NGS-Based Genetic Analysis in a Cohort of Italian Patients with Suspected Inherited Myopathies and/or HyperCKemia
Abstract
1. Introduction
2. Patients and Methods
2.1. Patients Cohort
2.2. Genetic Studies
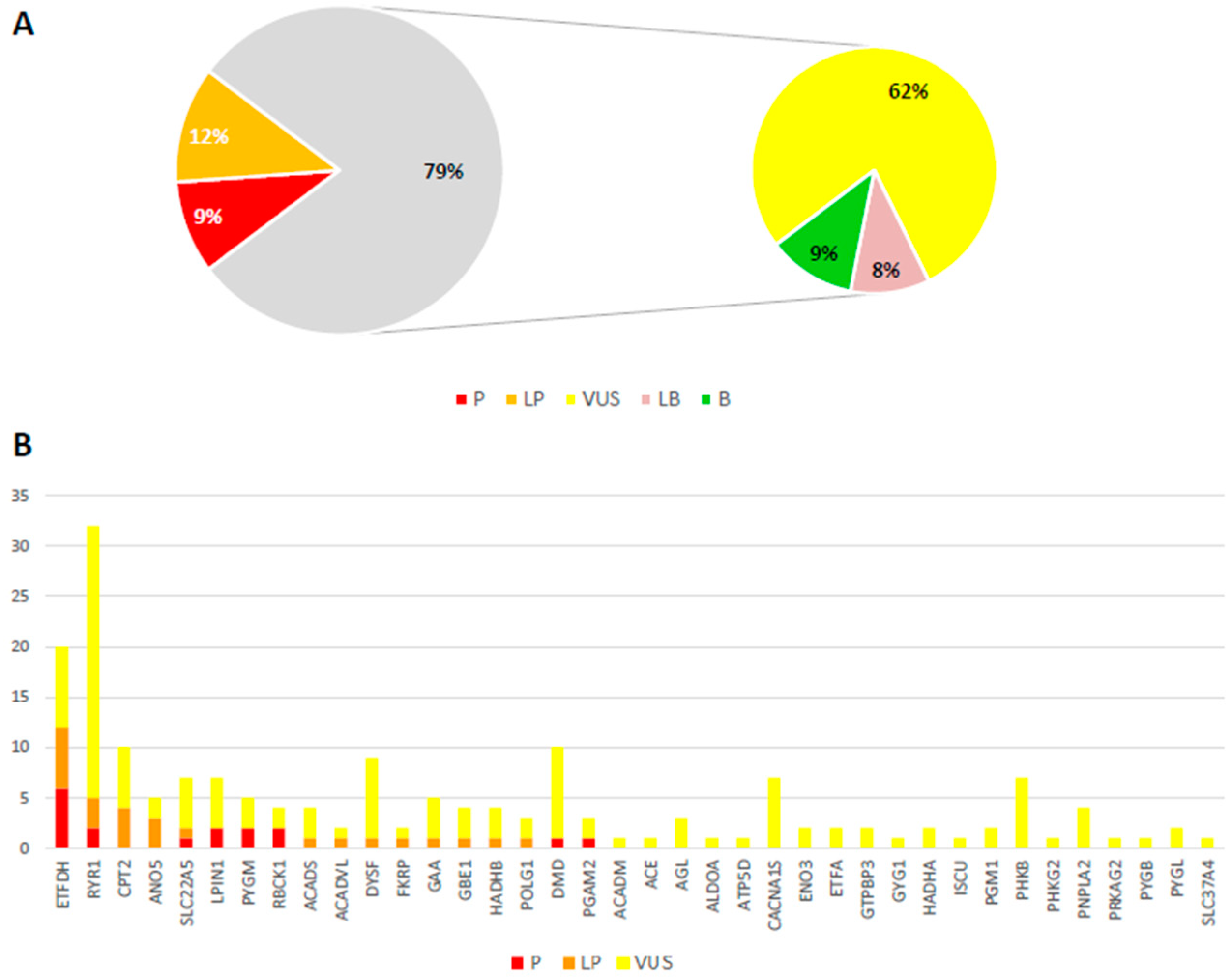
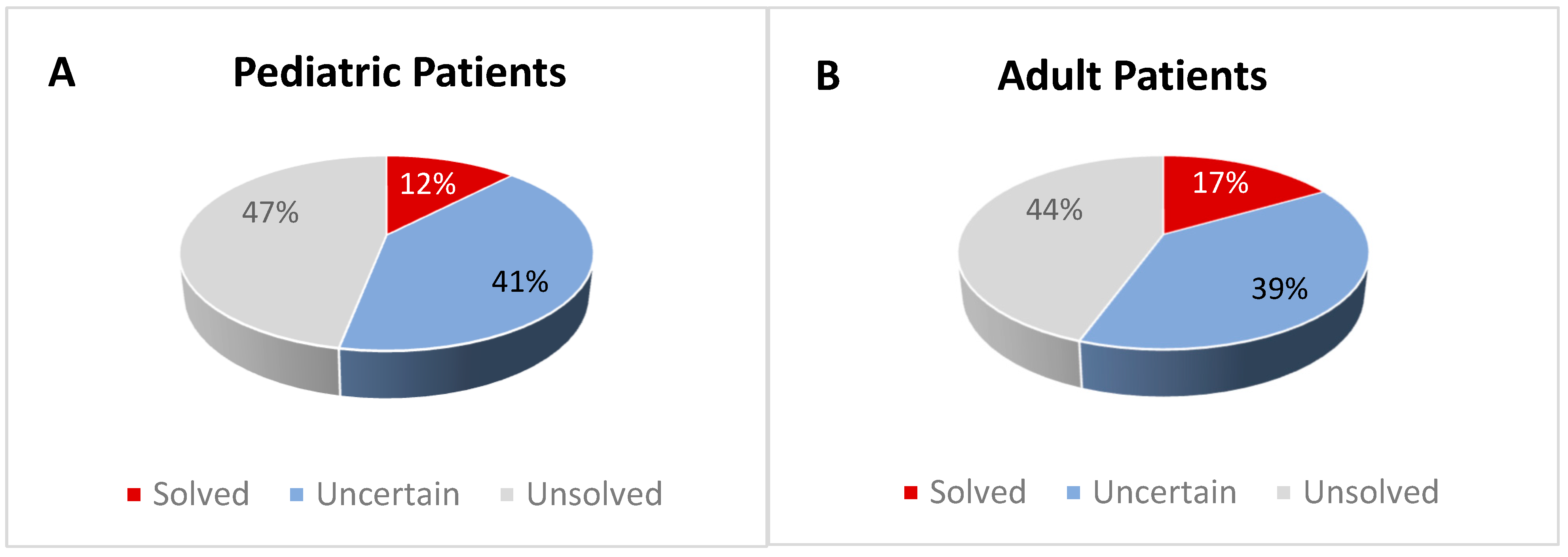
3. Results
3.1. Solved Patients
3.2. Patients with Uncertain Diagnosis
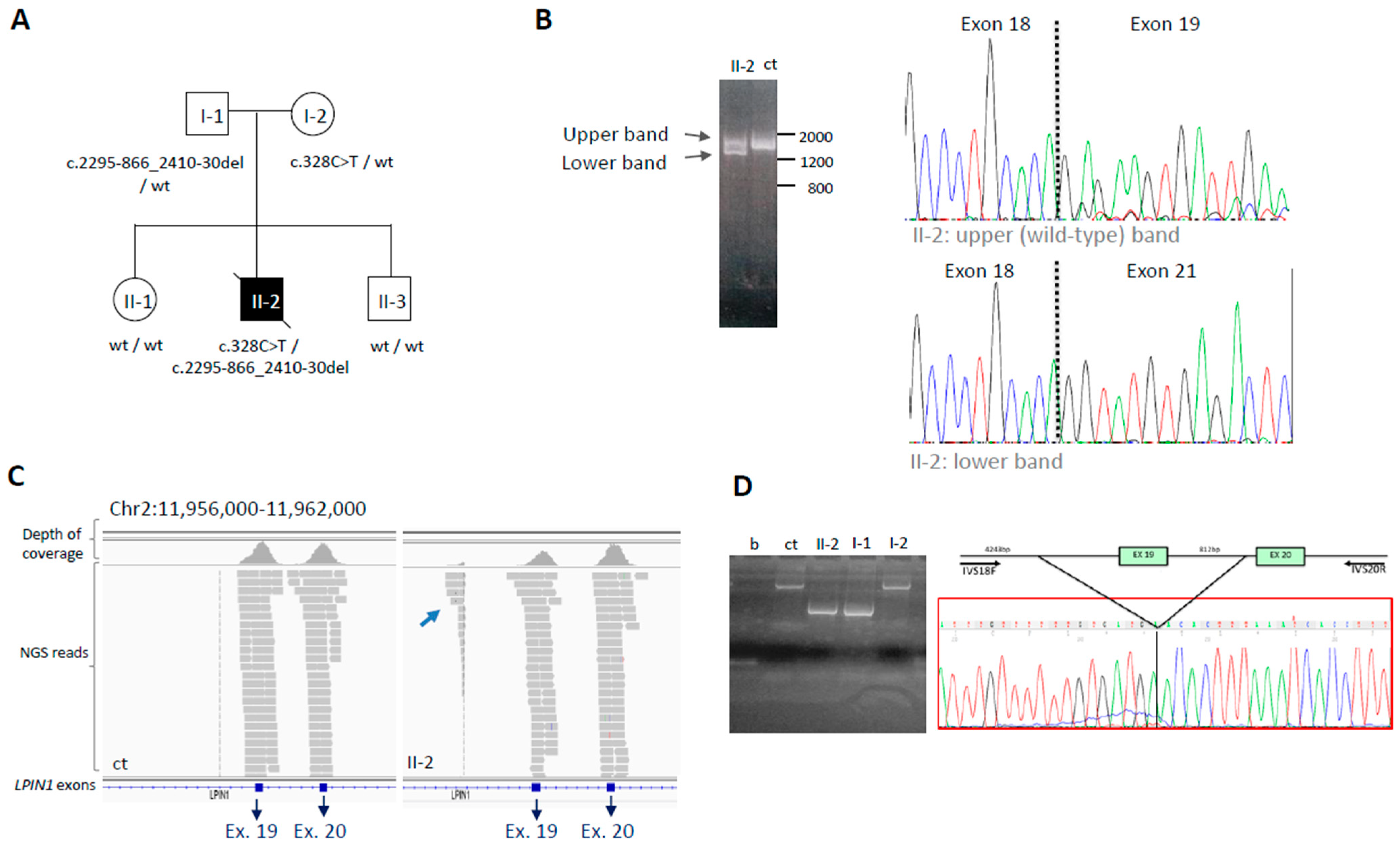
3.2.1. The LPIN1 Example: From Uncertain to Solved Case by RNA Analysis
3.2.2. The CPT2 Example: AR or AD?
3.3. Unsolved Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Munsat, T.L.; Baloh, R.; Pearson, C.M.; Fowler, W. Serum enzyme alterations in neuromuscular disorders. JAMA 1973, 226, 1536–1543. [Google Scholar] [CrossRef]
- Prelle, A.; Tancredi, L.; Sciacco, M.; Chiveri, L.; Comi, G.P.; Martinelli-Boneschi, F.M.; Bagnardi, V.; Battistel, A.; Ciscato, P.; Bordoni, A.; et al. Retrospective study of a large population of patients with asymptomatic or minimally symptomatic raised serum creatine kinase levels. J. Neurol. 2002, 249, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Rowland, L.P.; Willner, J.; DiMauro, S.; Miranda, A. Approaches to the membrane theory of Duchenne muscular dystrophy. In Muscular Dystrophy-Advances and New Trends; Angelini, C., Danieli, G.A., Fontanari, D., Eds.; Excerpta Medica: Amsterdam, The Netherlands, 1980; pp. 3–13. [Google Scholar]
- Khan, F.Y. Rhabdomyolysis: A review of the literature. Neth. J. Med. 2009, 67, 272–283. [Google Scholar] [PubMed]
- Scalco, R.S.; Gardiner, A.R.; Pitceathly, R.D.; Zanoteli, E.; Becker, J.; Holton, J.L.; Houlden, H.; Jungbluth, H.; Quinlivan, R. Rhabdomyolysis: A genetic perspective. Orphanet J. Rare Dis. 2015, 10, 51. [Google Scholar] [CrossRef]
- David, W.S. Myoglobinuria. Neurol. Clin. 2000, 18, 215–243. [Google Scholar] [CrossRef] [PubMed]
- Voermans, N.; Snoeck, M.; Jungbluth, H. RYR1-related rhabdomyolysis: A common but probably underdiagnosed manifestation of skeletal muscle ryanodine receptor dysfunction. Rev. Neurol. 2016, 172, 546–558. [Google Scholar] [CrossRef] [PubMed]
- Barca, E.; Emmanuele, V.; DiMauro, S. Metabolic Myoglobinuria. Curr. Neurol. Neurosci. Rep. 2015, 15, 69. [Google Scholar] [CrossRef]
- Mochel, F.; Knight, M.A.; Tong, W.-H.; Hernandez, D.; Ayyad, K.; Taivassalo, T.; Andersen, P.M.; Singleton, A.; Rouault, T.A.; Fischbeck, K.H.; et al. Splice Mutation in the Iron-Sulfur Cluster Scaffold Protein ISCU Causes Myopathy with Exercise Intolerance. Am. J. Hum. Genet. 2008, 82, 652–660. [Google Scholar] [CrossRef]
- Legati, A.; Reyes, A.; Nasca, A.; Invernizzi, F.; Lamantea, E.; Tiranti, V.; Garavaglia, B.; Lamperti, C.; Ardissone, A.; Moroni, I.; et al. New genes and pathomechanisms in mitochondrial disorders unraveled by NGS technologies. Biochim. Biophys. Acta 2016, 1857, 1326–1335. [Google Scholar] [CrossRef]
- Nykamp, K.; Anderson, M.; Powers, M.; Garcia, J.; Herrera, B.; Ho, Y.-Y.; Kobayashi, Y.; Patil, N.; Thusberg, J.; Westbrook, M.; et al. Sherloc: A comprehensive refinement of the ACMG–AMP variant classification criteria. Genet. Med. 2017, 19, 1105–1117. [Google Scholar] [CrossRef]
- Nilsson, J.; Schoser, B.; Laforet, P.; Kalev, O.; Lindberg, C.; Romero, N.B.; López, M.D.; Akman, H.O.; Wahbi, K.; Iglseder, S.; et al. Polyglucosan body myopathy caused by defective ubiquitin ligase RBCK1. Ann. Neurol. 2013, 74, 914–919. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Kishnani, P.; Wu, J.Y.; Chen, Y.T. Hepatic and neuromuscular forms of glycogen storage disease type IV caused by mutations in the same glycogen-branching enzyme gene. J. Clin. Investig. 1996, 97, 941–948. [Google Scholar] [CrossRef]
- Michot, C.; Hubert, L.; Brivet, M.; De Meirleir, L.; Valayannopoulos, V.; Müller-Felber, W.; Venkateswaran, R.; Ogier, H.; Desguerre, I.; Altuzarra, C.; et al. LPIN1 gene mutations: A major cause of severe rhabdomyolysis in early childhood. Hum. Mutat. 2010, 31, E1564–E1573. [Google Scholar] [CrossRef] [PubMed]
- Ørngreen, M.C.; Dunø, M.; Ejstrup, R.; Christensen, E.; Schwartz, M.; Sacchetti, M.; Vissing, J. Fuel utilization in subjects with carnitine palmitoyltransferase 2 gene mutations. Ann. Neurol. 2005, 57, 60–66. [Google Scholar] [CrossRef]
- Scalco, R.S.; Snoeck, M.; Quinlivan, R.; Treves, S.; Laforét, P.; Jungbluth, H.; Voermans, N.C. Exertional rhabdomyolysis: Physiological response or manifestation of an underlying myopathy? BMJ Open Sport Exerc. Med. 2016, 2, e000151. [Google Scholar] [CrossRef]
- Nance, J.R.; Mammen, A.L. Diagnostic evaluation of rhabdomyolysis. Muscle Nerve 2015, 51, 793–810. [Google Scholar] [CrossRef] [PubMed]
- Dosi, C.; Rubegni, A.; Baldacci, J.; Galatolo, D.; Doccini, S.; Astrea, G.; Berardinelli, A.; Bruno, C.; Bruno, G.; Comi, G.P.; et al. Using Cluster Analysis to Overcome the Limits of Traditional Phenotype–Genotype Correlations: The Example of RYR1-Related Myopathies. Genes 2023, 14, 298. [Google Scholar] [CrossRef]
- Rubegni, A.; Malandrini, A.; Dosi, C.; Astrea, G.; Baldacci, J.; Battisti, C.; Bertocci, G.; Donati, M.A.; Dotti, M.T.; Federico, A.; et al. Next-generation sequencing approach to hyperCKemia. Neurol. Genet. 2019, 5, e352. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, M.; Fossati, B.; Vitale, R.; Brigonzi, E.; Ricigliano, V.A.G.; Saraceno, L.; Cardani, R.; Pappone, C.; Meola, G. Flecainide-Induced Brugada Syndrome in a Patient with Skeletal Muscle Sodium Channelopathy: A Case Report with Critical Therapeutical Implications and Review of the Literature. Front. Neurol. 2018, 9, 385. [Google Scholar] [CrossRef]
- Vivante, A.; Ityel, H.; Pode-Shakked, B.; Chen, J.; Shril, S.; van der Ven, A.T.; Mann, N.; Schmidt, J.M.; Segel, R.; Aran, A.; et al. Exome sequencing in Jewish and Arab patients with rhabdomyolysis reveals single-gene etiology in 43% of cases. Pediatr. Nephrol. 2017, 32, 2273–2282. [Google Scholar] [CrossRef]
- Sambuughin, N.; Mungunsukh, O.; Ren, M.; Capacchione, J.F.; Horkayne-Szakaly, I.; Chuang, K.; Muldoon, S.M.; Smith, J.; O’Connor, F.G.; Deuster, P.A. Pathogenic and rare deleterious variants in multiple genes suggest oligogenic inheritance in recurrent exertional rhabdomyolysis. Mol. Genet. Metab. Rep. 2018, 16, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Legati, A.; Zanetti, N.; Nasca, A.; Peron, C.; Lamperti, C.; Lamantea, E.; Ghezzi, D. Current and New Next-Generation Sequencing Approaches to Study Mitochondrial DNA. J. Mol. Diagn. 2021, 23, 732–741. [Google Scholar] [CrossRef] [PubMed]
- Gemelli, C.; Traverso, M.; Trevisan, L.; Fabbri, S.; Scarsi, E.; Carlini, B.; Prada, V.; Mongini, T.; Ruggiero, L.; Patrone, S.; et al. An integrated approach to the evaluation of patients with asymptomatic or minimally symptomatic hyperCKemia. Muscle Nerve 2022, 65, 96–104. [Google Scholar] [CrossRef]
- Kruijt, N.; van den Bersselaar, L.R.; Kamsteeg, E.J.; Verbeeck, W.; Snoeck, M.M.J.; Everaerd, D.S.; Abdo, W.F.; Jansen, D.R.M.; Erasmus, C.E.; Jungbluth, H.; et al. The etiology of rhabdomyolysis: An interaction between genetic susceptibility and external triggers. Eur. J. Neurol. 2021, 28, 647–659. [Google Scholar] [CrossRef] [PubMed]
| (A) Disorders of fatty acid oxidation/lipid metabolism | |||
| Gene | Protein | Inheritance | Disease |
| ACADM | Medium-chain acyl-CoA Dehydrogenase | AR | Deficiency of medium chain acyl-CoA dehydrogenase |
| ACADS | Short-chain acyl-CoA Dehydrogenase | AR | Deficiency of short chain acyl-CoA dehydrogenase |
| ACADVL | Very-long-chain acyl-CoA dehydrogenase | AR | Deficiency of very-long-chain acyl-CoA dehydrogenase |
| CPT2 | Carnitine palmitoyl-transferase II | AR | Deficiency of Carnitine palmitoyl-transferase 2 |
| ETFA | Electron transfer flavoprotein-asubunits | AR | Multiple acyl-coenzyme A dehydrogenase deficiency-Glutaric aciduria type IIA |
| ETFB | Electron transfer flavoprotein-bsubunits | AR | Multiple acyl-coenzyme A dehydrogenase deficiency-Glutaric aciduria type IIB |
| ETFDH | Electron transfer flavoprotein: ubiquinone oxidoreductase | AR | Multiple acyl-coenzyme A dehydrogenase deficiency-Glutaric aciduria IIC |
| FLAD1 | Flavin adenine dinucleotide synthetase | AR | Lipid storage myopathy due to flavin adenine dinucleotide synthetase deficiency |
| HADHA | Hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase, alpha subunit | AR | Mitochondrial trifunctional protein deficiency |
| HADHB | Hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase, beta subunit | AR | Mitochondrial trifunctional protein deficiency |
| PNPLA2 | Adipose Triglyceride lipase | AR | Neutral lipid storage disease |
| SLC22A5 | Organic cation transporter 2 | AR | Carnitine deficiency, systemic primary |
| (B) Disorders of glycogen metabolism | |||
| Gene | Protein | Inheritance | Disease |
| AGL | Amylo-1,6-Glucosidase, 4-Alpha-Glucanotransferase | AR | Glycogen storage disease IIIa/IIIb, glycogen debrancher enzyme |
| ALDOA | Fructose-1,6-bisphosphate aldolase | AR | Glycogen storage disease XII |
| ENO3 | Enolase b | AR | Glycogen storage disease XIII |
| G6PC | Glucose-6-phosphatase (G6Pase) | AR | Glycogen storage disease Ia |
| GAA | Acid Alpha-1,4-Glucosidase (Acid maltase) | AR | Glycogen storage disease II |
| GBE1 | 1,4-Alpha-Glucan Branching Enzyme Amylo-(1,4 to 1,6) Transglucosidase Amylo-(1,4 to 1,6) Transglycosylase | AR | Glycogen storage disease IV, Polyglucosan body disease (adult form) |
| GYG1 | Glycogenin-1 (glycosyltransferase) | AR | Glycogen storage disease XV, Polyglucosan body myopathy 2 |
| GYS1 | Glycogen synthase (mucle) | AR | Glycogen storage disease 0 |
| GYS2 | Glycogen synthase (liver) | AR | Glycogen storage disease 0 |
| LDHA | Lactate dehydrogenase (muscle, A subunit) | AR | Glycogen storage disease XI |
| PFKM | Phosphofructokinase (muscle) | AR | Glycogen storage disease VII, Tarui disease |
| PYGB | Glycogen phosphorylase (brain) | AR | Glucogen storage disease V |
| PYGL | Glycogen phosphorylase (liver) | AR | Glycogen storage disease VI |
| PYGM | Glycogen phosphorylase (muscle) | AR | Glycogen storage disease V, McArdle disease |
| PGAM2 | Phosphoglycerate mutase-2 (mucle) | AR | Glycogen storage disease X |
| PGK1 | Phosphoglycerate kinase-1 | X-linked | Phosphoglycerate kinase 1 deficiency |
| PGM1 | Phosphoglucomutase-1 | AR | Congenital disorder of glycosylation, type 1t Glycogen storage disease XIV |
| PHKA1 | Phosphorylase kinase-a1 (muscle) | X-linked | Glycogen storage disease IXd |
| PHKA2 | Phosphorylase kinase-a2 (liver) | X-linked | Glycogen storage disease IXa2 |
| PHKB | Phosphorylase kinase-b | AR | Glycogen storage disease IXb |
| PHKG2 | Phosphorylase kinase-g (liver, testis) | AR | Glycogen storage disease IXc |
| PRKAG2 | Noncatalytic gamma subunit of AMP-activated protein kinase | AD | Glycogen storage disease of heart, lethal congenital |
| SLC2A2 | Glucose transporter-like (GLUT2) | AR | Fanconi-Bickel syndrome - Glycogen storage disease XI |
| SLC37A4 | Glucose-6-PhosphateTransporter 1 | AR | Glycogen storage disease Ib-Ic |
| (C) Mitochondrial functions | |||
| Gene | Protein | Inheritance | Disease |
| ATP5D | Mitochondrial ATP synthase F1 complex-delta subunit | AR | Mitochondrial complex V (ATP synthase) deficiency |
| DGUOK | Mitochondrial deoxyguanosine kinase | AR | Mitochondrial DNA depletion syndrome 3 (hepatocerebral type) |
| FDX1L | Ferredoxin 1-like ptotein | AR | Mitochondrial myopathy, episodic, with optic atrophy and reversible leukoencephalopathy |
| ISCU | Iron-sulfur (Fe-S) clusters scaffold protein | AR | Iron-sulphur cluster deficiency myopathy (mitochondrial disorder) |
| HSD17B10 | 2-methyl-3-hydroxybutyryl Co-A dehydrogenase | X-linked | Neurodegenerative disorder, chorioathetosis with mental retardation and abnormal behavior |
| LPIN1 | Phosphatidic acid phosphohydrolase 1 | AR | Phosphatidic acid phosphatase deficiency |
| POLG | Polymerase gamma | AR/AD | Mitochondrial DNA depletion syndrome Progressive external ophthalmoplegia |
| (D) Muscular dystrophies/congenital myopathies | |||
| Gene | Protein | Inheritance | Disease |
| ANO5 | Transmembrane protein 16E Anoctamin 5 | AR | Miyoshi muscular dystrophy 3 Muscular dystrophy, limb-girdle, autosomal recessive 12 |
| CACNA1S | Calcium channel | AD | Malignant hyperthermia susceptibility 5 Thyrotoxic periodic paralysis Hypokalemic periodic paralysis, type 1 |
| CAV3 | Caveolin-3 | AD | Myopathy, distal, Tateyama type |
| CHKB | Choline kinase | AR | Muscular dystrophy, congenital, megaconial type |
| DMD | Dystrophin | X-linked | Duchenne muscular dystrophy, Becker muscular dystrophy |
| DYSF | Dysferlin | AR | LGMD2B, Miyoshi myopathy |
| FKRP | Fukutin-related protein | AR | LGMD2I |
| FKTN | Fukutin | AR | Fukuyama congenital muscular dystrophy |
| RBCK1 | RANBP-Type and C3HC4-Type Zinc Finger-Containing 1 | AR | Polyglucosan body myopathy 1 with or without immunodeficiency |
| SIL1 | Nucleotide Exchange Factor | AR | Marinesco-Sjogren syndrome |
| (E) Disorders of intramuscular calcium release and excitation-contraction coupling | |||
| Gene | Protein | Inheritance | Disease |
| RYR1 | Skeletal muscle ryanodine receptor 1 | AD/AR | Malignant hyperthermia-susceptibility, Exertional rhabdomyolysis, Congenital myopathy |
| N°/Child or Adult | Mutated Gene/Inheritance | RefSeq Match | Reference Group Genes * | cDNA | Protein | ACMG/Franklin Classification | Zygosity | Familiarity |
|---|---|---|---|---|---|---|---|---|
| 1/C | ETFDH/AR | NM_004453.4 | A | c.176-2A>T | splice acceptor variant | P | Homozygous | Not investigated |
| 2/C | HADHB/AR | NM_000183.3 | A | c.1280G>A c.1370C>T | p.Gly427Glu p.Ala457Val | LP VUS | Compound heterozygous | Mother p.Gly427Glu Father p.Ala457Val |
| 3/C | RYR1/AD-AR | NM_000540.3 | E | c.10010G>A | p.Arg3337Gln | LP | Heterozygous | Father (s) p.Arg3337Gln Mother negative |
| 4/C | RYR1/AD-AR | NM_000540.3 | E | c.14918C>T | p.Pro4973Leu | P | Heterozygous | Mother (s) p.Pro4973Leu |
| 5/C | RYR1/AD-AR | NM_000540.3 | E | c.13490C>G c.4759G>C | p.Pro4497Arg p.Ala1587Pro | VUS VUS | Compound heterozygous | Mother p.Pro4497Arg Father p.Ala1587Pro |
| 6/C | LPIN1/AR | NM_001261428 | C | c.328C>T c.2395-866_2410-30del | p.Arg110 * p.Glu766_Ser838del | P P | Compound heterozygous | Mother p.Arg110 * Father p.Glu766_Ser838del |
| 7/A | ANO5/AR | NM_213599.3 | D | c.902G>T c.2516T>G | p.Gly301Val 21 p.Met839Arg | LP LP | Possibly compound heterozygous | Not investigated |
| 8/A | CPT2/AR-AD | NM_000098.3 | A | c.338C>T | p.Ser113Leu | LP | Homozygous | Not investigated |
| 9/A | CPT2/AR-AD | NM_000098.3 | A | c.338C>T c.887G>T | p.Ser113Leu p.Arg296Leu | LP VUS | Possibly compound heterozygous | Not investigated |
| 10/A | ETFDH/AR | NM_004453.4 | A | c.1531G>A c.1832G>A | p.Asp511Asn p.Gly611Glu | VUS LP | Possibly compound heterozygous | Not investigated |
| 11/A | ETFDH/AR | NM_004453.4 | A | c.250G>A | p.Ala84Thr | P | Homozygous | Not investigated |
| 12/A | ETFDH/AR | NM_004453.4 | A | c.1249C>T c.1531G>A | p.Gln417Ter p.Asp511Asn | P P | Possibly compound heterozygous | Not investigated |
| 13/A | ETFDH/AR | NM_004453.4 | A | c.74dupA c.256C>T | p.Tyr25 * p.Arg86Cys | LP VUS | Possibly compound heterozygous | Not investigated |
| 14/A | ETFDH/AR | NM_004453.4 | A | c.250G>A | p.Ala84Thr | LP | Homozygous | Not investigated |
| 15/A | ETFDH/AR | NM_004453.4 | A | c.1531G>A | p.Asp511Asn | LP | Homozygous | Not investigated |
| 16/A | PYGM/AR | NM_005609.4 | B | c.1A>G | p.? | P | Homozygous | Not investigated |
| 17/A | RBCK1/AR | NM_031229.4 | D | c.896_899delAGTG | p.Glu299Valfs * 46 | P | Homozygous | Not investigated |
| 18/A | RYR1/AD-AR | NM_000540.3 | E | c.14545G>A | p.Val4849Ile | LP | Heterozygous | Father negative Mother (s) p.Val4849Ile |
| 19/A | RYR1/AD-AR | NM_000540.3 | E | c.11708G>A | p.Arg3903Gln | P | Heterozygous | Not investigated |
| 20/A | RYR1/AD-AR | NM_000540.3 | E | c.8594T>C c.6226_6228delA AG | p.Val2865Ala p.Lys2076del | VUS LP | Possibly compound heterozygous | Not investigated |
| 21/A | RYR1/AD-AR | NM_000540.3 | E | c.12700G>A c.4910C>T | p.Val4234Met p.Ala1637Val | LP VUS | Possibly compound heterozygous | Not investigated |
| N°/ Child or Adult | Age/Sex | Alive | CK | RM/Myoglobinuria | CNS Involvement | Muscle Findings | Other |
|---|---|---|---|---|---|---|---|
| 1/C | 13y/M | No | Elevated | No/No | No | Lipid accumulation (muscle and heart) | EMG: myopathy |
| 2/C | 11y/F | Yes | Mild increase | No/No | No | Neurogenic | Previous episodes of weakness after exertion. An episode of severe limb-girdle and axial weakness with onset due to fever |
| 3/C | 16y/M | Yes | No | Yes/No | No | Not done | Myalgia, cramps |
| 4/C | 16y/M | Yes | Mild increase | No/No | No | Absence of alterations | Myalgia and cramps after excercise, family history of hyperckemia and myalgias |
| 5/C | 12y/F | Yes | Elevated | Yes/No | No | Not done | Myopathy |
| 6/C | 3y/M | No | Elevated | No/Yes | No | Not done | MR: symmetrical muscular inflammation (legs) |
| 7/A | 28y/M | Yes | Elevated | No/No | No | Not done | Myalgia, cramps. Proximal Hypostenia (No osteotendinous reflexes) |
| 8/A | 25y/M | Yes | Elevated | Yes/Yes | No | Not done | Myalgia and fatigue |
| 9/A | 51y/M | Yes | Elevated | Yes/Yes | No | Not done | Myalgia, muscle weakness |
| 10/A | 21y/F | Yes | Elevated | No/No | No | Vacuolar myopathy, lipid accumulation | Dicarboxylic aciduria, fatigue and myalgia |
| 11/A | 42y/M | Yes | Elevated | No/No | No | Lipid accumulation, altered mitochondria | EMG: myopathy |
| 12/A | 29y/F | Yes | Mild increase | No/No | No | Lipid accumulation | EMG: neurogenic signs |
| 13/A | 77y/M | Yes | Mild increase | No/No | No | Lipid accumulation | EMG: neurogenic signs |
| 14/A | 22y/M | Yes | No | No/Yes | No | Not done | Hypostenia, hypotonia (increased liver enzymes, steatosis) |
| 15/A | unreported/M | Yes | Elevated | No/No | No | Not done | Myalgia, cramps |
| 16/A | 43y/M | Yes | Elevated | No/Yes | No | Inflammatory necrotizing | Fatigue and cramps |
| 17/A | 27y/F | Yes | Mild increase | No/No | No | PAS+ | Cardiomyopathy and muscle weakness |
| 18/A | 25y/M | Yes | Elevated | Yes/No | No | Not done | Cramps, muscle weakness |
| 19/A | 40y/M | Yes | Elevated | Yes/No | No | Not done | Myalgia, muscle weakness |
| 20/A | 48y/M | Yes | Elevated | Yes/No | No | Not done | EMG: myopathy |
| 21/A | 24y/M | Yes | Mild increase | No/No | No | Myogenic signs. Normal electron microscopy | Myalgia, dyspnoea, muscle pain. Psychomotor delay |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Invernizzi, F.; Izzo, R.; Colangelo, I.; Legati, A.; Zanetti, N.; Garavaglia, B.; Lamantea, E.; Peverelli, L.; Ardissone, A.; Moroni, I.; et al. NGS-Based Genetic Analysis in a Cohort of Italian Patients with Suspected Inherited Myopathies and/or HyperCKemia. Genes 2023, 14, 1393. https://doi.org/10.3390/genes14071393
Invernizzi F, Izzo R, Colangelo I, Legati A, Zanetti N, Garavaglia B, Lamantea E, Peverelli L, Ardissone A, Moroni I, et al. NGS-Based Genetic Analysis in a Cohort of Italian Patients with Suspected Inherited Myopathies and/or HyperCKemia. Genes. 2023; 14(7):1393. https://doi.org/10.3390/genes14071393
Chicago/Turabian StyleInvernizzi, Federica, Rossella Izzo, Isabel Colangelo, Andrea Legati, Nadia Zanetti, Barbara Garavaglia, Eleonora Lamantea, Lorenzo Peverelli, Anna Ardissone, Isabella Moroni, and et al. 2023. "NGS-Based Genetic Analysis in a Cohort of Italian Patients with Suspected Inherited Myopathies and/or HyperCKemia" Genes 14, no. 7: 1393. https://doi.org/10.3390/genes14071393
APA StyleInvernizzi, F., Izzo, R., Colangelo, I., Legati, A., Zanetti, N., Garavaglia, B., Lamantea, E., Peverelli, L., Ardissone, A., Moroni, I., Maggi, L., Bonanno, S., Fiori, L., Velardo, D., Magri, F., Comi, G. P., Ronchi, D., Ghezzi, D., & Lamperti, C. (2023). NGS-Based Genetic Analysis in a Cohort of Italian Patients with Suspected Inherited Myopathies and/or HyperCKemia. Genes, 14(7), 1393. https://doi.org/10.3390/genes14071393








