miRNAs: Potential as Biomarkers and Therapeutic Targets for Cancer
Abstract
1. Introduction
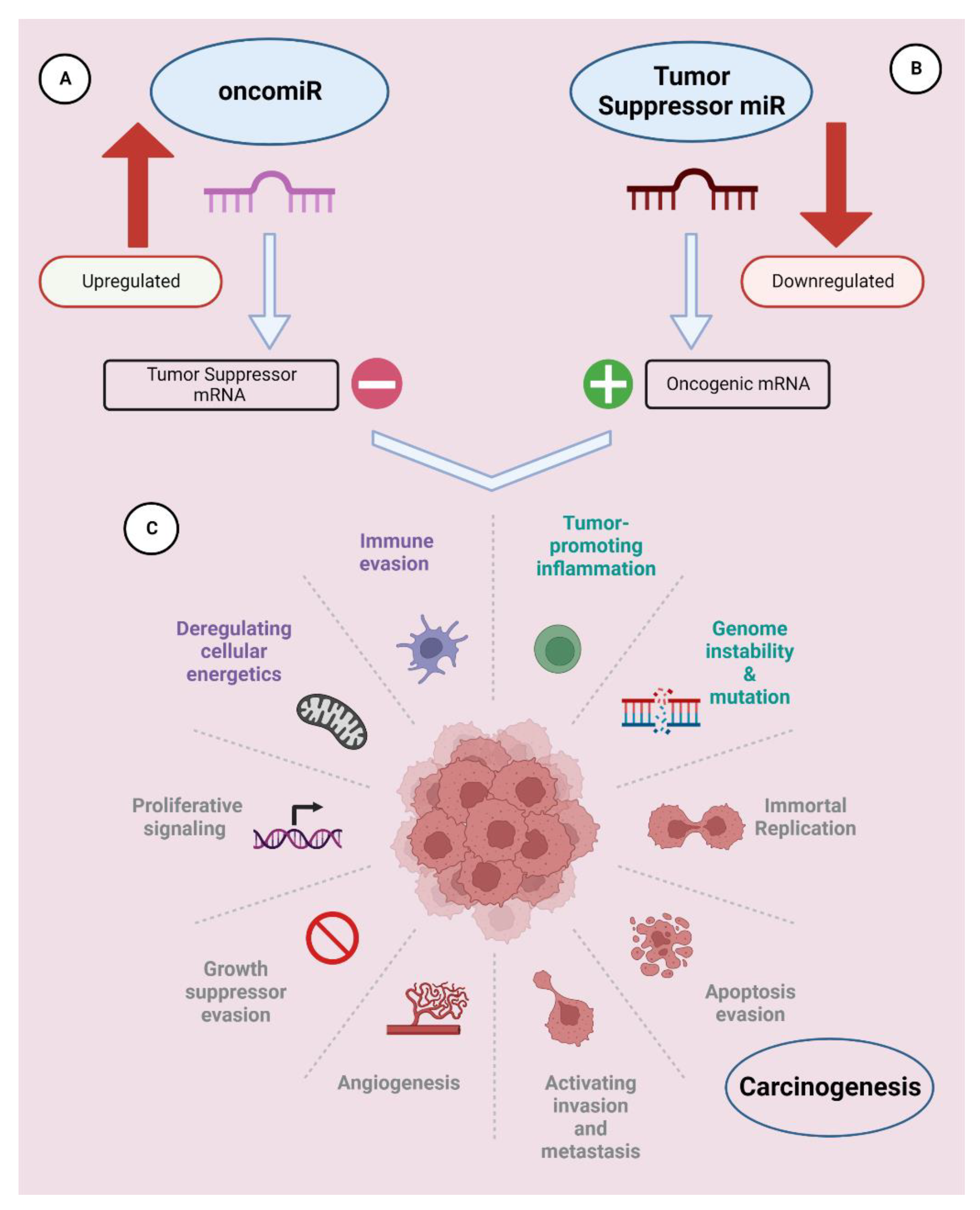
2. Role of miRNAs in Cancer
2.1. Humans
2.1.1. OncomiRs
2.1.2. Tumor-Suppressor miRNAs (TS-miRNAs)
2.2. Dogs
3. Role of miRNAs in Cancer Metastasis
3.1. Humans
3.2. Dogs
4. miRNAs as Potential Cancer Biomarkers
4.1. Humans
4.1.1. Diagnosis
4.1.2. Prognosis
4.1.3. Tumor Staging
4.1.4. Treatment Resistance
4.2. Dogs
5. Role of miRNAs in Cancer Treatment
5.1. Humans
5.2. Dogs
6. Future Perspective and Summary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- www.miRBase.org. Available online: https://www.mirbase.org/cgi-bin/browse.pl (accessed on 8 November 2022).
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef]
- Broughton, J.P.; Lovci, M.T.; Huang, J.L.; Yeo, G.W.; Pasquinelli, A.E. Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol. Cell 2016, 64, 320–333. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H. The clinical relevance of circulating, exosomal miRNAs as biomarkers for cancer. Expert Rev. Mol. Diagn. 2015, 15, 1159–1169. [Google Scholar] [CrossRef]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, N.; Haider, S.; Jagannathan, S.; Anaissie, E.; Driscoll, J.J. MicroRNA theragnostics for the clinical management of multiple myeloma. Leukemia 2014, 28, 732–738. [Google Scholar] [CrossRef]
- Svoronos, A.A.; Engelman, D.M.; Slack, F.J. OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer. Cancer Res. 2016, 76, 3666–3670. [Google Scholar] [CrossRef] [PubMed]
- Mogilyansky, E.; Rigoutsos, I. The miR-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013, 20, 1603–1614. [Google Scholar] [CrossRef]
- Zhu, H.; Han, C.; Wu, T. MiR-17-92 cluster promotes hepatocarcinogenesis. Carcinogenesis 2015, 36, 1213–1222. [Google Scholar] [CrossRef]
- Tsuchida, A.; Ohno, S.; Wu, W.; Borjigin, N.; Fujita, K.; Aoki, T.; Ueda, S.; Takanashi, M.; Kuroda, M. miR-92 is a key oncogenic component of the miR-17-92 cluster in colon cancer. Cancer Sci. 2011, 102, 2264–2271. [Google Scholar] [CrossRef]
- Takakura, S.; Mitsutake, N.; Nakashima, M.; Namba, H.; Saenko, V.A.; Rogounovitch, T.I.; Nakazawa, Y.; Hayashi, T.; Ohtsuru, A.; Yamashita, S. Oncogenic role of miR-17-92 cluster in anaplastic thyroid cancer cells. Cancer Sci. 2008, 99, 1147–1154. [Google Scholar] [CrossRef]
- Hayashita, Y.; Osada, H.; Tatematsu, Y.; Yamada, H.; Yanagisawa, K.; Tomida, S.; Yatabe, Y.; Kawahara, K.; Sekido, Y.; Takahashi, T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005, 65, 9628–9632. [Google Scholar] [CrossRef]
- Martens-de Kemp, S.R.; Komor, M.A.; Hegi, R.; Bolijn, A.S.; Tijssen, M.; de Groen, F.L.M.; Depla, A.; van Leerdam, M.; Meijer, G.A.; Fijneman, R.J.A.; et al. Overexpression of the miR-17-92 cluster in colorectal adenoma organoids causes a carcinoma-like gene expression signature. Neoplasia 2022, 32, 100820. [Google Scholar] [CrossRef] [PubMed]
- Lu, F.; Zhao, X.; Zhang, Z.; Xiong, M.; Wang, Y.; Sun, Y.; He, B.; Zhu, J. The diagnostic and prognostic value of the miR-17-92 cluster in hepatocellular carcinoma: A meta-analysis. Front. Genet. 2022, 13, 927079. [Google Scholar] [CrossRef]
- Menon, A.; Abd-Aziz, N.; Khalid, K.; Poh, C.L.; Naidu, R. miRNA: A Promising Therapeutic Target in Cancer. Int. J. Mol. Sci. 2022, 23, 11502. [Google Scholar] [CrossRef]
- Ozgun, A.; Karagoz, B.; Bilgi, O.; Tuncel, T.; Baloglu, H.; Kandemir, E.G. MicroRNA-21 as an indicator of aggressive phenotype in breast cancer. Onkologie 2013, 36, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Echevarria-Vargas, I.M.; Valiyeva, F.; Vivas-Mejia, P.E. Upregulation of miR-21 in cisplatin resistant ovarian cancer via JNK-1/c-Jun pathway. PLoS ONE 2014, 9, e97094. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, C.; Shan, S.; Liu, X.; Jiang, Z.; Ren, T. TLR4/ROS/miRNA-21 pathway underlies lipopolysaccharide instructed primary tumor outgrowth in lung cancer patients. Oncotarget 2016, 7, 42172–42182. [Google Scholar] [CrossRef]
- Si, H.; Sun, X.; Chen, Y.; Cao, Y.; Chen, S.; Wang, H.; Hu, C. Circulating microRNA-92a and microRNA-21 as novel minimally invasive biomarkers for primary breast cancer. J. Cancer Res. Clin. Oncol. 2013, 139, 223–229. [Google Scholar] [CrossRef]
- Baez-Vega, P.M.; Echevarria Vargas, I.M.; Valiyeva, F.; Encarnacion-Rosado, J.; Roman, A.; Flores, J.; Marcos-Martinez, M.J.; Vivas-Mejia, P.E. Targeting miR-21-3p inhibits proliferation and invasion of ovarian cancer cells. Oncotarget 2016, 7, 36321–36337. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, Z.; Gao, C.; Chen, P.; Chen, J.; Liu, W.; Xiao, S.; Lu, H. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab. Investig. 2008, 88, 1358–1366. [Google Scholar] [CrossRef]
- Yaman Agaoglu, F.; Kovancilar, M.; Dizdar, Y.; Darendeliler, E.; Holdenrieder, S.; Dalay, N.; Gezer, U. Investigation of miR-21, miR-141, and miR-221 in blood circulation of patients with prostate cancer. Tumor Biol. 2011, 32, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Toiyama, Y.; Takahashi, M.; Hur, K.; Nagasaka, T.; Tanaka, K.; Inoue, Y.; Kusunoki, M.; Boland, C.R.; Goel, A. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J. Natl. Cancer Inst. 2013, 105, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Simonian, M.; Mosallayi, M.; Mirzaei, H. Circulating miR-21 as novel biomarker in gastric cancer: Diagnostic and prognostic biomarker. J. Cancer Res. Ther. 2018, 14, 475. [Google Scholar] [CrossRef] [PubMed]
- Sicard, F.; Gayral, M.; Lulka, H.; Buscail, L.; Cordelier, P. Targeting miR-21 for the therapy of pancreatic cancer. Mol. Ther. 2013, 21, 986–994. [Google Scholar] [CrossRef] [PubMed]
- Si, M.L.; Zhu, S.; Wu, H.; Lu, Z.; Wu, F.; Mo, Y.Y. miR-21-mediated tumor growth. Oncogene 2007, 26, 2799–2803. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.K.; Blansit, K.; Kiet, T.; Sherman, A.; Wong, G.; Earle, C.; Bourguignon, L.Y. The inhibition of miR-21 promotes apoptosis and chemosensitivity in ovarian cancer. Gynecol. Oncol. 2014, 132, 739–744. [Google Scholar] [CrossRef]
- Tian, W.; Pang, X.; Luan, F. Diagnosis value of miR-181, miR-652, and CA72-4 for gastric cancer. J. Clin. Lab. Anal. 2022, 36, e24411. [Google Scholar] [CrossRef]
- Tian, J.; Shen, R.; Yan, Y.; Deng, L. miR-186 promotes tumor growth in cutaneous squamous cell carcinoma by inhibiting apoptotic protease activating factor-1. Exp. Ther. Med. 2018, 16, 4010–4018. [Google Scholar] [CrossRef]
- Zhai, Z.; Mu, T.; Zhao, L.; Li, Y.; Zhu, D.; Pan, Y. MiR-181a-5p facilitates proliferation, invasion, and glycolysis of breast cancer through NDRG2-mediated activation of PTEN/AKT pathway. Bioengineered 2022, 13, 83–95. [Google Scholar] [CrossRef]
- Strotbek, M.; Schmid, S.; Sanchez-Gonzalez, I.; Boerries, M.; Busch, H.; Olayioye, M.A. miR-181 elevates Akt signaling by co-targeting PHLPP2 and INPP4B phosphatases in luminal breast cancer. Int. J. Cancer 2017, 140, 2310–2320. [Google Scholar] [CrossRef]
- Tong, S.J.; Liu, J.; Wang, X.; Qu, L.X. microRNA-181 promotes prostate cancer cell proliferation by regulating DAX-1 expression. Exp. Ther. Med. 2014, 8, 1296–1300. [Google Scholar] [CrossRef]
- Barbato, A.; Iuliano, A.; Volpe, M.; D’Alterio, R.; Brillante, S.; Massa, F.; De Cegli, R.; Carrella, S.; Salati, M.; Russo, A.; et al. Integrated Genomics Identifies miR-181/TFAM Pathway as a Critical Driver of Drug Resistance in Melanoma. Int. J. Mol. Sci. 2021, 22, 1801. [Google Scholar] [CrossRef]
- Abrahamsson, A.; Dabrosin, C. Tissue specific expression of extracellular microRNA in human breast cancers and normal human breast tissue in vivo. Oncotarget 2015, 6, 22959–22969. [Google Scholar] [CrossRef] [PubMed]
- Tao, K.; Yang, J.; Guo, Z.; Hu, Y.; Sheng, H.; Gao, H.; Yu, H. Prognostic value of miR-221-3p, miR-342-3p and miR-491-5p expression in colon cancer. Am. J. Transl. Res. 2014, 6, 391–401. [Google Scholar]
- Pineau, P.; Volinia, S.; McJunkin, K.; Marchio, A.; Battiston, C.; Terris, B.; Mazzaferro, V.; Lowe, S.W.; Croce, C.M.; Dejean, A. miR-221 overexpression contributes to liver tumorigenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.X.; Hu, Q.; Qiu, M.T.; Zhong, S.L.; Xu, J.J.; Tang, J.H.; Zhao, J.H. miR-221/222: Promising biomarkers for breast cancer. Tumor Biol. 2013, 34, 1361–1370. [Google Scholar] [CrossRef]
- Calin, G.A.; Sevignani, C.; Dumitru, C.D.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M.; et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 2999–3004. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, L.A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef]
- Yang, W.; Lee, D.Y.; Ben-David, Y. The roles of microRNAs in tumorigenesis and angiogenesis. Int. J. Physiol. Pathophysiol. Pharmacol. 2011, 3, 140–155. [Google Scholar]
- Qu, H.; Zheng, L.; Song, H.; Jiao, W.; Li, D.; Fang, E.; Wang, X.; Mei, H.; Pu, J.; Huang, K.; et al. microRNA-558 facilitates the expression of hypoxia-inducible factor 2 α through binding to 5’-untranslated region in neuroblastoma. Oncotarget 2016, 7, 40657–40673. [Google Scholar] [CrossRef]
- Baradaran, B.; Shahbazi, R.; Khordadmehr, M. Dysregulation of key microRNAs in pancreatic cancer development. Biomed. Pharmacother. 2019, 109, 1008–1015. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Fu, J.; Zhang, W.; Guo, M. Genetic and epigenetic changes in lung carcinoma and their clinical implications. Mod. Pathol. 2011, 24, 932–943. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Duan, Y.T.; Lu, P.; Zhang, Z.J.; Zheng, X.K.; Wang, J.L.; Feng, W.S. Epigenetic Targets and their Inhibitors in Cancer Therapy. Curr. Top. Med. Chem. 2018, 18, 2395–2419. [Google Scholar] [CrossRef]
- Link, S.; Grund, S.E.; Diederichs, S. Alternative splicing affects the subcellular localization of Drosha. Nucleic Acids Res. 2016, 44, 5330–5343. [Google Scholar] [CrossRef]
- Xia, L.; Zhang, D.; Du, R.; Pan, Y.; Zhao, L.; Sun, S.; Hong, L.; Liu, J.; Fan, D. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int. J. Cancer 2008, 123, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Wan, R.; Hu, G.; Yang, L.; Xiong, J.; Wang, F.; Shen, J.; He, S.; Guo, X.; Ni, J.; et al. miR-15b and miR-16 induce the apoptosis of rat activated pancreatic stellate cells by targeting Bcl-2 in vitro. Pancreatology 2012, 12, 91–99. [Google Scholar] [CrossRef]
- Musumeci, M.; Coppola, V.; Addario, A.; Patrizii, M.; Maugeri-Sacca, M.; Memeo, L.; Colarossi, C.; Francescangeli, F.; Biffoni, M.; Collura, D.; et al. Control of tumor and microenvironment cross-talk by miR-15a and miR-16 in prostate cancer. Oncogene 2011, 30, 4231–4242. [Google Scholar] [CrossRef]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS is regulated by the let-7 microRNA family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef]
- Kumar, M.S.; Erkeland, S.J.; Pester, R.E.; Chen, C.Y.; Ebert, M.S.; Sharp, P.A.; Jacks, T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc. Natl. Acad. Sci. USA 2008, 105, 3903–3908. [Google Scholar] [CrossRef] [PubMed]
- Buechner, J.; Tomte, E.; Haug, B.H.; Henriksen, J.R.; Lokke, C.; Flaegstad, T.; Einvik, C. Tumour-suppressor microRNAs let-7 and mir-101 target the proto-oncogene MYCN and inhibit cell proliferation in MYCN-amplified neuroblastoma. Br. J. Cancer 2011, 105, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Qin, S.; Fan, C.; Xu, C.; Du, N.; Ren, H. Let-7: A regulator of the ERalpha signaling pathway in human breast tumors and breast cancer stem cells. Oncol. Rep. 2013, 29, 2079–2087. [Google Scholar] [CrossRef]
- Slusarz, A.; Pulakat, L. The two faces of miR-29. J. Cardiovasc. Med. 2015, 16, 480–490. [Google Scholar] [CrossRef]
- Zhang, W.; Wu, Q.; Liu, Y.; Wang, X.; Ma, C.; Zhu, W. LncRNA HOTAIR Promotes Chemoresistance by Facilitating Epithelial to Mesenchymal Transition through miR-29b/PTEN/PI3K Signaling in Cervical Cancer. Cells Tissues Organs 2022, 211, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Vera, O.; Bok, I.; Jasani, N.; Nakamura, K.; Xu, X.; Mecozzi, N.; Angarita, A.; Wang, K.; Tsai, K.Y.; Karreth, F.A. A MAPK/miR-29 Axis Suppresses Melanoma by Targeting MAFG and MYBL2. Cancers 2021, 13, 1408. [Google Scholar] [CrossRef]
- Hozaka, Y.; Seki, N.; Tanaka, T.; Asai, S.; Moriya, S.; Idichi, T.; Wada, M.; Tanoue, K.; Kawasaki, Y.; Mataki, Y.; et al. Molecular Pathogenesis and Regulation of the miR-29-3p-Family: Involvement of ITGA6 and ITGB1 in Intra-Hepatic Cholangiocarcinoma. Cancers 2021, 13, 2804. [Google Scholar] [CrossRef]
- Grassilli, S.; Bertagnolo, V.; Brugnoli, F. Mir-29b in Breast Cancer: A Promising Target for Therapeutic Approaches. Diagnostics 2022, 12, 2139. [Google Scholar] [CrossRef]
- Zhang, W.; Qian, J.X.; Yi, H.L.; Yang, Z.D.; Wang, C.F.; Chen, J.Y.; Wei, X.Z.; Fu, Q.; Ma, H. The microRNA-29 plays a central role in osteosarcoma pathogenesis and progression. Mol. Biol. 2012, 46, 622–627. [Google Scholar] [CrossRef]
- Yu, P.N.; Yan, M.D.; Lai, H.C.; Huang, R.L.; Chou, Y.C.; Lin, W.C.; Yeh, L.T.; Lin, Y.W. Downregulation of miR-29 contributes to cisplatin resistance of ovarian cancer cells. Int. J. Cancer 2014, 134, 542–551. [Google Scholar] [CrossRef]
- Li, M.H.; Wu, Z.Y.; Wang, Y.; Chen, F.Z.; Liu, Y. Expression of miR-29 and STAT3 in osteosarcoma and its effect on proliferation regulation of osteosarcoma cells. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7275–7282. [Google Scholar] [CrossRef]
- Ji, Q.; Hao, X.; Zhang, M.; Tang, W.; Yang, M.; Li, L.; Xiang, D.; Desano, J.T.; Bommer, G.T.; Fan, D.; et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS ONE 2009, 4, e6816. [Google Scholar] [CrossRef]
- Corney, D.C.; Hwang, C.I.; Matoso, A.; Vogt, M.; Flesken-Nikitin, A.; Godwin, A.K.; Kamat, A.A.; Sood, A.K.; Ellenson, L.H.; Hermeking, H.; et al. Frequent downregulation of miR-34 family in human ovarian cancers. Clin. Cancer Res. 2010, 16, 1119–1128. [Google Scholar] [CrossRef]
- Wu, X.; Cheng, Y.L.; Matthen, M.; Yoon, A.; Schwartz, G.K.; Bala, S.; Taylor, A.M.; Momen-Heravi, F. Down-regulation of the tumor suppressor miR-34a contributes to head and neck cancer by up-regulating the MET oncogene and modulating tumor immune evasion. J. Exp. Clin. Cancer Res. 2021, 40, 70. [Google Scholar] [CrossRef]
- Sun, D.; Wu, Y.; Zhang, S.; Han, Y.; Shen, J.; Zheng, W.; Wei, L.; Liu, Y.; Ren, L.; Gu, Z.; et al. Distinct roles of miR-34 family members on suppression of lung squamous cell carcinoma. Biomed. Pharmacother. 2021, 142, 111967. [Google Scholar] [CrossRef]
- Shi, L.; Fan, B.; Chen, D.; Guo, C.; Xiang, H.; Nie, Y.; Zhong, D.; Shi, X. Human cytomegalovirus protein UL136 activates the IL-6/STAT3 signal through MiR-138 and MiR-34c in gastric cancer cells. Int. J. Clin. Oncol. 2020, 25, 1936–1944. [Google Scholar] [CrossRef] [PubMed]
- Dehghan, R.; Najafi, R.; Azizi Jalilian, F.; Saidijam, M.; Radaei, Z.; Zamani, A.; Ezati, R.; Asna-Ashari, F.; Amini, R. A promising effect of zerumbone with improved anti-tumor-promoting inflammation activity of miR-34a in colorectal cancer cell lines. Mol. Biol. Rep. 2021, 48, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Christensen, L.L.; Tobiasen, H.; Holm, A.; Schepeler, T.; Ostenfeld, M.S.; Thorsen, K.; Rasmussen, M.H.; Birkenkamp-Demtroeder, K.; Sieber, O.M.; Gibbs, P.; et al. MiRNA-362-3p induces cell cycle arrest through targeting of E2F1, USF2 and PTPN1 and is associated with recurrence of colorectal cancer. Int. J. Cancer 2013, 133, 67–78. [Google Scholar] [CrossRef]
- Wahdan-Alaswad, R.S.; Cochrane, D.R.; Spoelstra, N.S.; Howe, E.N.; Edgerton, S.M.; Anderson, S.M.; Thor, A.D.; Richer, J.K. Metformin-Induced Killing of Triple-Negative Breast Cancer Cells Is Mediated by Reduction in Fatty Acid Synthase via miRNA-193b. Horm. Cancer 2014, 5, 374–389. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Kong, F.; Wu, K.; Song, K.; He, J.; Sun, W. miR-193b directly targets STMN1 and uPA genes and suppresses tumor growth and metastasis in pancreatic cancer. Mol. Med. Rep. 2014, 10, 2613–2620. [Google Scholar] [CrossRef]
- Hashemi, M.; Moradi, N.; Ziaee, S.A.; Narouie, B.; Soltani, M.H.; Rezaei, M.; Shahkar, G.; Taheri, M. Association between single nucleotide polymorphism in miR-499, miR-196a2, miR-146a and miR-149 and prostate cancer risk in a sample of Iranian population. J. Adv. Res. 2016, 7, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.G.; Zhou, X.M.; Cui, Z.G.; Hou, G. Effects of common polymorphisms in miR-146a and miR-196a2 on lung cancer susceptibility: A meta-analysis. J. Thorac. Dis. 2016, 8, 1297–1305. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhu, G.; Di, H.; Li, H.; Liu, X.; Zhao, M.; Zhang, Z.; Yang, Y. Associations between genetic variants located in mature microRNAs and risk of lung cancer. Oncotarget 2016, 7, 41715–41724. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Wang, N.; Duan, G. Long non-coding RNA XIST regulates ovarian cancer progression via modulating miR-335/BCL2L2 axis. World J. Surg. Oncol. 2021, 19, 165. [Google Scholar] [CrossRef]
- Eckstein, S. Dogs and Cancer: Get the Facts. Available online: https://pets.webmd.com/dogs/guide/dogs-and-cancer-get-the-facts#1 (accessed on 20 May 2023).
- Uhl, E.; Krimer, P.; Schliekelman, P.; Tompkins, S.M.; Suter, S. Identification of altered MicroRNA expression in canine lymphoid cell lines and cases of B- and T-Cell lymphomas. Genes Chromosomes Cancer 2011, 50, 950–967. [Google Scholar] [CrossRef]
- Fenger, J.M.; Bear, M.D.; Volinia, S.; Lin, T.Y.; Harrington, B.K.; London, C.A.; Kisseberth, W.C. Overexpression of miR-9 in mast cells is associated with invasive behavior and spontaneous metastasis. BMC Cancer 2014, 14, 84. [Google Scholar] [CrossRef]
- Noguchi, S.; Mori, T.; Hoshino, Y.; Yamada, N.; Maruo, K.; Akao, Y. MicroRNAs as tumour suppressors in canine and human melanoma cells and as a prognostic factor in canine melanomas. Vet. Comp. Oncol. 2013, 11, 113–123. [Google Scholar] [CrossRef]
- Noguchi, S.; Mori, T.; Hoshino, Y.; Yamada, N.; Nakagawa, T.; Sasaki, N.; Akao, Y.; Maruo, K. Comparative study of anti-oncogenic microRNA-145 in canine and human malignant melanoma. J. Vet. Med. Sci. 2012, 74, 1–8. [Google Scholar] [CrossRef]
- Gioia, G.; Mortarino, M.; Gelain, M.E.; Albonico, F.; Ciusani, E.; Forno, I.; Marconato, L.; Martini, V.; Comazzi, S. Immunophenotype-related microRNA expression in canine chronic lymphocytic leukemia. Vet. Immunol. Immunopathol. 2011, 142, 228–235. [Google Scholar] [CrossRef]
- Grimes, J.A.; Prasad, N.; Levy, S.; Cattley, R.; Lindley, S.; Boothe, H.W.; Henderson, R.A.; Smith, B.F. A comparison of microRNA expression profiles from splenic hemangiosarcoma, splenic nodular hyperplasia, and normal spleens of dogs. BMC Vet. Res. 2016, 12, 272. [Google Scholar] [CrossRef]
- Heishima, K.; Mori, T.; Sakai, H.; Sugito, N.; Murakami, M.; Yamada, N.; Akao, Y.; Maruo, K. MicroRNA-214 Promotes Apoptosis in Canine Hemangiosarcoma by Targeting the COP1-p53 Axis. PLoS ONE 2015, 10, e0137361. [Google Scholar] [CrossRef]
- Lutful Kabir, F.M.; DeInnocentes, P.; Bird, R.C. Altered microRNA Expression Profiles and Regulation of INK4A/CDKN2A Tumor Suppressor Genes in Canine Breast Cancer Models. J. Cell. Biochem. 2015, 116, 2956–2969. [Google Scholar] [CrossRef]
- Boggs, R.M.; Wright, Z.M.; Stickney, M.J.; Porter, W.W.; Murphy, K.E. MicroRNA expression in canine mammary cancer. Mamm. Genome 2008, 19, 561–569. [Google Scholar] [CrossRef]
- von Deetzen, M.C.; Schmeck, B.T.; Gruber, A.D.; Klopfleisch, R. Malignancy Associated MicroRNA Expression Changes in Canine Mammary Cancer of Different Malignancies. ISRN Vet. Sci. 2014, 2014, 148597. [Google Scholar] [CrossRef] [PubMed]
- Negrini, M.; Calin, G.A. Breast cancer metastasis: A microRNA story. Breast Cancer Res. 2008, 10, 203. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Reinhardt, F.; Pan, E.; Soutschek, J.; Bhat, B.; Marcusson, E.G.; Teruya-Feldstein, J.; Bell, G.W.; Weinberg, R.A. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat. Biotechnol. 2010, 28, 341–347. [Google Scholar] [CrossRef]
- Ma, T.; Yang, L.; Zhang, J. MiRNA5423p downregulation promotes trastuzumab resistance in breast cancer cells via AKT activation. Oncol. Rep. 2015, 33, 1215–1220. [Google Scholar] [CrossRef]
- Wang, W.; Zhao, L.; Wei, X.; Wang, L.; Liu, S.; Yang, Y.; Wang, F.; Sun, G.; Zhang, J.; Ma, Y.; et al. MicroRNA-320a promotes 5-FU resistance in human pancreatic cancer cells. Sci. Rep. 2016, 6, 27641. [Google Scholar] [CrossRef] [PubMed]
- Teplyuk, N.M.; Uhlmann, E.J.; Gabriely, G.; Volfovsky, N.; Wang, Y.; Teng, J.; Karmali, P.; Marcusson, E.; Peter, M.; Mohan, A.; et al. Therapeutic potential of targeting microRNA-10b in established intracranial glioblastoma: First steps toward the clinic. EMBO Mol. Med. 2016, 8, 268–287. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Wang, J.G.; Zhang, L.; Yang, H.P.; Wang, L.; Ding, D.; Chen, Q.; Yang, W.L.; Ren, K.H.; Zhou, D.M.; et al. MicroRNA-320a inhibits breast cancer metastasis by targeting metadherin. Oncotarget 2016, 7, 38612–38625. [Google Scholar] [CrossRef]
- Lynch, J.; Fay, J.; Meehan, M.; Bryan, K.; Watters, K.M.; Murphy, D.M.; Stallings, R.L. MiRNA-335 suppresses neuroblastoma cell invasiveness by direct targeting of multiple genes from the non-canonical TGF-β signalling pathway. Carcinogenesis 2012, 33, 976–985. [Google Scholar] [CrossRef]
- Bulkowska, M.; Rybicka, A.; Senses, K.M.; Ulewicz, K.; Witt, K.; Szymanska, J.; Taciak, B.; Klopfleisch, R.; Hellmen, E.; Dolka, I.; et al. MicroRNA expression patterns in canine mammary cancer show significant differences between metastatic and non-metastatic tumours. BMC Cancer 2017, 17, 728. [Google Scholar] [CrossRef]
- Ren, X.; Fan, Y.; Shi, D.; Xu, E.; Liu, Y. MicroRNA-124 inhibits canine mammary carcinoma cell proliferation, migration and invasion by targeting CDH2. Res. Vet. Sci. 2022, 146, 5–14. [Google Scholar] [CrossRef]
- Hino, Y.; Rahman, M.M.; Lai, Y.C.; Husna, A.A.; Chen, H.W.; Hasan, M.N.; Nakagawa, T.; Miura, N. Hypoxic miRNAs expression are different between primary and metastatic melanoma cells. Gene 2021, 782, 145552. [Google Scholar] [CrossRef]
- Sarver, A.L.; Thayanithy, V.; Scott, M.C.; Cleton-Jansen, A.M.; Hogendoorn, P.C.; Modiano, J.F.; Subramanian, S. MicroRNAs at the human 14q32 locus have prognostic significance in osteosarcoma. Orphanet J. Rare Dis. 2013, 8, 7. [Google Scholar] [CrossRef]
- Leonardo, L.; Laura, P.; Serena, B.M. miR-1 and miR-133b expression in canine osteosarcoma. Res. Vet. Sci. 2018, 117, 133–137. [Google Scholar] [CrossRef]
- Lopez, C.M.; Yu, P.Y.; Zhang, X.; Yilmaz, A.S.; London, C.A.; Fenger, J.M. MiR-34a regulates the invasive capacity of canine osteosarcoma cell lines. PLoS ONE 2018, 13, e0190086. [Google Scholar] [CrossRef]
- Fenger, J.M.; Roberts, R.D.; Iwenofu, O.H.; Bear, M.D.; Zhang, X.; Couto, J.I.; Modiano, J.F.; Kisseberth, W.C.; London, C.A. MiR-9 is overexpressed in spontaneous canine osteosarcoma and promotes a metastatic phenotype including invasion and migration in osteoblasts and osteosarcoma cell lines. BMC Cancer 2016, 16, 784. [Google Scholar] [CrossRef]
- Xu, X.; Zhu, Y.; Liang, Z.; Li, S.; Xu, X.; Wang, X.; Wu, J.; Hu, Z.; Meng, S.; Liu, B.; et al. c-Met and CREB1 are involved in miR-433-mediated inhibition of the epithelial-mesenchymal transition in bladder cancer by regulating Akt/GSK-3beta/Snail signaling. Cell Death Dis. 2016, 7, e2088. [Google Scholar] [CrossRef]
- Takasaki, S. Roles of microRNAs in cancers and development. Methods Mol. Biol. 2015, 1218, 375–413. [Google Scholar] [CrossRef]
- Agarwal, P.; Crepps, M.P.; Stahr, N.A.; Kretzschmar, W.P.; Harris, H.C.; Prasad, N.; Levy, S.E.; Smith, B.F. Identification of canine circulating miRNAs as tumor biospecific markers using Next-Generation Sequencing and Q-RT-PCR. Biochem. Biophys. Rep. 2021, 28, 101106. [Google Scholar] [CrossRef]
- Yao, S.; Zhao, X.; Wang, L.; Chen, F.; Gong, H.; Chen, C.; Cai, C. pH-activated DNA nanomachine for miRNA-21 imaging to accurately identify cancer cell. Mikrochim. Acta 2022, 189, 266. [Google Scholar] [CrossRef]
- Liu, C.G.; Calin, G.A.; Meloon, B.; Gamliel, N.; Sevignani, C.; Ferracin, M.; Dumitru, C.D.; Shimizu, M.; Zupo, S.; Dono, M.; et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc. Natl. Acad. Sci. USA 2004, 101, 9740–9744. [Google Scholar] [CrossRef]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef]
- Yanaihara, N.; Caplen, N.; Bowman, E.; Seike, M.; Kumamoto, K.; Yi, M.; Stephens, R.M.; Okamoto, A.; Yokota, J.; Tanaka, T.; et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006, 9, 189–198. [Google Scholar] [CrossRef]
- Rosenfeld, N.; Aharonov, R.; Meiri, E.; Rosenwald, S.; Spector, Y.; Zepeniuk, M.; Benjamin, H.; Shabes, N.; Tabak, S.; Levy, A.; et al. MicroRNAs accurately identify cancer tissue origin. Nat. Biotechnol. 2008, 26, 462–469. [Google Scholar] [CrossRef]
- Iwasaki, H.; Shimura, T.; Kitagawa, M.; Yamada, T.; Nishigaki, R.; Fukusada, S.; Okuda, Y.; Katano, T.; Horike, S.I.; Kataoka, H. A Novel Urinary miRNA Biomarker for Early Detection of Colorectal Cancer. Cancers 2022, 14, 461. [Google Scholar] [CrossRef]
- Bidarra, D.; Constancio, V.; Barros-Silva, D.; Ramalho-Carvalho, J.; Moreira-Barbosa, C.; Antunes, L.; Mauricio, J.; Oliveira, J.; Henrique, R.; Jeronimo, C. Circulating MicroRNAs as Biomarkers for Prostate Cancer Detection and Metastasis Development Prediction. Front. Oncol. 2019, 9, 900. [Google Scholar] [CrossRef]
- Downs-Holmes, C.; Silverman, P. Breast cancer: Overview & updates. Nurse Pr. 2011, 36, 20–26, quiz 7. [Google Scholar] [CrossRef]
- Vanacore, D.; Boccellino, M.; Rossetti, S.; Cavaliere, C.; D’Aniello, C.; Di Franco, R.; Romano, F.J.; Montanari, M.; La Mantia, E.; Piscitelli, R.; et al. Micrornas in prostate cancer: An overview. Oncotarget 2017, 8, 50240–50251. [Google Scholar] [CrossRef]
- Guo, X.; Han, T.; Hu, P.; Guo, X.; Zhu, C.; Wang, Y.; Chang, S. Five microRNAs in serum as potential biomarkers for prostate cancer risk assessment and therapeutic intervention. Int. Urol. Nephrol. 2018, 50, 2193–2200. [Google Scholar] [CrossRef]
- Jackson, B.L.; Grabowska, A.; Ratan, H.L. MicroRNA in prostate cancer: Functional importance and potential as circulating biomarkers. BMC Cancer 2014, 14, 930. [Google Scholar] [CrossRef]
- Lyu, J.; Zhao, L.; Wang, F.; Ji, J.; Cao, Z.; Xu, H.; Shi, X.; Zhu, Y.; Zhang, C.; Guo, F.; et al. Discovery and Validation of Serum MicroRNAs as Early Diagnostic Biomarkers for Prostate Cancer in Chinese Population. Biomed. Res. Int. 2019, 2019, 9306803. [Google Scholar] [CrossRef]
- Ogata-Kawata, H.; Izumiya, M.; Kurioka, D.; Honma, Y.; Yamada, Y.; Furuta, K.; Gunji, T.; Ohta, H.; Okamoto, H.; Sonoda, H.; et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE 2014, 9, e92921. [Google Scholar] [CrossRef]
- Fogli, S.; Polini, B.; Carpi, S.; Pardini, B.; Naccarati, A.; Dubbini, N.; Lanza, M.; Breschi, M.C.; Romanini, A.; Nieri, P. Identification of plasma microRNAs as new potential biomarkers with high diagnostic power in human cutaneous melanoma. Tumor Biol. 2017, 39, 1010428317701646. [Google Scholar] [CrossRef]
- Xie, Z.; Chen, G.; Zhang, X.; Li, D.; Huang, J.; Yang, C.; Zhang, P.; Qin, Y.; Duan, Y.; Gong, B.; et al. Salivary microRNAs as promising biomarkers for detection of esophageal cancer. PLoS ONE 2013, 8, e57502. [Google Scholar] [CrossRef]
- Humeau, M.; Vignolle-Vidoni, A.; Sicard, F.; Martins, F.; Bournet, B.; Buscail, L.; Torrisani, J.; Cordelier, P. Salivary MicroRNA in Pancreatic Cancer Patients. PLoS ONE 2015, 10, e0130996. [Google Scholar] [CrossRef]
- Yu, S.L.; Chen, H.Y.; Chang, G.C.; Chen, C.Y.; Chen, H.W.; Singh, S.; Cheng, C.L.; Yu, C.J.; Lee, Y.C.; Chen, H.S.; et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell 2008, 13, 48–57. [Google Scholar] [CrossRef]
- Schetter, A.J.; Leung, S.Y.; Sohn, J.J.; Zanetti, K.A.; Bowman, E.D.; Yanaihara, N.; Yuen, S.T.; Chan, T.L.; Kwong, D.L.; Au, G.K.; et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 2008, 299, 425–436. [Google Scholar] [CrossRef]
- Gahlawat, A.W.; Witte, T.; Haarhuis, L.; Schott, S. A novel circulating miRNA panel for non-invasive ovarian cancer diagnosis and prognosis. Br. J. Cancer 2022, 127, 1550–1556. [Google Scholar] [CrossRef]
- Farran, B.; Dyson, G.; Craig, D.; Dombkowski, A.; Beebe-Dimmer, J.L.; Powell, I.J.; Podgorski, I.; Heilbrun, L.; Bolton, S.; Bock, C.H. A study of circulating microRNAs identifies a new potential biomarker panel to distinguish aggressive prostate cancer. Carcinogenesis 2018, 39, 556–561. [Google Scholar] [CrossRef]
- Shukla, K.K.; Misra, S.; Pareek, P.; Mishra, V.; Singhal, B.; Sharma, P. Recent scenario of microRNA as diagnostic and prognostic biomarkers of prostate cancer. Urol. Oncol. 2017, 35, 92–101. [Google Scholar] [CrossRef]
- Maierthaler, M.; Benner, A.; Hoffmeister, M.; Surowy, H.; Jansen, L.; Knebel, P.; Chang-Claude, J.; Brenner, H.; Burwinkel, B. Plasma miR-122 and miR-200 family are prognostic markers in colorectal cancer. Int. J. Cancer 2017, 140, 176–187. [Google Scholar] [CrossRef]
- Monzo, M.; Santasusagna, S.; Moreno, I.; Martinez, F.; Hernandez, R.; Munoz, C.; Castellano, J.J.; Moreno, J.; Navarro, A. Exosomal microRNAs isolated from plasma of mesenteric veins linked to liver metastases in resected patients with colon cancer. Oncotarget 2017, 8, 30859–30869. [Google Scholar] [CrossRef]
- Chen, J.; Wang, W.; Zhang, Y.; Chen, Y.; Hu, T. Predicting distant metastasis and chemoresistance using plasma miRNAs. Med. Oncol. 2014, 31, 799. [Google Scholar] [CrossRef]
- Bhatnagar, N.; Li, X.; Padi, S.K.; Zhang, Q.; Tang, M.S.; Guo, B. Downregulation of miR-205 and miR-31 confers resistance to chemotherapy-induced apoptosis in prostate cancer cells. Cell Death Dis. 2010, 1, e105. [Google Scholar] [CrossRef]
- Li, B.; Shi, X.B.; Nori, D.; Chao, C.K.; Chen, A.M.; Valicenti, R.; White Rde, V. Down-regulation of microRNA 106b is involved in p21-mediated cell cycle arrest in response to radiation in prostate cancer cells. Prostate 2011, 71, 567–574. [Google Scholar] [CrossRef]
- Mao, A.; Zhao, Q.; Zhou, X.; Sun, C.; Si, J.; Zhou, R.; Gan, L.; Zhang, H. MicroRNA-449a enhances radiosensitivity by downregulation of c-Myc in prostate cancer cells. Sci. Rep. 2016, 6, 27346. [Google Scholar] [CrossRef]
- Nagesh, P.K.B.; Chowdhury, P.; Hatami, E.; Boya, V.K.N.; Kashyap, V.K.; Khan, S.; Hafeez, B.B.; Chauhan, S.C.; Jaggi, M.; Yallapu, M.M. miRNA-205 Nanoformulation Sensitizes Prostate Cancer Cells to Chemotherapy. Cancers 2018, 10, 289. [Google Scholar] [CrossRef]
- Ni, J.; Bucci, J.; Chang, L.; Malouf, D.; Graham, P.; Li, Y. Targeting MicroRNAs in Prostate Cancer Radiotherapy. Theranostics 2017, 7, 3243–3259. [Google Scholar] [CrossRef]
- Thieu, W.; Tilki, D.; de Vere White, R.; Evans, C.P. The role of microRNA in castration-resistant prostate cancer. Urol. Oncol. 2014, 32, 517–523. [Google Scholar] [CrossRef]
- Wang, Y.; Lieberman, R.; Pan, J.; Zhang, Q.; Du, M.; Zhang, P.; Nevalainen, M.; Kohli, M.; Shenoy, N.K.; Meng, H.; et al. miR-375 induces docetaxel resistance in prostate cancer by targeting SEC23A and YAP1. Mol. Cancer 2016, 15, 70. [Google Scholar] [CrossRef]
- Hu, Q.; Gong, J.P.; Li, J.; Zhong, S.L.; Chen, W.X.; Zhang, J.Y.; Ma, T.F.; Ji, H.; Lv, M.M.; Zhao, J.H.; et al. Down-regulation of miRNA-452 is associated with adriamycin-resistance in breast cancer cells. Asian Pac. J. Cancer Prev. 2014, 15, 5137–5142. [Google Scholar] [CrossRef]
- Safaei, S.; Amini, M.; Najjary, S.; Mokhtarzadeh, A.; Bolandi, N.; Saeedi, H.; Alizadeh, N.; Javadrashid, D.; Baradaran, B. miR-200c increases the sensitivity of breast cancer cells to Doxorubicin through downregulating MDR1 gene. Exp. Mol. Pathol. 2022, 125, 104753. [Google Scholar] [CrossRef]
- Hu, Y.; Qiu, Y.; Yagüe, E.; Ji, W.; Liu, J.; Zhang, J. miRNA-205 targets VEGFA and FGF2 and regulates resistance to chemotherapeutics in breast cancer. Cell Death Dis. 2016, 7, e2291. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.H.; Tao, Z.H.; Zhang, J.; Li, T.; Ni, C.; Xie, J.; Zhang, J.F.; Hu, X.C. MiRNA-21 induces epithelial to mesenchymal transition and gemcitabine resistance via the PTEN/AKT pathway in breast cancer. Tumor Biol. 2016, 37, 7245–7254. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.; Xue, A.; Chi, Y.; Xue, J.; Wang, W.; Zhao, Z.; Fan, M.; Yang, C.H.; Shao, Z.M.; Pfeffer, L.M.; et al. Induction of miRNA-181a by genotoxic treatments promotes chemotherapeutic resistance and metastasis in breast cancer. Oncogene 2016, 35, 1302–1313. [Google Scholar] [CrossRef]
- Liu, H.; Wang, Y.; Li, X.; Zhang, Y.J.; Li, J.; Zheng, Y.Q.; Liu, M.; Song, X.; Li, X.R. Expression and regulatory function of miRNA-182 in triple-negative breast cancer cells through its targeting of profilin 1. Tumor Biol. 2013, 34, 1713–1722. [Google Scholar] [CrossRef]
- Xue, J.; Chi, Y.; Chen, Y.; Huang, S.; Ye, X.; Niu, J.; Wang, W.; Pfeffer, L.M.; Shao, Z.M.; Wu, Z.H.; et al. MiRNA-621 sensitizes breast cancer to chemotherapy by suppressing FBXO11 and enhancing p53 activity. Oncogene 2016, 35, 448–458. [Google Scholar] [CrossRef]
- Mohammadi Yeganeh, S.; Vasei, M.; Tavakoli, R.; Kia, V.; Paryan, M. The effect of miR-340 over-expression on cell-cycle-related genes in triple-negative breast cancer cells. Eur. J. Cancer Care 2017, 26, e12496. [Google Scholar] [CrossRef] [PubMed]
- Nordentoft, I.; Birkenkamp-Demtroder, K.; Agerbaek, M.; Theodorescu, D.; Ostenfeld, M.S.; Hartmann, A.; Borre, M.; Orntoft, T.F.; Dyrskjot, L. miRNAs associated with chemo-sensitivity in cell lines and in advanced bladder cancer. BMC Med. Genom. 2012, 5, 40. [Google Scholar] [CrossRef] [PubMed]
- Heishima, K.; Ichikawa, Y.; Yoshida, K.; Iwasaki, R.; Sakai, H.; Nakagawa, T.; Tanaka, Y.; Hoshino, Y.; Okamura, Y.; Murakami, M.; et al. Circulating microRNA-214 and -126 as potential biomarkers for canine neoplastic disease. Sci. Rep. 2017, 7, 2301. [Google Scholar] [CrossRef] [PubMed]
- Cleveland, M.J.; Casale, S. Incidence of malignancy and outcomes for dogs undergoing splenectomy for incidentally detected nonruptured splenic nodules or masses: 105 cases (2009–2013). J. Am. Vet. Med. Assoc. 2016, 248, 1267–1273. [Google Scholar] [CrossRef]
- Kent, M.S.; Zwingenberger, A.; Westropp, J.L.; Barrett, L.E.; Durbin-Johnson, B.P.; Ghosh, P.; Vinall, R.L. MicroRNA profiling of dogs with transitional cell carcinoma of the bladder using blood and urine samples. BMC Vet. Res. 2017, 13, 339. [Google Scholar] [CrossRef]
- Vinall, R.L.; Kent, M.S.; deVere White, R.W. Expression of microRNAs in urinary bladder samples obtained from dogs with grossly normal bladders, inflammatory bladder disease, or transitional cell carcinoma. Am. J. Vet. Res. 2012, 73, 1626–1633. [Google Scholar] [CrossRef]
- Zamarian, V.; Ferrari, R.; Stefanello, D.; Ceciliani, F.; Grieco, V.; Minozzi, G.; Chiti, L.E.; Arigoni, M.; Calogero, R.; Lecchi, C. miRNA profiles of canine cutaneous mast cell tumours with early nodal metastasis and evaluation as potential biomarkers. Sci. Rep. 2020, 10, 18918. [Google Scholar] [CrossRef]
- Fish, E.J.; Martinez-Romero, E.G.; DeInnocentes, P.; Koehler, J.W.; Prasad, N.; Smith, A.N.; Bird, R.C. Circulating microRNA as biomarkers of canine mammary carcinoma in dogs. J. Vet. Intern. Med. 2020, 34, 1282–1290. [Google Scholar] [CrossRef]
- Garnica, T.K.; Lesbon, J.C.C.; Avila, A.; Rochetti, A.L.; Matiz, O.R.S.; Ribeiro, R.C.S.; Zoppa, A.; Nishiya, A.T.; Costa, M.T.; de Nardi, A.B.; et al. Liquid biopsy based on small extracellular vesicles predicts chemotherapy response of canine multicentric lymphomas. Sci. Rep. 2020, 10, 20371. [Google Scholar] [CrossRef]
- Beaumier, A.; Robinson, S.R.; Robinson, N.; Lopez, K.E.; Meola, D.M.; Barber, L.G.; Bulmer, B.J.; Calvalido, J.; Rush, J.E.; Yeri, A.; et al. Extracellular vesicular microRNAs as potential biomarker for early detection of doxorubicin-induced cardiotoxicity. J. Vet. Intern. Med. 2020, 34, 1260–1271. [Google Scholar] [CrossRef]
- Husna, A.A.; Rahman, M.M.; Lai, Y.C.; Chen, H.W.; Hasan, M.N.; Nakagawa, T.; Miura, N. Identification of melanoma-specific exosomal miRNAs as the potential biomarker for canine oral melanoma. Pigment Cell Melanoma Res. 2021, 34, 1062–1073. [Google Scholar] [CrossRef] [PubMed]
- Narita, M.; Nishida, H.; Asahina, R.; Nakata, K.; Yano, H.; Dickinson, P.J.; Tanaka, T.; Akiyoshi, H.; Maeda, S.; Kamishina, H. Expression of microRNAs in plasma and in extracellular vesicles derived from plasma for dogs with glioma and dogs with other brain diseases. Am. J. Vet. Res. 2020, 81, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Starkey, M.P.; Compston-Garnett, L.; Malho, P.; Dunn, K.; Dubielzig, R. Metastasis-associated microRNA expression in canine uveal melanoma. Vet. Comp. Oncol. 2018, 16, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Leonardi, L.; Scotlandi, K.; Pettinari, I.; Benassi, M.S.; Porcellato, I.; Pazzaglia, L. MiRNAs in Canine and Human Osteosarcoma: A Highlight Review on Comparative Biomolecular Aspects. Cells 2021, 10, 428. [Google Scholar] [CrossRef]
- Sethi, S.; Ali, S.; Sethi, S.; Sarkar, F.H. MicroRNAs in personalized cancer therapy. Clin. Genet. 2014, 86, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Costa, P.M.; Pedroso de Lima, M.C. MicroRNAs as Molecular Targets for Cancer Therapy: On the Modulation of MicroRNA Expression. Pharmaceuticals 2013, 6, 1195–1220. [Google Scholar] [CrossRef]
- Liu, Y.; Zheng, M.; Jiao, M.; Yan, C.; Xu, S.; Du, Q.; Morsch, M.; Yin, J.; Shi, B. Polymeric nanoparticle mediated inhibition of miR-21 with enhanced miR-124 expression for combinatorial glioblastoma therapy. Biomaterials 2021, 276, 121036. [Google Scholar] [CrossRef]
- Yan, S.; Liu, G.; Pei, C.; Chen, W.; Li, P.; Wang, Q.; Jin, X.; Zhu, J.; Wang, M.; Liu, X. Inhibition of NADPH oxidase protects against metastasis of human lung cancer by decreasing microRNA-21. Anticancer Drugs 2015, 26, 388–398. [Google Scholar] [CrossRef]
- Dong, C.G.; Wu, W.K.; Feng, S.Y.; Wang, X.J.; Shao, J.F.; Qiao, J. Co-inhibition of microRNA-10b and microRNA-21 exerts synergistic inhibition on the proliferation and invasion of human glioma cells. Int. J. Oncol. 2012, 41, 1005–1012. [Google Scholar] [CrossRef]
- Cheng, C.J.; Bahal, R.; Babar, I.A.; Pincus, Z.; Barrera, F.; Liu, C.; Svoronos, A.; Braddock, D.T.; Glazer, P.M.; Engelman, D.M.; et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature 2015, 518, 107–110. [Google Scholar] [CrossRef]
- Song, H.; Oh, B.; Choi, M.; Oh, J.; Lee, M. Delivery of anti-microRNA-21 antisense-oligodeoxynucleotide using amphiphilic peptides for glioblastoma gene therapy. J. Drug Target. 2015, 23, 360–370. [Google Scholar] [CrossRef]
- Bahreyni, A.; Liu, H.; Mohamud, Y.; Xue, Y.C.; Zhang, J.; Luo, H. A new miRNA-Modified coxsackievirus B3 inhibits triple negative breast cancer growth with improved safety profile in immunocompetent mice. Cancer Lett. 2022, 548, 215849. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Zhou, X.; Mei, M.; Yuan, X.B.; Han, L.; Wang, G.X.; Jia, Z.F.; Xu, P.; Pu, P.Y.; Kang, C.S. MicroRNA-21 inhibitor sensitizes human glioblastoma cells U251 (PTEN-mutant) and LN229 (PTEN-wild type) to taxol. BMC Cancer 2010, 10, 27. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, R.; Maeda, A.; Ueno, Y.; Sakai, H.; Kimura, S.; Sawadaishi, T.; Kohgo, S.; Yamada, K.; Mori, T. Intraperitoneal administration of synthetic microRNA-214 elicits tumor suppression in an intraperitoneal dissemination mouse model of canine hemangiosarcoma. Vet. Res. Commun. 2022, 46, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, R.; Mori, T.; Noguchi, S.; Akao, Y.; Maruo, K.; Kitade, Y. Synthetic microRNA-205 exhibited tumour suppression in spontaneous canine malignant melanoma by intratumoral injection. Vet. Comp. Oncol. 2019, 17, 407–412. [Google Scholar] [CrossRef]
- Alegre, F.; Ormonde, A.R.; Snider, K.M.; Woolard, K.; Yu, A.M.; Wittenburg, L.A. A genetically engineered microRNA-34a prodrug demonstrates anti-tumor activity in a canine model of osteosarcoma. PLoS ONE 2018, 13, e0209941. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chakrabortty, A.; Patton, D.J.; Smith, B.F.; Agarwal, P. miRNAs: Potential as Biomarkers and Therapeutic Targets for Cancer. Genes 2023, 14, 1375. https://doi.org/10.3390/genes14071375
Chakrabortty A, Patton DJ, Smith BF, Agarwal P. miRNAs: Potential as Biomarkers and Therapeutic Targets for Cancer. Genes. 2023; 14(7):1375. https://doi.org/10.3390/genes14071375
Chicago/Turabian StyleChakrabortty, Atonu, Daniel J. Patton, Bruce F. Smith, and Payal Agarwal. 2023. "miRNAs: Potential as Biomarkers and Therapeutic Targets for Cancer" Genes 14, no. 7: 1375. https://doi.org/10.3390/genes14071375
APA StyleChakrabortty, A., Patton, D. J., Smith, B. F., & Agarwal, P. (2023). miRNAs: Potential as Biomarkers and Therapeutic Targets for Cancer. Genes, 14(7), 1375. https://doi.org/10.3390/genes14071375




