. Genomic Analyses Reveal Demographic History and Temperate Adaptation of the Newly Discovered Honey Bee Subspecies Apis mellifera sinisxinyuan n. ssp. Mol. Biol. Evol. 2016, 33, 1337–1348. [Google Scholar] [CrossRef]
- Zhou, Y.Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Su, J.L.; Yuan, Y.L. Hydrology in China’s Offshore Waters; Ocean Press: Beijing, China, 2005; pp. 20–21. [Google Scholar]
- Zhang, Q.Y.; Hong, W.S. Resource status and remediation strategy for large yellow croaker in Guanjingyang Bay. Mar. Fish. 2015, 27, 179–186. [Google Scholar] [CrossRef]
- Wang, D.; Wu, F.X. China Fishery Statistical Yearbook; China Agriculture Press: Beijing, China, 2022. [Google Scholar]
- Wang, J.; Su, Y.Q.; Quan, C.G.; Zhang, D.W. Genetic diversity of the wild and reared Pseudosciaena crocea. Chin. J. Oceanol. Limnol. 2001, 19, 152–156. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, X.W.; Wu, X.F.; Xue, L.Y.; Xiao, S.J.; Wang, Z.Y. Analysis of genetic diversity in cultured populations of daiqu and min—Yue groups of Larimichthys crocea. J. Southwest Univ. Nat. Sci. Ed. 2015, 37, 6–12. (In Chinese). Available online: https://www.cnki.net/kcms/doi/10.13718/j.cnki.xdzk.2015.08.002.html (accessed on 17 February 2023).
- Chen, J.; Li, Z.B.; Fang, X.; Lei, G.; Zhang, G.L.; Zhao, B.L.; Wang, Z.L. The genetic structure of wild and cultivated populations of Pseudosciaena crocea. Mar. Sci. 2010, 34, 45–48. [Google Scholar] [CrossRef]
- Wang, L.; Shi, X.; Su, Y.; Meng, Z.; Lin, H. Loss of genetic diversity in the cultured stocks of the large yellow croaker, Larimichthys crocea, revealed by microsatellites. Int. J. Mol. Sci. 2012, 13, 5584–5597. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Wang, Y.L.; Lin, L.M.; Khoo, S.K.; Okamoto, N. Genetic polymorphisms in wild and cultured large yellow croaker Pseudosciaena crocea using AFLP fingerprinting. J. Fish. Sci. China 2002, 9, 198–202. (In Chinese) [Google Scholar] [CrossRef]
- Blanton, M.L.; Specker, J.L. The hypothalamic-pituitary-thyroid (HPT) axis in fish and its role in fish development and reproduction. Crit. Rev. Toxicol. 2007, 37, 97–115. [Google Scholar] [CrossRef] [PubMed]
- Jantzen, C.E.; Toor, F.; Annunziato, K.A.; Cooper, K.R. Effects of chronic perfluorooctanoic acid (PFOA) at low concentration on morphometrics, gene expression, and fecundity in zebrafish (Danio rerio). Reprod. Toxicol. 2017, 69, 34–42. [Google Scholar] [CrossRef]
- He, X.Y.; Fang, X.; Luo, B.Y.; Qiu, G.F. Identification and characterization of a new germline-specific marker vasa gene and its promoter in the giant freshwater prawn Macrobrachium rosenbergii. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2022, 259, 110716. [Google Scholar] [CrossRef]
- Sun, X.Y.; Tao, B.B.; Wang, Y.X.; Hu, W.; Sun, Y.H. Isolation and characterization of germline stem cells in protogynous hermaphroditic Monopterus albus. Int. J. Mol. Sci. 2022, 23, 5861. [Google Scholar] [CrossRef]
- Hartung, O.; Forbes, M.M.; Marlow, F.L. Zebrafish vasa is required for germ-cell differentiation and maintenance. Mol. Reprod. Dev. 2014, 81, 946–961. [Google Scholar] [CrossRef] [PubMed]
- Ye, M.L.; Chen, Y. Zebrafish as an emerging model to study gonad development. Comput. Struct. Biotechnol. J. 2020, 18, 2373–2380. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.M.; Wang, Z.K.; Jiang, J.J.; Gao, J.N.; Wang, J.; Zhou, X.S.; Zhang, Q.Q. Cloning, expression promoter analysis of vasa gene in Japanese flounder (Paralichthys olivaceus). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2014, 167, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Raghuveer, K.; Senthilkumaran, B. Cloning and differential expression pattern of vasa in the developing and recrudescing gonads of catfish, Clarias gariepinus. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2010, 157, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Bielczyk Maczyńska, E.; Serbanovic Canic, J.; Ferreira, L.; Soranzo, N.; Stemple, D.L.; Ouwehand, W.H.; Cvejic, A. A loss of function screen of identified genome-wide association study Loci reveals new genes controlling hematopoiesis. PLoS Genet. 2014, 10, e1004450. [Google Scholar] [CrossRef] [PubMed]
- Dockray, G.J. The expanding family of-RFamide peptides and their effects on feeding behaviour. Exp. Physiol. 2004, 89, 229–235. [Google Scholar] [CrossRef]
- Bechtold, D.A.; Luckman, S.M. The role of RFamide peptides in feeding. J. Endocrinol. 2007, 192, 3–15. [Google Scholar] [CrossRef]
- Tocher, D.R.; Zheng, X.; Schlechtriem, C.; Hastings, N.; Dick, J.R.; Teale, A.J. Highly unsaturated fatty acid synthesis in marine fish: Cloning, functional characterization, and nutritional regulation of fatty acyl Δ6 desaturase of Atlantic cod (Gadus morhua L.). Lipids 2006, 41, 1003–1016. [Google Scholar] [CrossRef]
- Nara, T.Y.; He, W.S.; Tang, C.; Clarke, S.D.; Nakamura, M.T. The E-box like sterol regulatory element mediates the suppression of human Δ-6 desaturase gene by highly unsaturated fatty acids. Biochem. Biophys. Res. Commun. 2002, 296, 111–117. [Google Scholar] [CrossRef]
- Guo, Q.Y.; Xing, X.L.; Jiang, C.J.; Yang, X. Quality characteristics and differences of wild and cultured large yellow croakers (Larimichthys crocea). Mod. Food Sci. Technol. 2019, 35, 92–101. [Google Scholar] [CrossRef]
- Gu, Z.; Mu, H.; Shen, H.; Deng, K.; Liu, D.; Yang, M.; Zhang, Y.; Zhang, W.; Mai, K. High level of dietary soybean oil affects the glucose and lipid metabolism in large yellow croaker Larimichthys crocea through the insulin-mediated PI3K/AKT signaling pathway. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2019, 231, 34–41. [Google Scholar] [CrossRef] [PubMed]
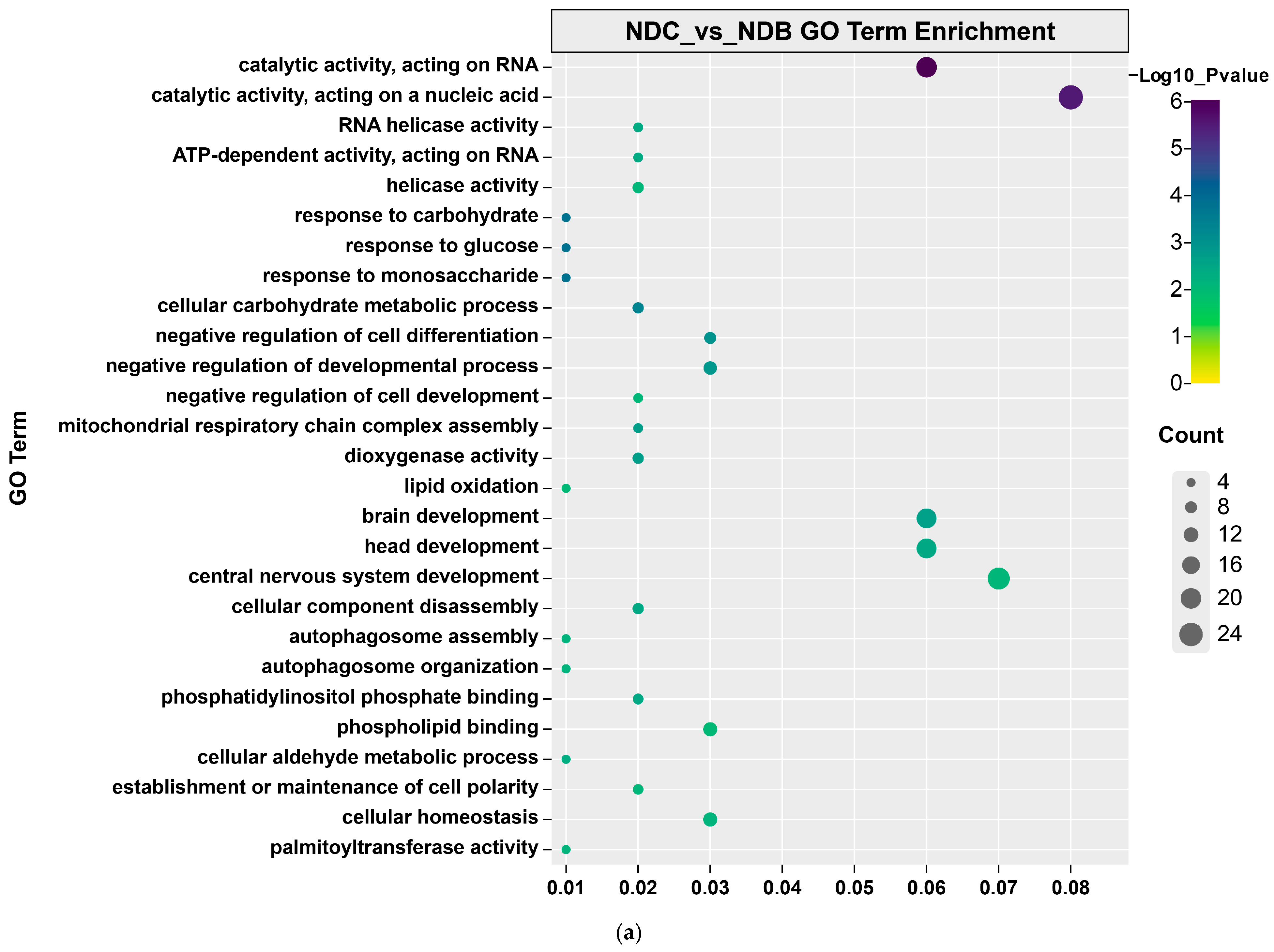
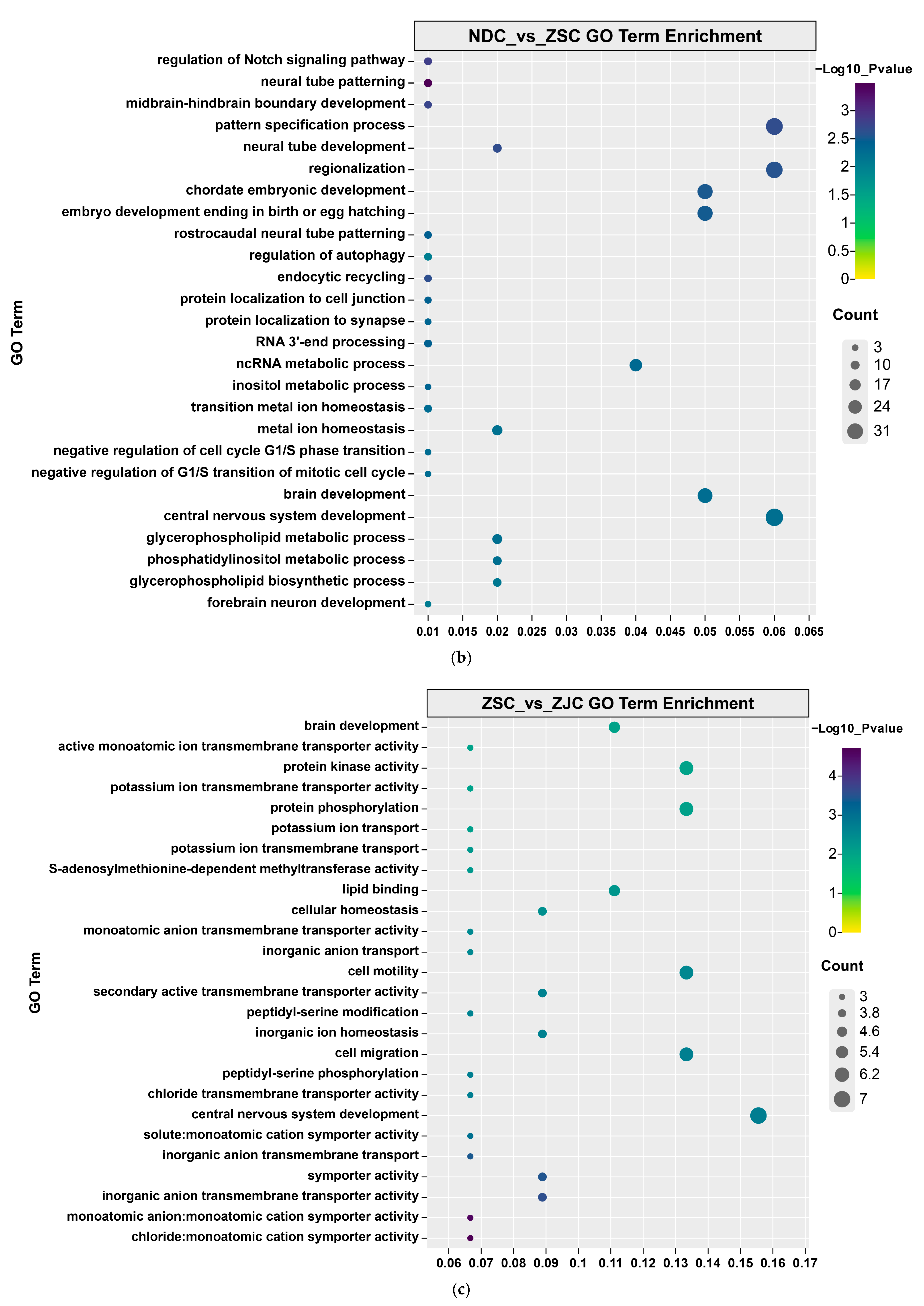
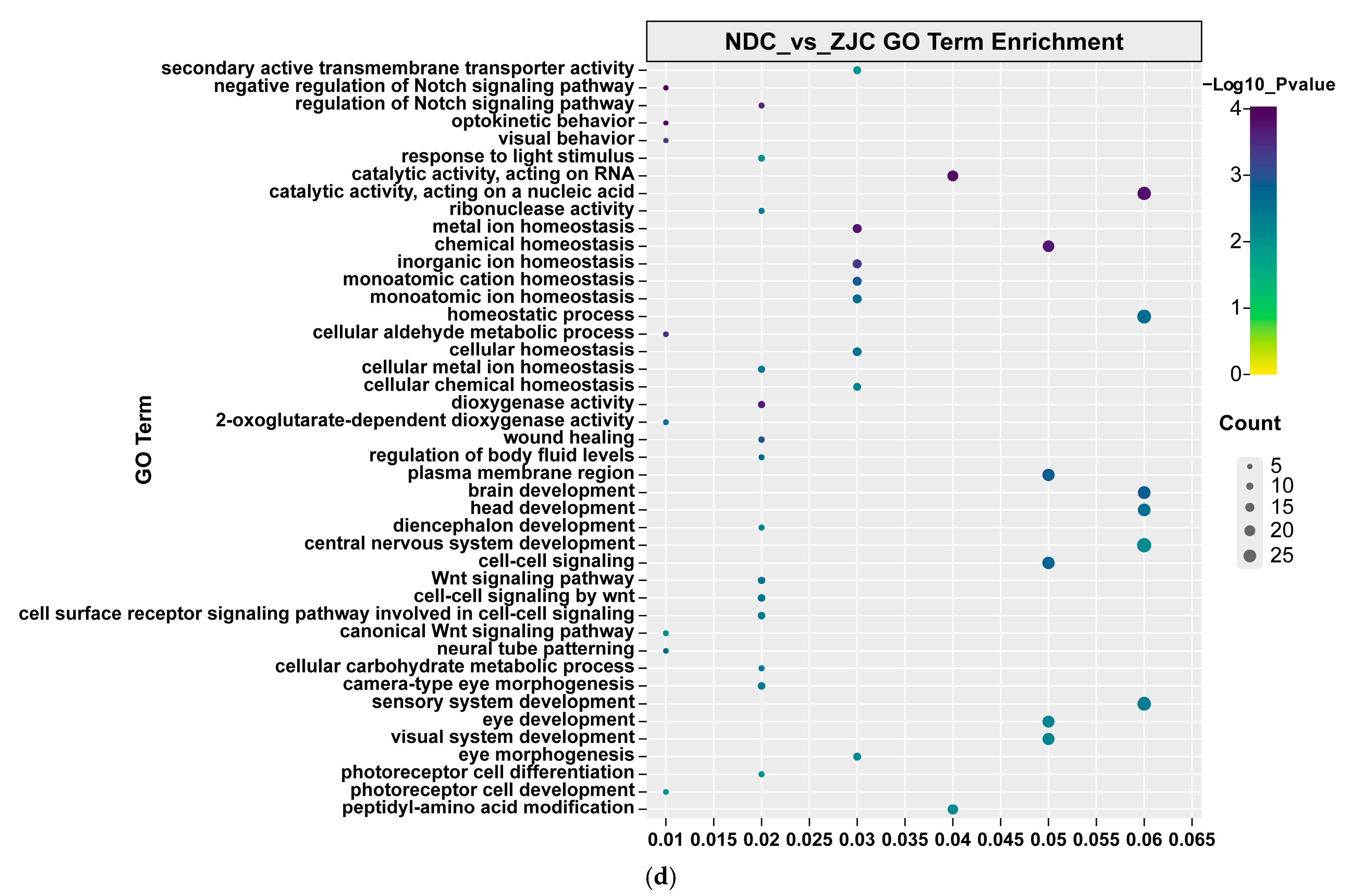
| Population ID | Observed Heterozygosity | Expected Heterozygosity | Nucleotide Diversity (π) | Inbreeding Coefficients FIS |
|---|---|---|---|---|
| ZSC | 0.097 | 0.142 | 0.155 | 0.179 |
| NDC | 0.172 | 0.222 | 0.199 | 0.106 |
| NDB | 0.093 | 0.122 | 0.129 | 0.107 |
| ZJC | 0.081 | 0.123 | 0.136 | 0.133 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, K.; Zhou, Y.; Song, W.; Jiang, L.; Yan, X. Genome-Wide RADseq Reveals Genetic Differentiation of Wild and Cultured Populations of Large Yellow Croaker. Genes 2023, 14, 1508. https://doi.org/10.3390/genes14071508
Zhang K, Zhou Y, Song W, Jiang L, Yan X. Genome-Wide RADseq Reveals Genetic Differentiation of Wild and Cultured Populations of Large Yellow Croaker. Genes. 2023; 14(7):1508. https://doi.org/10.3390/genes14071508
Chicago/Turabian StyleZhang, Kaifen, Yongdong Zhou, Weihua Song, Lihua Jiang, and Xiaojun Yan. 2023. "Genome-Wide RADseq Reveals Genetic Differentiation of Wild and Cultured Populations of Large Yellow Croaker" Genes 14, no. 7: 1508. https://doi.org/10.3390/genes14071508
APA StyleZhang, K., Zhou, Y., Song, W., Jiang, L., & Yan, X. (2023). Genome-Wide RADseq Reveals Genetic Differentiation of Wild and Cultured Populations of Large Yellow Croaker. Genes, 14(7), 1508. https://doi.org/10.3390/genes14071508






