Cloning, Identification, and Functional Analysis of the Chalcone Isomerase Gene from Astragalus sinicus
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Extraction of Total RNA and Genomic DNA
2.3. Cloning of CHI and Bioinformatics Analyses
2.4. Quantitative Real-Time PCR (qRT-PCR) and Determination of Anthocyanins Content
2.5. Transformation and Phenotypic Analysis of Transgenic Arabidopsis
2.6. Statistical Analysis
3. Results
3.1. Full-Length Cloning of AsiCHI and Sequence Analysis
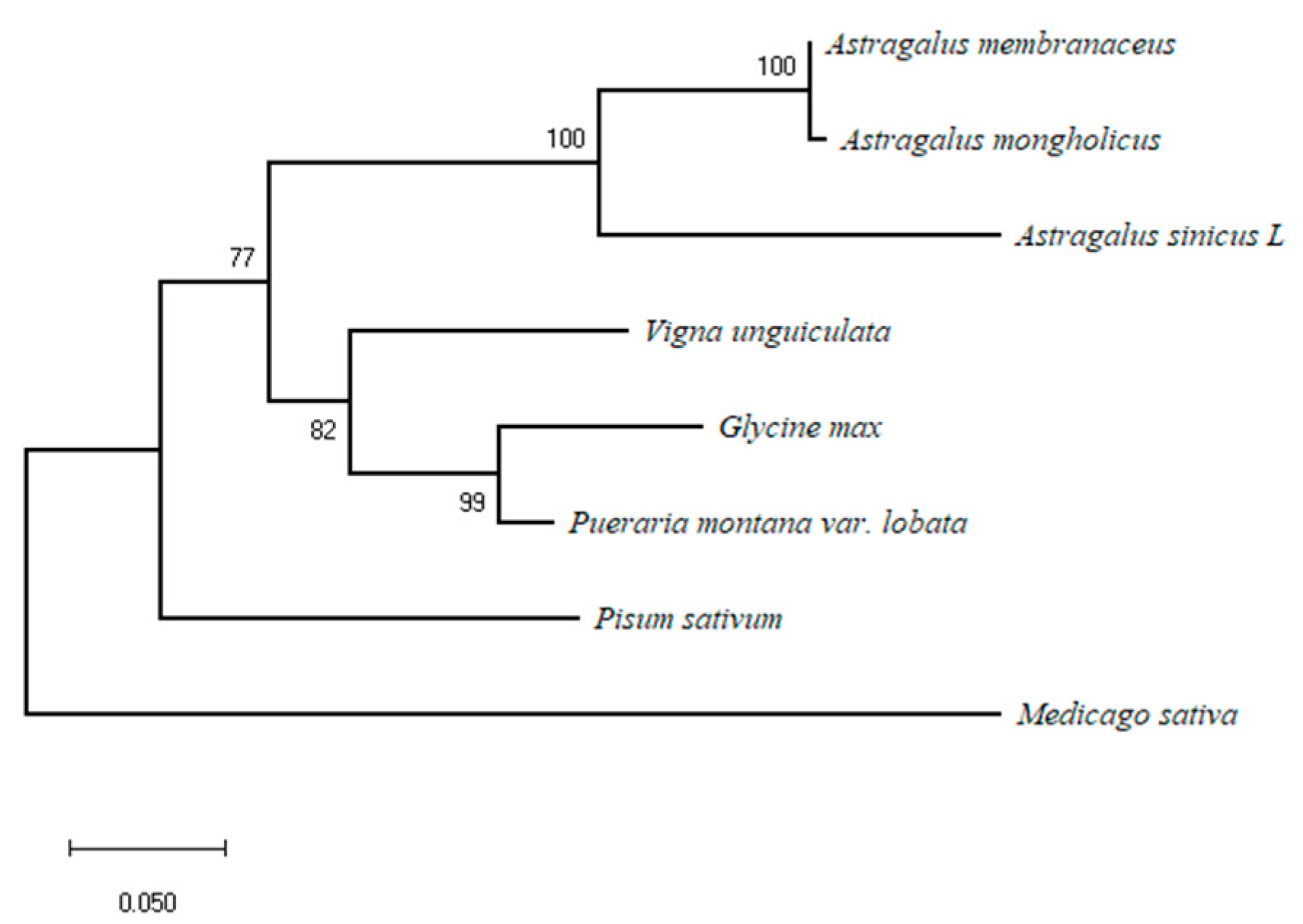
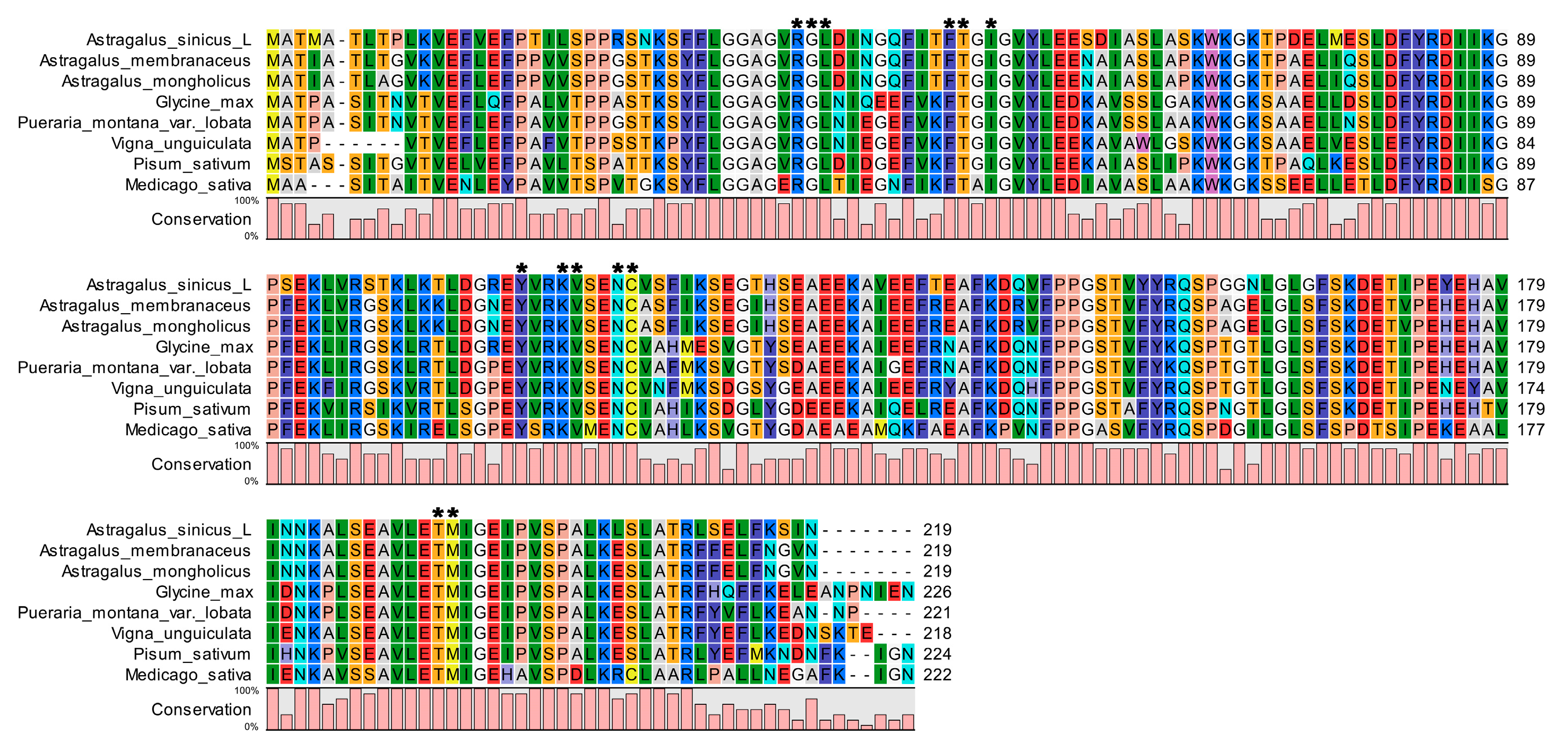
3.2. Sequence Alignment and Phylogenetic Analysis of AsiCHI Protein
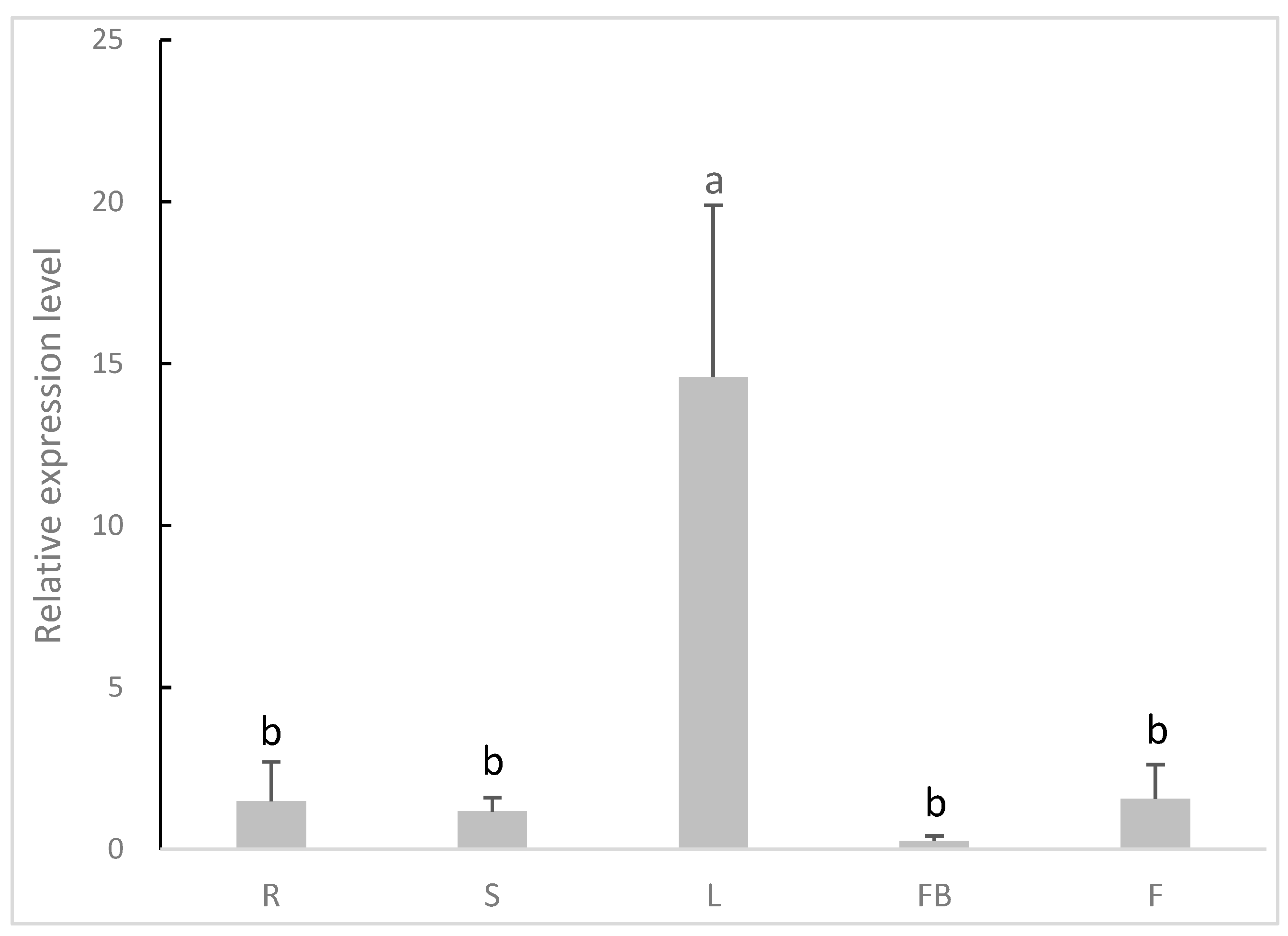
3.3. Gene Expression Profiles of AsiCHI by qRT-PCR
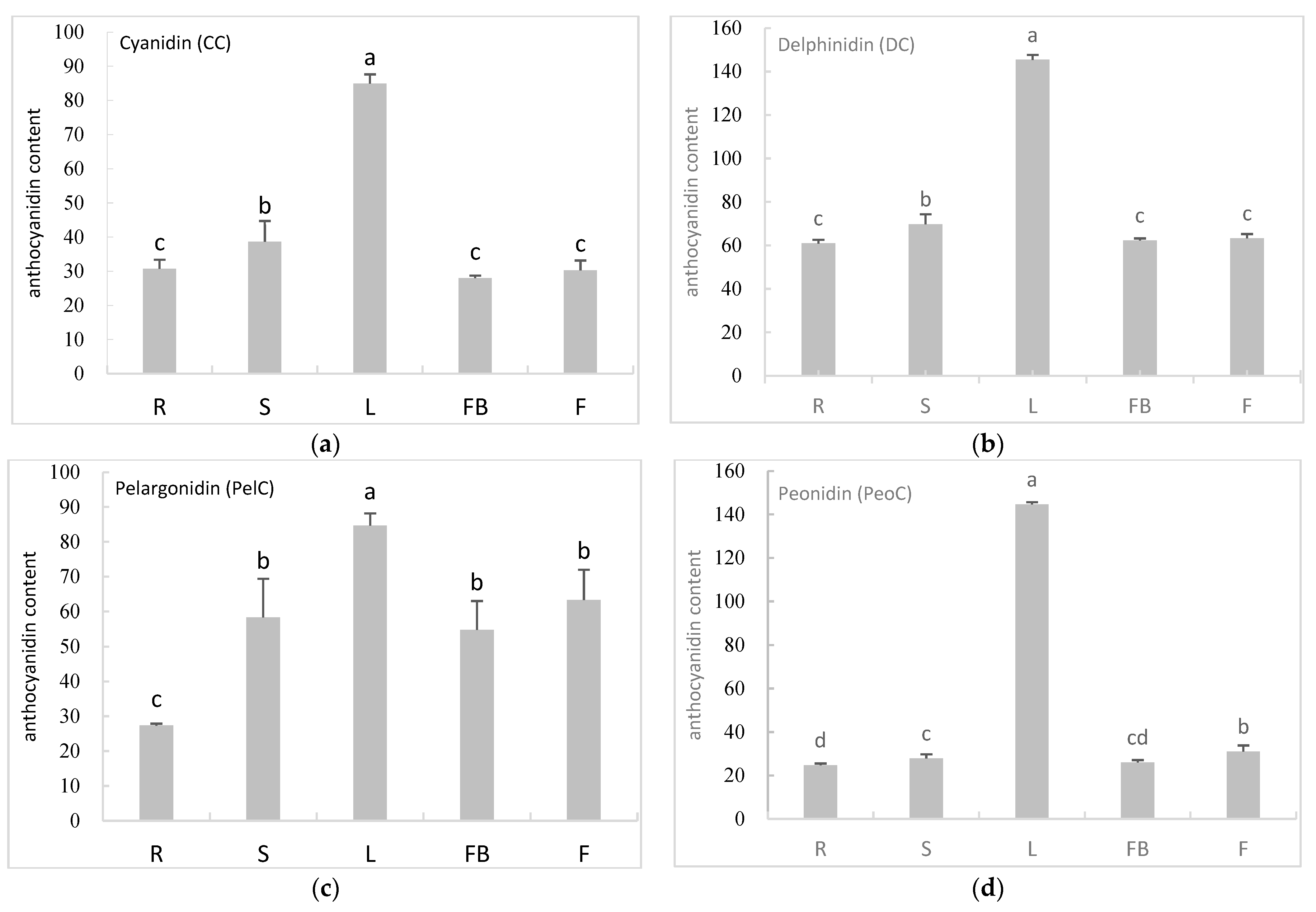
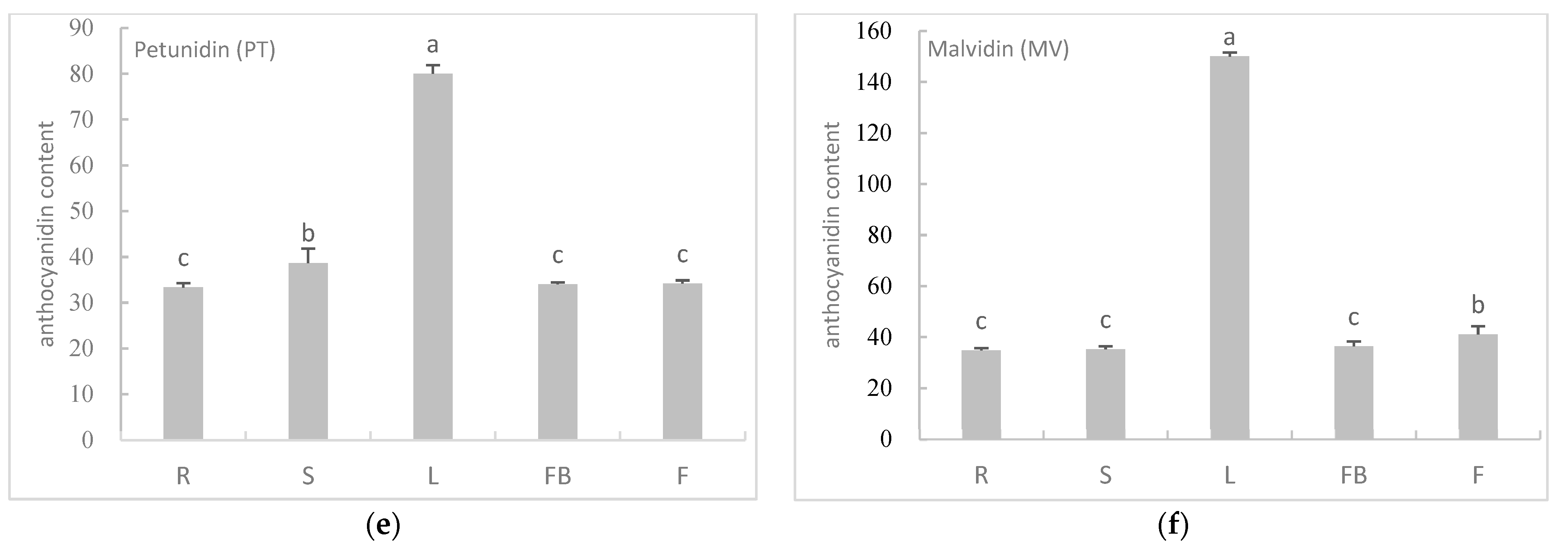
3.4. The Content of Anthocyanidins
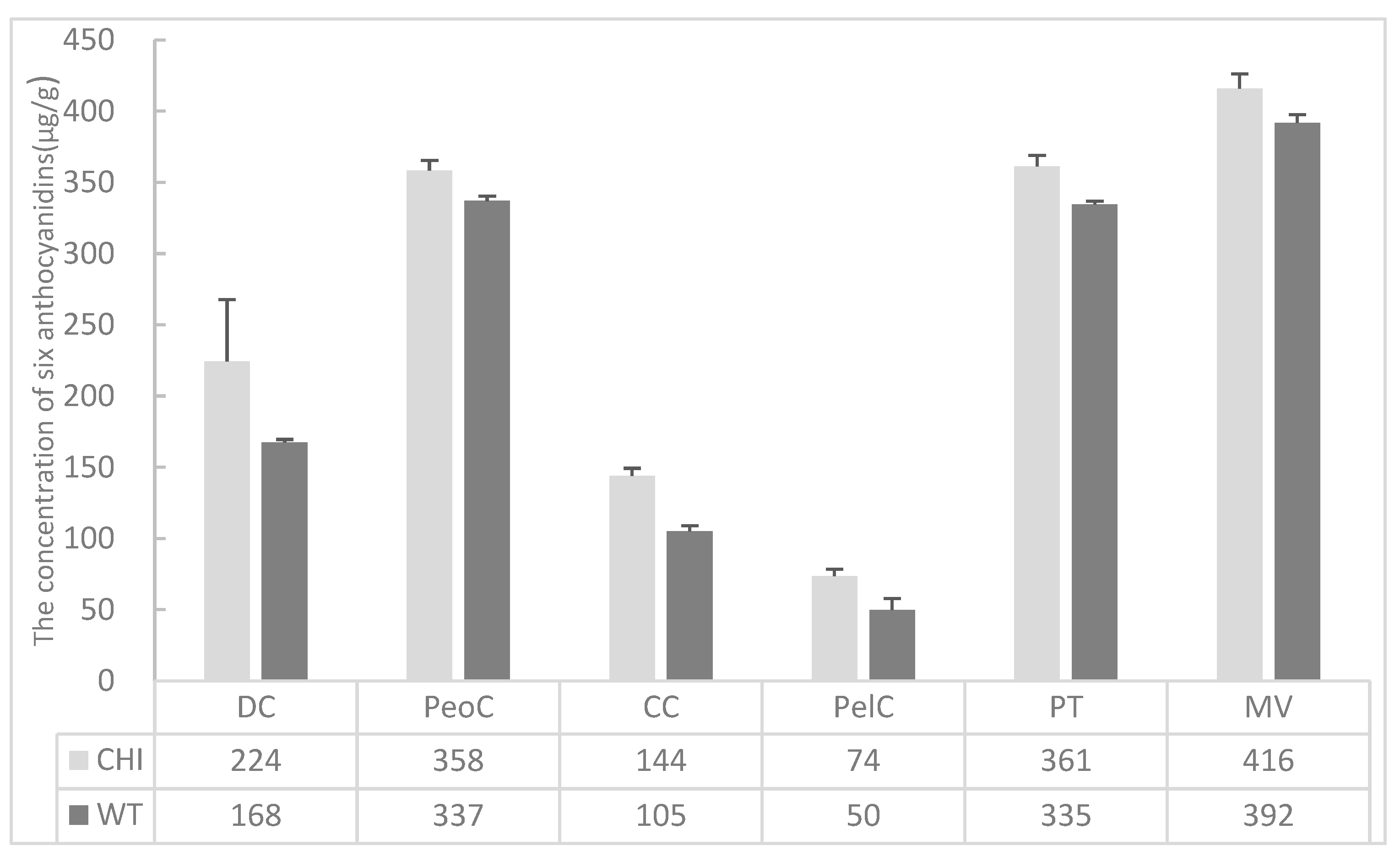
3.5. Anthocyanidins Analysis in Transgenic Arabidopsis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lloyd, A.M.; Walbot, V. Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science 1992, 258, 1773–1775. [Google Scholar] [CrossRef]
- Chen, M.; Zhu, W.J. Isolation and characterization of a chalcone isomerase gene promoter from potato cultivars. Genet. Mol. Res. 2015, 14, 18872–18885. [Google Scholar] [CrossRef]
- Kumar, A.; Sharma, M. Homology modeling and molecular dynamics based insights into Chalcone synthase and Chalcone isomerase in Phyllanthus emblica L. 3 Biotech 2020, 10, 373. [Google Scholar] [CrossRef] [PubMed]
- Song, C.P.; Xia, S.B. Cloning and Expression Analysis of Chalcone Synthase and Chalcone Isomerase Encoding Genes in Gossypium hirsutum. Agric. Biotechnol. 2018, 7, 15–21+26. [Google Scholar]
- Wu, X.L.; Prior, R.L. Systematic identification and characterization of anthocyanins by HPLC-ESI-MS/MS in common foods in the United States: Fruits and berries. J. Agric. Food Chem. 2005, 53, 2589–2599. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, W. Tissue-specific accumulation and subcellular localization of chalcone isomerase (CHI) in grapevine. Plant Cell Tissue Organ Cult. 2019, 137, 125–137. [Google Scholar] [CrossRef]
- Li, X.G.; Wang, J. Cloning of an anthocyanidin synthase gene homolog from blackcurrant (Ribes nigrum L.) and its expression at different fruit stages. Genet. Mol. Res. 2015, 14, 2726–2734. [Google Scholar] [CrossRef]
- Jez, J.; Bowman, M. Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase. Nat. Struct. Mol. Biol. 2000, 7, 786–791. [Google Scholar]
- Liu, X.M.; Ahmad, N. Molecular cloning and functional characterization of chalcone isomerase from Carthamus tinctorius. AMB Express 2019, 9, 132. [Google Scholar] [CrossRef]
- Keykha, F.; Bagheri, A. Analysis of Chalcone Synthase and Chalcone Isomerase Gene Expression in Pigment Production Pathway at Different Flower Colors of Petunia Hybrida. J. Cell. Mol. Med. 2016, 8, 8–14. [Google Scholar]
- Cheng, H.; Li, L. Molecular cloning and function assay of a chalcone isomerase gene (GbCHI) from Ginkgo biloba. Plant Cell Rep. 2011, 30, 49–62. [Google Scholar] [CrossRef]
- Liu, Y.; Xue, X. Cloning and Functional Characterization of Chalcone Isomerase Genes Involved in Anthocyanin Biosynthesis in Clivia miniata. Ornam. Plant Res. 2021, 1, 2. [Google Scholar] [CrossRef]
- Yin, Y.C.; Zhang, X.D. The Research Progress of Chalcone Isomerase (CHI) in Plants. Mol. Biotechnol. 2019, 61, 32–52. [Google Scholar] [CrossRef]
- Lim, W.S.; Li, J.R. Co-expression of onion chalcone isomerasein Del/Ros1-expressing tomato enhances anthocyanin and flavonol production. Plant Cell Tissue Organ Cult. 2017, 128, 113–124. [Google Scholar] [CrossRef]
- Chang, D.N.; Gao, S.J. The chromosome-level genome assembly of Astragalus sinicus and comparative genomic analyses provide new resources and insights for understanding legume-rhizobial interactions. Plant Commun. 2021, 3, 100263. [Google Scholar] [CrossRef]
- Cho, H.J.; Widholm, J.M. Agrobacterium rhizogenes-mediated transformation and regeneration of the legume Astragalus sinicus (Chinese milk vetch). Plant Sci. 1998, 138, 53–65. [Google Scholar] [CrossRef]
- Chou, M.X.; Wei, X.Y. A novel nodule-enhanced gene encoding a putative universal stress protein from Astragalus sinicus. J. Plant Physiol. 2007, 164, 764–772. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Zhou, C. The role of Chinese Milk Vetch as cover crop in complex soil nitrogen dynamics in rice rotation system of South China. Sci. Rep. 2018, 8, 12061. [Google Scholar] [CrossRef]
- Fan, Q.Y.; Xu, C.X. Application of milk vetch (Astragalus sinicus L.) with reduced chemical fertilizer improves rice yield and nitrogen, phosphorus, and potassium use efficiency in southern China. Eur. J. Agron. 2023, 144, 126762. [Google Scholar] [CrossRef]
- Vandamme, E.; Renkens, M. Root hairs explain P uptake efficiency of soybean genotypes grown in a P-deficient Ferralsol. Plant Soil 2013, 369, 269–282. [Google Scholar] [CrossRef]
- Zhang, Y.B.; Wang, L. Revealing the underlying molecular basis of phosphorus recycling in the green manure crop Astragalus sinicus. J. Clean. Prod. 2022, 341, 130924. [Google Scholar] [CrossRef]
- Guo, Z.H.; Ruan, W.Y. Vacuolar phosphate transporters account for variation in phosphate accumulation in Astragalus sinicus cultivars. Crop J. 2021, 9, 227–237. [Google Scholar] [CrossRef]
- Dare, A.P.; Tomes, S. Overexpression of chalcone isomerase in apple reduces phloridzin accumulation and increases susceptibility to herbivory by two-spotted mites. Plant J. 2020, 103, 293–307. [Google Scholar] [CrossRef]
- Yang, M.; Li, J. Characterization and expression analysis of a chalcone isomerase-like gene in relation to petal color of Actinidia chrysantha. Biologia 2017, 72, 753–763. [Google Scholar] [CrossRef]
- Elarabi, N.I.; Abdelhadi, A.A. Overexpression of chalcone isomerase A gene in Astragalus trigonus for stimulating apigenin. Sci. Rep. 2021, 11, 24176. [Google Scholar] [CrossRef] [PubMed]
- Shimada, N.; Aoki, T. A cluster of genes encodes the two types of chalcone isomerase involved in the biosynthesis of general flavonoids and legume-specific 5-deoxy(iso)flavonoids in Lotus japonicus. Plant Physiol. 2003, 131, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jiang, Y. Genome-Wide Classification and Evolutionary Analysis Reveal Diverged Patterns of Chalcone Isomerase in Plants. Biomolecules 2022, 12, 961. [Google Scholar] [CrossRef]
- Yu, S.; Li, J. Identification of Chalcone Isomerase Family Genes and Roles of CnCHI4 in Flavonoid Metabolism in Camellia nitidissima. Biomolecules 2023, 13, 41. [Google Scholar] [CrossRef]
- Ni, R.; Zhu, T.T. Identification and evolutionary analysis of chalcone isomerase-fold proteins in ferns. J. Exp. Bot. 2020, 71, 290–304. [Google Scholar] [CrossRef]
- Xu, H.; Lan, Y. AfCHIL, a Type IV Chalcone Isomerase, Enhances the Biosynthesis of Naringenin in Metabolic Engineering. Front. Plant Sci. 2022, 13, 891066. [Google Scholar] [CrossRef]
- Morita, Y.; Takagi, K. A chalcone isomerase-like protein enhances flavonoid production and flower pigmentation. Plant J. 2014, 78, 294–304. [Google Scholar] [CrossRef] [PubMed]
- Waki, T.; Mameda, R. A conserved strategy of chalcone isomerase-like protein to rectify promiscuous chalcone synthase specificity. Nat. Commun. 2020, 11, 870. [Google Scholar] [CrossRef] [PubMed]
- Dastmalchi, M.; Dhaubhadel, S. Soybean chalcone isomerase: Evolution of the fold, and the differential expression and localization of the gene family. Planta 2015, 241, 507–523. [Google Scholar] [CrossRef] [PubMed]
- Ralston, L.; Subramanian, S. Partial Reconstruction of Flavonoid and Isoflavonoid Biosynthesis in Yeast Using Soybean Type I and Type II Chalcone Isomerases. Plant Physiol. 2005, 137, 1375–1388. [Google Scholar] [CrossRef] [PubMed]
- Hur, S.; Newby, Z.E. Transition state stabilization by general acid catalysis, water expulsion, and enzyme reorganization in Medicago savita chalcone isomerase. Biochemistry 2004, 101, 2730–2735. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.J.; Khan, A. Identification of chalcone synthase genes and their expression patterns reveal pollen abortion in cotton. Saudi J. Biol. Sci. 2020, 27, 3691–3699. [Google Scholar] [CrossRef]
- Zu, Q.L.; Qu, Y.Y. The Chalcone Isomerase Family in Cotton: Whole-Genome Bioinformatic and Expression Analyses of the Gossypium barbadense L. Response to Fusarium Wilt Infection. Genes 2019, 10, 1006. [Google Scholar] [CrossRef]
- McKhann, H.I.; Hirsch, A.M. Isolation of chalcone synthase and chalcone isomerase cDNAs from alfalfa (Medicago sativa L.): Highest transcript levels occur in young roots and root tips. Plant Mol. Biol. 1994, 24, 767–777. [Google Scholar] [CrossRef]
- Nan, G.; Wu, H. Cloning, identification, and functional analysis of chalcone isomerase gene and its promoter from Tartary buckwheat. Acta Physiol. Plant 2022, 44, 78. [Google Scholar] [CrossRef]
- Guo, J.Y.; Zhou, W. Isolation and Functional Analysis of Chalcone Isomerase Gene from Purple-Fleshed Sweet Potato. Plant Mol. Biol. Rep. 2015, 33, 1451–1463. [Google Scholar] [CrossRef]
- Jiang, M.; Liu, Y. Molecular cloning and characterization of anthocyanin biosynthesis genes in eggplant (Solanum melongena L.). Acta Physiol. Plant 2016, 38, 163. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, Y. Overexpression of Ps-CHI1, a homologue of the chalcone isomerase gene from tree peony (Paeonia suffruticosa), reduces the intensity of flower pigmentation in transgenic tobacco. Plant Cell Tissue Organ Cult. 2014, 116, 285–295. [Google Scholar] [CrossRef]
- Keykha, F.; Bagheri, A. RNAi-induced silencing in floral tissues of Petunia hybrida by agroinfiltration: A rapid assay for chalcone isomerase gene function analysis. Cell. Mol. Biol. 2016, 62, 26–31. [Google Scholar] [PubMed]
- Chao, N.; Wang, R.F. Functional characterization of two chalcone isomerase (CHI) revealing their responsibility for anthocyanins accumulation in mulberry. Plant Physiol. Biochem. 2021, 161, 65–73. [Google Scholar] [CrossRef] [PubMed]
| Primer Name | Sequence (5′~3′) | Used to |
|---|---|---|
| rCHI-R1 | GTCTCCAACACAACCTCAGAAAGTGCCTTGT | 5′-RACE amplification |
| rCHI-R2 | CCTGGGGGAAATACTTGATCCTTGAATGCTT | |
| rCHI-F1 | GGGAACACACAGTGAAGCTGAAGA | 3′-RACE amplification |
| rCHI-F2 | CCCCCAGGATCTACTGTTTACTACAGACAA | |
| qCHI-F | AGTCCTTTTTCCTCGGTG | qRT-PCR primers |
| qCHI-R | GTGATGCTATGTCGCTTT | |
| Tubulin F | CACCGAGGGAGCAGAGTTGAT | Reference gene for qRT-PCR analysis |
| Tubulin R | CTGAAACCCTTGAAGGCAGTCA | |
| HPT-F | GGTCGCGGAGGCTATGGATGC | Detection of transgenic lines |
| HPT-R | GCTTCTGCGGGCGATTTGTGT | |
| CHI-F | GGGAACACACAGTGAAGCTGAA | Detection of transgenic lines |
| CHI-R | GCTCAGATAAGCGAGTAGCCAAA |
| Species | NCBI Reference Sequences | No. of Residues | Identity | E Value | PI | Molecular Weight (kDa) |
|---|---|---|---|---|---|---|
| Astragalus sinicus L. | OQ870547 | 219 | 100 | 0 | 5.11 | 24.14 |
| Astragalus membranaceus | ATY39974.1 | 219 | 82.65 | 0 | 5.4 | 24.02 |
| Astragalus mongholicus | ABA55017.2 | 219 | 82.19 | 0 | 5.41 | 23.99 |
| Glycine max | Q53B70.1 | 226 | 71.1 | 0 | 5.27 | 24.98 |
| Pueraria montana var. lobata | ADV71377.1 | 221 | 71.69 | 0 | 5.59 | 24.13 |
| Pisum sativum | XP_050892596.1 | 224 | 69.86 | 0 | 5.69 | 24.73 |
| Vigna unguiculata | QCE05742.1 | 218 | 72.64 | 0 | 4.91 | 24.36 |
| Medicago sativa | AAB41524.1 | 222 | 59.72 | 0 | 5.25 | 23.83 |
| Cyanidin (CC) | Delphinidin (DC) | Pelargonidin (PelC) | Peonidin (PeoC) | Petunidin (PT) | Malvidin (MV) | |
|---|---|---|---|---|---|---|
| AsiCHI | 0.943 ** | 0.963 ** | 0.671 ** | 0.973 ** | 0.958 ** | 0.974 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Xu, J.; Si, L.; Cao, K.; Wang, Y.; Li, H.; Wang, J. Cloning, Identification, and Functional Analysis of the Chalcone Isomerase Gene from Astragalus sinicus. Genes 2023, 14, 1400. https://doi.org/10.3390/genes14071400
Zhang X, Xu J, Si L, Cao K, Wang Y, Li H, Wang J. Cloning, Identification, and Functional Analysis of the Chalcone Isomerase Gene from Astragalus sinicus. Genes. 2023; 14(7):1400. https://doi.org/10.3390/genes14071400
Chicago/Turabian StyleZhang, Xian, Jing Xu, Linlin Si, Kai Cao, Yuge Wang, Hua Li, and Jianhong Wang. 2023. "Cloning, Identification, and Functional Analysis of the Chalcone Isomerase Gene from Astragalus sinicus" Genes 14, no. 7: 1400. https://doi.org/10.3390/genes14071400
APA StyleZhang, X., Xu, J., Si, L., Cao, K., Wang, Y., Li, H., & Wang, J. (2023). Cloning, Identification, and Functional Analysis of the Chalcone Isomerase Gene from Astragalus sinicus. Genes, 14(7), 1400. https://doi.org/10.3390/genes14071400





