Abstract
Catechol-O-methyl transferase (COMT) gene variants are involved in different neuropsychiatric disorders and cognitive impairments, associated with altered dopamine function. This study investigated the genotypic and haplotypic association of COMT rs4680 and rs4618 polymorphisms with the severity of cognitive and other clinical symptoms in 544 male and 385 female subjects with schizophrenia. COMT rs4818 G carriers were more frequent in male patients with mild abstract thinking difficulties, compared to CC homozygotes or C allele carriers. Male carriers of COMT rs4680 A allele had worse abstract thinking (N5) scores than GG carriers, whereas AA homozygotes were more frequent in male subjects with lower scores on the intensity of the somatic concern (G1) item, compared to G carriers. Male carriers of COMT rs4818–rs4680 GA haplotype had the highest scores on the G1 item (somatic concern), whereas GG haplotype carriers had the lowest scores on G2 (anxiety) and G6 (depression) items. COMT GG haplotype was less frequent in female patients with severe disturbance of volition (G13 item) compared to the group with mild symptoms, while CG haplotype was more frequent in female patients with severe then mild symptoms. These findings suggest the sex-specific genotypic and haplotypic association of COMT variants with a severity of cognitive and other clinical symptoms of schizophrenia.
Keywords:
COMT; clinical symptoms; cognition; haplotype; polymorphisms; schizophrenia; sex-differences 1. Introduction
Schizophrenia, one of the most severe mental disorders, is associated with the imbalance between resilience and vulnerability in relation with life stressors [1,2,3]. Schizophrenia is characterized by positive, negative and cognitive symptoms, according to the DSM-5 criteria [4]. Cognitive symptoms include disturbances in executive function, working and verbal memory, language, learning, attention, processing speed, vigilance, and problem solving [4,5,6,7], leading to decreased social functioning and adjustment and employment problems [4,5,7,8]. Cognition is affected by different neurotransmitter systems [9], with the important role of various proteins and genes [10].
Dopamine is associated with neurobiological underpinning of schizophrenia [11], and with cognitive processing [12,13]. Catechol-O-methyl transferase or COMT is an enzyme that degrades dopamine, noradrenaline, and adrenaline, and acts as an important modulator of the brain function, especially in the prefrontal cortex [14,15], the brain region engaged in most of the cognitive processes. COMT is an enzyme involved in the inactivation of compounds having a catechol structure, including catecholamine neurotransmitters such as dopamine, by introducing a methyl group, donated by S-adenosyl methionine, to the catecholamine. The active site of COMT consists of the S-adenosyl-l-methionine (SAM) binding domain and the catalytic site. The catalytic site contains a metal ion (Mg2+) and amino acids important for substrate binding and catalysis of the methylation reaction. The polymorphic residue according to rs4680 is buried in a hydrophobic residue, around 16A° away from the SAM-binding site [14], and in a complementary hydrophobic methyl binding pocket according to rs4818 [15].
One of the most frequently investigated genes, associated with cognitive phenotypes, is the COMT gene. Therefore, COMT gene and its polymorphisms have been studied in different neuropsychiatric disorders and cognitive impairments, associated with altered dopamine function [16]. The most often studied is a functional polymorphism COMT Val158Met (rs4680) [17].
Specifically, COMT rs4680 polymorphism, or a G/A substitution, leads to valine’s (Val) replacement with methionine (Met) at codon 158 of the membrane-bound COMT (MB-COMT) and at codon 108 of the soluble short form (S-COMT) [18], and results in a significant (three- to four-fold) fall in the COMT activity in the A (Met) carriers. Another COMT polymorphism, COMT rs4818 polymorphism is located on exon 4, and consists of a C/G substitution (Leu/Leu) at codon 86 of the S-COMT and at codon 136 of the MB-COMT [19]. Since GG carriers have higher COMT activity than the CC carriers of this polymorphism, presence of the G variant is related to greater COMT activity and reduced prefrontal dopamine activity [19]. It is assumed that COMT activity is more under influence of the COMT rs4818 than COMT rs4680 polymorphism [20]. Genotypic [21,22] and haplotypic [23] associations of the COMT were shown for particular features in schizophrenia. Besides GWAS, that need thousands of samples and multicenter studies [24], haplotype-based studies might detect association or modulation of specific domains of symptoms of schizophrenia, compared to negative association studies using a single SNP or individual genotypes related to COMT rs4680 [25] or COMT rs4818 [26,27]. Haplotype associations have been reported for anhedonia [28], and for the treatment response [29,30].
Inconsistent findings across studies might also be associated, among many other factors, with the sexually dimorphic influence of COMT upon various psychiatric symptoms [31] and sex related differences in schizophrenia [28].
In the present study, we controlled COMT rs4680 and rs4818 polymorphisms data for the possible effects of sex, age, and smoking, and evaluated all 929 subjects (544 male and 385 female participants) with chronic schizophrenia in unrelated Caucasian subjects, using the Positive and Negative Syndrome Scale (PANSS) [32]. We determined this association with PANSS cognition subscale [33] symptoms (i.e., with P2 = Conceptual disorganization, N5 = Abstract thinking, G10 = Disorientation, G11 = Attention problems) [34], The PANSS symptom level deconstruction established the utility of the use of the selective cognitive items and other negative or general psychopathology items listed from the PANSS [35].
Regarding genetic models, we assessed an association of symptoms in schizophrenia with COMT rs4680 or COMT 4818 polymorphisms using genotypic, allelic, dominant (AA + GA vs. GG for COMT rs4680 and CG + GG vs. GG for COMT rs4818) and recessive (AA vs. GA + GG for COMT rs4680 and CC vs. CG + GG for COMT rs4818) models, respectively. We expected that COMT variants would be associated with cognitive, negative or, general PANSS psychopathology in our patients. We hypothesized that the presence of the G allele of the COMT rs4680 or rs4818 polymorphism, both related to higher COMT activity and reduced dopaminergic function, will be associated with pronounced cognitive decline and more severe clinical symptoms in schizophrenia, compared to presence of the A or C allele of the COMT rs4680 or rs4818 polymorphisms, respectively.
2. Materials and Methods
2.1. Subjects
The study included 929 subjects (544 male and 385 female participants) with schizophrenia, and some of them were included in our previous studies [28,29,30]. Patients were included in the study if they had schizophrenia for at least 5 years, and if they were 18–65 years old. Before participation, patients were informed in detail about the procedures, and after they had signed the written informed consent, the study was carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki from 1975). Both in- and out-patients participated. Exclusion criteria were intellectual disabilities, first-episode psychosis, substance abuse and/or dependence in the previous three months, and if they had any comorbid severe somatic or neurological disorder.
They were diagnosed using the Structured Clinical Interview for DSM-IV (SCID) [36], while the severity of clinical symptoms was evaluated using the gold standard, the Positive and Negative Syndrome Scale (PANSS) [32], and the presence of cognitive decline was determined with PANSS cognitive subscale, that includes items P2 (conceptual disorganization), N5 (abstract thinking), G10 (disorientation), and G11 (attention problems) [33,34]. Additionally, subjects were subdivided into those with mild and moderate to severe symptoms with cut off points: 4 for particular PANSS items, i.e., 16 for PANSS cognitive subscale, 28 for PANSS positive and negative subscale, and 64 for PANSS general subscale, as reported previously [37]. Average PANSS score was 103 (88; 124). All subjects were recruited from the University Hospitals from the Zagreb County (Department for Psychiatry and Psychological Medicine at the University Hospital Centre Zagreb and from Department for Biological Psychiatry and Psychogeriatrics, University Psychiatric Hospital Vrapce, Zagreb, Croatia). The study was conducted with the approval of the Ethics Committees of the University Hospital Center Zagreb and University Psychiatric Hospital Vrapce, Zagreb, Croatia.
2.2. Genotyping
All subjects were sampled during regular check-ups in the morning, and blood was processed on the same day. Genomic DNA was isolated from the peripheral blood using the salting-out method [38]. The COMT rs4818 (assay ID: C__2538750_10) and COMT rs4680 (assay ID: C__25746809_50) polymorphisms genotypes were determined using the primers and probes from Applied Biosystems as TaqMan® Drug Metabolism Genotyping Assays (Applied Biosystems, Foster City, CA, USA) on ABI Prism 7300 Real time PCR System apparatus (Applied Biosystems, Foster City, CA, USA), according to the procedures described by Applied Biosystems. Thermal cycler conditions were 10 min at 95 °C followed by 50 cycles of denaturation at 92 °C for 15 s and elongation at 60 °C for 90 s. The 10 μL reaction volume contained around 30 ng of DNA. Besides the codominant model, which includes all three genotypes, dominant, and recessive models for both COMT polymorphisms, as well as the allelic model, were used.
2.3. Statistical Analyses
The data were analyzed using Prism version 7.00 (GraphPad Software, Inc., San Diego, CA, USA). Due to the deviation from a normal distribution (determined with Kolmogorov–Smirnov test), nonparametric tests were used for all analyses. Multiple linear regression was performed to analyze the effect of sex, age, and smoking on severity of cognitive decline (PANSS cognitive scores) and severity of other individual items of the PANSS in patients with schizophrenia. The Mann–Whitney U test was used for independent pair comparisons and Kruskal–Wallis ANOVA, with post hoc Dunn test was used for multiple comparisons. Box-plot diagrams were used for visual representation of significant results, where the central box represented the interquartile range, the middle line represented the median, the vertical line extended from the minimum to the maximum value, while separate dots represented the outliers. Extreme values (more than 3 box-lengths outside of the box) were excluded from the analyses. Genotype, allele, and haplotype distribution among subjects with mild and severe symptoms of schizophrenia was calculated using the χ2-test, while the standardized residuals (R) were calculated to determine which parameter was the most significant contributor to the differences among groups [39].
The Hardy–Weinberg equilibrium for COMT rs4818 and rs4680 polymorphisms was calculated using χ2-test [40]. LD pairwise values for two COMT SNPs and haplotype frequencies were determined using the Haploview software v. 4.2 [41] and represented with D’ value. Loci are considered to be in linkage if D’ is >0.80. PLINK v. 1.07. software was used to assign best-estimate haplotype pairs to each individual using the expectation–maximization algorithm [42].
G*Power 3.1 Software (Germany) [43] was used to determine a priori sample size. For all two tailed analyses, the p-value (0.05/2 = 0.025) was corrected because two SNPs were compared and the results were considered significant if p < 0.025. Required sample size (with α = 0.025; power (1 − β) = 0.800; and a small effect size = 0.15) for Mann–Whitney test was N = 416, whereas for Kruskal–Wallis ANOVA (for 3 groups, the required sample size was N = 432; and for 4 groups, the required sample size was N = 492). As the study included N = 929 participants, it had adequate sample size and statistical power.
3. Results
Demographic Data
The study included a total of 929 subjects (544 male and 385 female participants) with schizophrenia. Male subjects were significantly younger (p < 0.001), and had more severe total (p < 0.001), positive (p < 0.001), negative (p < 0.001), and general (p < 0.001) symptoms of schizophrenia, as well as higher PANSS cognitive scores (p < 0.001), than female subjects (Table 1). Moreover, male subjects were more frequently smokers (69.1%, R = 2.1; p < 0.001) than female subjects (51.7%).

Table 1.
Demographic data of enrolled schizophrenia patients.
Multiple linear regression was performed to analyze the effect of sex, age, and smoking on severity of cognitive decline (PANSS cognitive scores) in patients with schizophrenia. Younger age (β = −0.211; p < 0.001) and male sex (β = −0.360; p < 0.001) were significant predictors of higher PANSS scores, while smoking (β = −0.006; p = 0.858) was not associated with PANSS cognitive scores (R2 = 0.228; F = 63.398; p < 0.001). Since male subjects were significantly younger than female, and there were significant differences in PANSS scores between two sexes, all analyses were performed in male and female subjects separately.
Genotype and allele frequencies of the rs4818 and rs4680 polymorphisms located in the COMT gene have been determined in total 929 subjects. Minor allele frequency (MAF) and corresponding Hardy–Weinberg equilibrium significance value for each polymorphism, as well as linkage disequilibrium between COMT rs4818 and rs4680 polymorphisms have been determined using Haploview software v. 4.2. MAFs for rs4818 and rs4680 were 0.386 (HW p value = 0.859) and 0.498 (HW p value = 0.658), respectively. Haplotype analysis showed strong LD between COMT rs4818 and rs4680 polymorphisms (D’ × 100 = 88; LOD > 2) (Figure 1), hence haplotypes for COMT rs4818 and rs4680 block were determined for each subject using expectation–maximization algorithm. The most common haplotype was CA, which was present in 48.0% of subjects, followed by GG (36.3%) and CG (13.4%) haplotypes. The rarest haplotype was GA (2.2%).
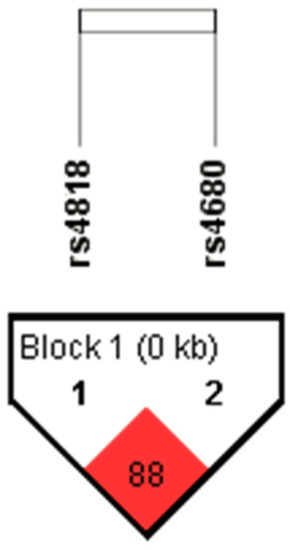
Figure 1.
The LD plot of the rs4818 and rs4680 polymorphisms located in the COMT gene. Pairwise LD value (×100), as denoted in a bright red rectangle (D’ = 88), indicates a strong link between COMT rs4818 and rs4680 polymorphisms.
There were no significant differences in the distribution of the COMT rs4818 and rs4680 genotypes, alleles, or haplotypes between male and female patients, as well as between smokers and non-smokers (Table 2).

Table 2.
The distribution of the genotypes and alleles of the COMT rs4818 and rs4680 polymorphisms, and COMT rs4818–rs4680 haplotype block in patients with schizophrenia divided by sex and smoking status.
The severity of the PANSS positive, negative, and general psychopathology symptoms of schizophrenia, and cognitive decline evaluated with PANSS cognitive subscale, that includes items P2 (conceptual disorganization), N5 (abstract thinking), G10 (disorientation), and G11 (attention problems), as well as scores on particular items of PANSS scale, were evaluated in regard to COMT rs4818 and rs4680 polymorphisms and COMT rs4818–rs4680 haplotype in male and female subjects separately.
Polymorphism COMT rs4818 was not associated with any of the symptoms of schizophrenia in male or in female subjects when the total scores were evaluated (Table 3 and Table 4). However, when subjects were divided according to the severity of symptoms into those with mild symptoms and those with moderate and severe symptoms (Table 5), G carriers were more often present (25.6%) in male subjects with mild difficulties in abstract thinking (item N5) compared to male CC carriers (17.2%) (R = 1.2; χ2 = 5.210; p = 0.022), or male C allele carriers (20.2%) (R = 1.6; χ2 = 5.042; p = 0.025). Additionally, nominal association of COMT rs4818 polymorphism and scores on G1 item (corresponding to the intensity of somatic concern) was observed, where G allele was under-represented (19.3%) in male subjects with mild symptoms (R = −1.4; χ2 = 4.186; p = 0.041) compared to C allele carriers (24.7%); however, after correction for multiple testing, this association did not reach statistical significance.

Table 3.
The scores on PANSS total, positive, negative, general psychopathology and cognitive subscales, as well as on particular PANSS items, in male subjects with schizophrenia divided by carriers of different genotypes, alleles and haplotypes of the COMT rs4818 and rs4680 polymorphisms.

Table 4.
The scores on PANSS total, positive, negative, general psychopathology, and cognitive subscales, as well as on particular PANSS items, in female subjects with schizophrenia divided by carriers of different genotypes, alleles and haplotypes of the COMT rs4818 and rs4680 polymorphisms.

Table 5.
The distribution of the genotypes and alleles of the COMT rs4818 and rs4680 polymorphisms, and their haplotype block, in male subjects with mild and severe symptoms of schizophrenia divided by scores on PANSS total, positive, negative, general psychopathology and cognitive subscales as well as on particular PANSS items.
Polymorphism COMT rs4680 was associated with N5 (abstract thinking) scores in male subjects, where A carriers had higher scores than GG carriers (U = 23575.5; p = 0.023 (Table 3, Figure 2a). As shown in Table 5, when subjects were divided according to the severity of symptoms, COMT rs4680 AA homozygotes were more frequent (18.1%) in subjects with lower scores on G1 (somatic concern) item (R = 1.7; χ2 = 5.061; p = 0.024), compared to G carriers (23.7%). However, this association was not confirmed in female patients (Table 6).
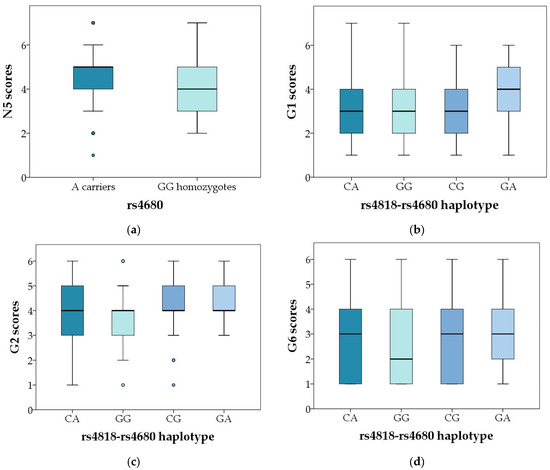
Figure 2.
Significant genotypic and haplotypic association of COMT rs4818 and rs4680 polymorphisms with particular symptoms of schizophrenia. (a) Scores in the PANSS N5 item in male carriers of different COMT rs4680 genotypes and alleles, p = 0.023; (b) PANSS G1 scores in male carriers of different COMT rs4818–4680 haplotypes; p = 0.011; (c) PANSS G2 scores in male carriers of different COMT rs4818–4680 haplotypes; p = 0.003; and (d) PANSS G6 scores in male carriers of different COMT rs4818–4680 haplotypes; p = 0.020, in the male carriers of different COMT rs4818–4680 haplotypes. Central box represents the interquartile range, the middle line represents the median, the vertical line extends from the minimum to the maximum value, while separate dots represent the outliers.

Table 6.
The distribution of the genotypes and alleles of the COMT rs4818 and rs4680 polymorphisms, and their haplotype block, in female subjects with mild and severe symptoms of schizophrenia divided by scores on PANSS total, positive, negative, general, and cognitive subscales as well as on particular PANSS items.
Both COMT polymorphisms showed an association with the PANSS general scores in female patients when they were subdivided in mild and severe symptoms category (Table 6); however, very low number of subjects (<2) in certain groups could possibly lead to false significant results, and since they were not confirmed when total PANSS general scores were evaluated, they were considered artefact.
The carriers of the rarest COMT rs4818-rs4680 haplotype (GA) had the highest scores on G1 item (somatic concern), while GG haplotype had the lowest scores on G2 (anxiety) and G6 (depression) items, compared to other haplotypes in male subjects (Table 3, Figure 2b–d and Figure 3b–d). Similarly, as shown in Table 6, in female subjects the GG haplotype was less frequent (31.8%; R = −1.4) in the group with severe disturbance of volition (G13 item) compared to the group with mild symptoms (39.8%), while CG haplotype was over-represented in group with severe symptoms (18.2%; R = 1.8), compared to those with mild symptoms (11.8%) (χ2 = 9.356; p = 0.025).
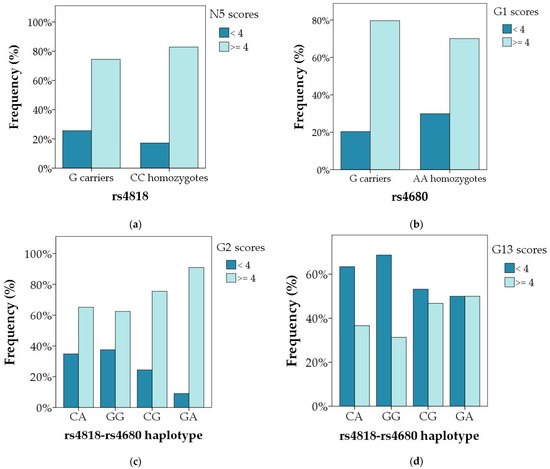
Figure 3.
Significant differences in the distribution of the genotypes and haplotypes of the COMT rs4818 and rs4680 polymorphisms between subjects with mild and severe particular symptoms of schizophrenia (a) N5 item in male carriers of different COMT rs4818 genotypes and alleles; p = 0.022; (b) G1 item in male carriers of different COMT rs4680 genotypes and alleles; p = 0.024; (c) G2 item in male carriers of different COMT rs4818–4680 haplotypes; p = 0.003; and (d) G13 item in female carriers of different COMT rs4818–4680 haplotypes; p = 0.025. Data are presented as percentage of patients with mild (score < 4) and severe (score ≥ 4) symptoms in each genotypic and haplotypic group.
4. Discussion
Our results have shown a significant genotypic and haplotypic association of COMT rs4680 and rs4818 gene polymorphisms with particular clinical symptoms evaluated by the individual PANSS subscales scores in male and female patients with schizophrenia. Our detailed analysis detected several associations between COMT rs4680 and rs4818 and the severity of particular individual PANSS items. In male patients with schizophrenia, the presence of the COMT rs4818 G allele was more often detected in those with mild “difficulties in abstract thinking” (N5), compared to CC homozygotes or C allele carriers. Regarding the other COMT SNP, in male patients, the presence of the COMT rs4680 A allele was associated with N5 scores compared to GG carriers, and the presence of the COMT rs4680 AA homozygous genotype was more frequently found participants with lower scores on the intensity of somatic concern (G1) item compared to G carriers. In females with schizophrenia, there were no significant associations between individual PANSS items (clinical symptoms of schizophrenia) and COMT rs4680 and rs4818 genetic variants. Concerning haplotype associations, male patients with schizophrenia who were COMT rs4818–rs4680 GA haplotype carriers had the highest scores on G1 item (somatic concern), whereas GG haplotype carriers had the lowest scores on G2 (anxiety) and G6 (depression) items. Female patients with schizophrenia had less frequent COMT GG haplotype associated with severe disturbance of volition (G13 item) compared to females with mild symptoms, while female patients with CG haplotype had more frequent severe then mild symptoms. All these associations were sex-specific.
In male patients with schizophrenia, lower N5 “Difficulties in abstract thinking” scores in COMT rs4680 GG carriers than in A carriers, and overrepresentation of COMT rs4818 G carriers compared to CC polymorphism in participants with less severe N5 scores, suggest an association between the presence higher COMT activity alleles and better preservation of abstract thinking. Actually, N5 was the only PANSS-negative item which severity was related to any of the two aforementioned COMT polymorphisms in males. Although abstract thinking belongs to the PANSS negative symptoms, according to a meta-analysis of the results of 45 factor analyses, it was not a part of the core negative items and was actually a symptom of disorganization/cognitive dimension [44]. The inability to engage in abstract thinking is a long-known typical symptom of schizophrenia [45]. It measures proverb interpretations and abstract similarities, by detecting the relationship between object, ideas, principles, and concepts. Its impairment is expressed as concrete form of cognition and speech, such as providing more concrete responses to proverbs [46]. Many studies have investigated abstract thinking in schizophrenia. Difficulty in abstract thinking was associated with the impaired decision making [47], distorted sensory experiences [48], and EEG findings of reduced alpha 1 spectral amplitude and lower Measurement and Treatment Research to Improve Cognition in Schizophrenia (MATRICS) Consensus Cognitive Battery 5 neuro-cognitive composite score [49].
Interestingly, baseline higher severity of PANSS N5 item was, along with P2 and G9 items, among three baseline PANSS items which predicted treatment resistance [50]. In another study, the PANSS item “abstract thinking” accounted for an association between negative symptoms and cognition [51], and was also found, together with conceptual disorganization, hostility, and uncooperativeness, to represent important mediating roles across different communities of schizophrenia symptoms [52]. In first-episode patients, baseline N5 and N7 were lower in remitters after one year of treatment, than in non-remitters, while other negative items were not [53]. In addition, P5 “abstractive thinking” was the only PANSS-negative symptom to predict depressive trajectory [54].
While these studies highlight the distinct position of N5 item among other PANSS negative symptoms [50,51,52], to best of our knowledge, only one study investigated all 30 PANSS items in relation with COMT polymorphism. This study found no associations between any individual PANSS item in males, including N5, with the COMT rs4680 polymorphism [55]. However, this study included a lower number of patients, and recruited only individuals with youth-onset schizophrenia with the mean age of 32. In our study, male patients were 38 years old. In turn, the age of onset of schizophrenia influences cognitive functioning, given that patients with youth-onset schizophrenia had severe cognitive deficits, in contrast to individuals with late-onset schizophrenia who maintained relatively preserved cognition [56]. Another study has focused on COMT polymorphism and abstraction in patients with schizophrenia. Our results disagree with the findings of better performance on abstraction test in individuals with schizophrenia who had greater number of COMT rs4680 A alleles in a dose–response fashion, although this effect was small [57]. However, the aforementioned study had small sample size for genetic study, included ethnically heterogeneous outpatient population with the average illness duration of 24 years, and abstraction was measured by a subtest from The Delis–Kaplan Executive Function System [57]. While those studies [55,57] and the present trial were cross-sectional, future studies are needed to establish the associations between COMT functional polymorphisms longitudinally, i.e., to explore if the relationships between COMT rs 4680 and rs4818 is detected in the acute phase change during successful treatment. Of note, treatment with antipsychotics decreases the scores on essentially all PANSS symptom, at least in non-treatment resistant patients [58], and it was depicted that abstract thinking in several diagnostic categories, including schizophrenia, improves over time [46].
The unique association of lesser severity of PANSS N5 component with the high-activity COMT rs4680 and rs4818 alleles in our study, indirectly suggests the involvement of lower prefrontal dopamine levels in better performance in abstract reasoning in males with schizophrenia. Namely, COMT genotypes at SNPs rs4680 and rs4818, but not rs737865 or rs165599, were associated with altered levels of S-COMT, but not MB-COMT, in the dorsolateral prefrontal cortex (DLPFC) of subjects who died from different causes, including suicide [59]. While it was reported that reduced activity in the left frontotemporal and the right orbitofrontal cortex could be a critical region for the deficit in abstract thinking in schizophrenia [60], low COMT activity in healthy men, resulting from the presence of the A allele of COMT rs4680 was associated with reductions in resting blood flow in frontal regions, compared to high COMT activity groups [61]. It may be speculated that males with lower-activity COMT polymorphisms in our study may have had changes in cortical neural activity and perfusion, which may have contributed to deficits in abstract thinking. Given the important role of abstract thinking in schizophrenia, it remains to be determined whether males with low-activity COMT alleles, who had worse N5 scores, differ in the degree of improvement of deficits in abstract thinking.
Both G2 “anxiety” and G6 “depression” items are a part of PANSS anxiety and depression dimension, according to the factor analysis [62]. Our findings suggest that high COMT activity related to low dopamine availability was associated with the lowest anxiety and depression dimension scores in males. In another study in Croatian population, COMT rs4680 was not associated with affective/depressive PANSS factors [63]. Our results do not agree with the findings of the higher levels of depression in patients with COMTrs4680 GG genotype, compared to A carriers, in patients with first-episode schizophrenia, whereby COMT rs4680 polymorphism moderated an association between the severity of depression and stressful life events [64]. That study had small sample size, and in both cited studies males and females were not separately analyzed [63,64]. Importantly, in patients with schizophrenia, COMT DNA methylation was inversely correlated with depressed subdomain of the PANSS, i.e., the higher the depressive symptoms, the lower the DNA methylation [65]. Future studies exploring the associations of COMT polymorphisms and depressive symptoms in schizophrenia need to consider childhood and life-time stressors, which may strongly influence this relationship.
The disturbance of volition (PANSS G13 item) refers to impaired will to move, and an altered timing of awareness of movement [66]. It belongs, along with G5 and several other items, to the disorganization factor [44]. In our female patients with schizophrenia, no associations between COMT genotypes or haplotypes and the severity of any PANSS sub-scores or individual items were observed. In line, no associations were found between PANSS item G13 and COMT rs4680 polymorphism in females, although female patients in that study had several other correlations [55]. In that cited study, females were about 19 years younger than our female patients, and had youth-onset schizophrenia [55]. However, when symptoms in the present study were categorized according to their severity, GG haplotype was less frequent in females with severe disturbance of volition, while the CG haplotype was over-represented in the group with severe symptoms, compared to those with mild symptoms. Disturbance of volition is commonly observed in individuals with schizophrenia. For example, it was the fifth-most-common PANSS single symptom in first-episode schizophrenia, reported in 65.4% of participants [67]. Moreover, the higher severity of disturbance of volition was, along with uncooperativeness, predictive of improved therapeutic benefit of paliperidone [68], but also during the initial asenapine treatment [69]. Despite being recognized as a common symptom in schizophrenia, disturbance of volition has received less attention than other PANSS items, and we were unable to find any data on the relationship between N5 item and either dopamine or COMT variants, but targeting such interactions appear as an attractive goal for future studies.
Sex-specific correlations of COMT polymorphism with different parameters have been previously reported in both preclinical and clinical studies. In preclinical studies, the administration of COMT inhibitor tolcapone produced more prominent changes in dopamine metabolism in female than male rat brains [70], while ovariectomy decreased, and estradiol replacement restored COMT levels, respectively [71]. In healthy women, increases in estradiol during the menstrual cycle were associated with improved working memory performance for participants who were GG carriers, and an association was in the opposite direction for AA carriers [72]. Sex divergent effects of COMT rs4680 polymorphism have been shown previously on the effects on the relationship between executive dysfunction and psychotic-like experience in college students [22]. In addition, among healthy individuals with COMT rs4633-rs4680 T allele, males had higher COMT specific activity in serum than females [73]. Moreover, the higher number of associations in males in our study may also be related to their substantially higher severity as well as wider range of severities in PANSS total scores and all other scales in male patients.
While the associations were sex-specific, in general 1) positive psychotic symptoms were not associated with any COMT genotypes or haplotype combinations, and 2) the presence of mainly high-activity COMT genotypes and haplotype was correlated with lower psychopathology on some indicators of PANSS negative and general psychopathology symptoms. Such findings somehow contribute to the general view that COMT is the most important regulator of dopamine function in PFC, but has a minor role in striatum and nucleus accumbens [74], whereas positive symptoms of schizophrenia are connected with hyperactivity of dopaminergic neurotransmission in striatum. However, an association between high-activity COMT and lower intensity of some negative and general symptoms is surprising, seeing that negative and cognitive symptoms are thought to arise from hypo-dopaminergic functioning in the frontal lobe and additional mesolimbic structures [75,76]. However, such findings were not detected in all patients with schizophrenia, and, given the high variance in dopaminergic measures in this population [77], it remains to be determined what is the role of COMT polymorphism in the complex landscape of symptoms in schizophrenia.
In addition, polygenic risk score (PGS) was associated with the genetic liability for schizophrenia, but it is not yet applied in precision medicine at the individual level. For example, it predicted treatment resistance in one study [78], but was not associated with the onset of treatment-resistance in another study [79]. Moreover, PGS predicted lower antipsychotic response in first-episode patients [80], but was not associated with the variance in cognitive test scores [81] or poorer cognitive outcome in schizophrenia [82]. Maybe the biggest obstacle for establishing precision medicine in schizophrenia is the lack of laboratory tools to predict the outcome. Moreover, schizophrenia is not a single disorder, but rather a group of distinct disorders with some overlapping symptoms [83]. Our correlations of COMT variants, implicated in higher dopamine degradation with lower severity of particular symptoms, may be an interesting topic for future longitudinal studies. Such studies should address the predictive value of different COMT genotypes and haplotypes on the trajectories of symptoms. Of note, genetics itself will probably never be a single factor that determines the severity of the outcome of schizophrenia, due to the presence of many other factors unrelated to genes, such as non-compliance, stressful life events, and substance abuse. Nevertheless, we hypothesize that different COMT variants may contribute to our understanding on the complexity of psychopathology in schizophrenia and our results may provide a small contribution to the field.
Taking into consideration that all our associations were sex-specific, we also recommend separate analyses for males and females in schizophrenia, or controlling the results for patient’s sex, especially in relation with studies addressing COMT variants. Our results add to the current knowledge on sexually dimorphic influence of COMT gene upon psychiatric phenotypes [84], that was also reported in healthy individuals [85]. In addition, it would be interesting to explore how different COMT genotypes and their haplotype combinations relate to psychopathology in different stages throughout the course of schizophrenia in male and female participants.
It is intriguing to put the COMT gene into evolutionary perspective. Due to its influence in the prefrontal cortex in complex cognitive functioning, and the role of COMT enzyme in modulating prefrontal dopamine levels, COMT functional polymorphisms may have impacted adaptation to new environments during human evolution. Of note, G allele of COMT rs4680 is the ancestral allele, while A allele is a result of mutation, which is considered advantageous for human evolution because A allele was unique to humans and related to higher intelligence [86] and better insight problem solving [85]. Schizophrenia is very rare in societies with hunter-gatherer lifestyles [83]. In turn, hunter-gatherers had lower frequency of A allele than farming or industrialized populations [86]. While it is unknown if COMT gene has any role in the evolutionary paradox in schizophrenia, we speculate that the relation between COMT polymorphisms and haplotype combinations (associated with higher enzymatic activity), with lower intensity of some symptoms in schizophrenia, may suggest distinct impact of the COMT genes on schizophrenia onset and schizophrenia severity.
Limitations and Strengths
This study was cross-sectional, and therefore the observed associations do not imply a causal relationship. The severity of several PANSS items/symptoms was associated with COMT genotypes and haplotypes in patients with schizophrenia.
Limitation is that only two polymorphisms were determined. However, it has been shown that COMT rs4680 is a functional polymorphism affecting COMT activity, protein abundance, and protein stability [17], while rs4680 and rs4818, but not rs737865 or rs165599 genotypes, were associated with altered levels of S-COMT, in the human dorsolateral prefrontal cortex (DLPFC) [59]. In a preclinical study, COMT knockout mice had 60% higher dopamine levels in PFC, but not in nucleus accumbens [87]. However, in a postmortem study, the mean abundance of COMT mRNA in DLPFC of psychiatric patients, including those with schizophrenia, did not differ by the COMT genotype [88]. Therefore, the relationship between COMT genetic variants and COMT activity in brain cortical areas is complex and incompletely understood. Many other factors may also influence COMT activity, such as COMT methylation level. For example, participants with schizophrenia who received atypical antipsychotics, most often risperidone, had lower incidence of COMT DNA methylation than patients on typical antipsychotics [65].
However, this study has strengths, such as large sample size and the ethnically homogenous Caucasian population, which is important given the pronounced differences in prevalence of COMT genotypes and haplotypes across different ethnic groups. Apart from genotype frequencies, psychopathology ratings may also differ across different ethnic groups. For example, patients from China scored 20–30% higher on 5 PANSS items, including disturbance of volition, whereas they had 20% lower scores on another 11 items, including abstract thinking, compared to American inpatients [89]. Moreover, while the vast majority of studies on COMT investigated the rs4680 genotype, the present study included COMT rs4680 and rs4818 polymorphisms, which is important because it was suggested that the COMT rs4818 polymorphism may be responsible for an even larger variation in the COMT activity than the rs4680 polymorphism [20], and that COMT haplotypes are associated with schizophrenia susceptibility [90]. In addition, the frequency of the COMT rs4680 and rs4818 genetic variants was controlled for the effect of smoking and sex.
5. Conclusions
Our findings demonstrated sex-specific associations of COMT high-activity rs4680 and rs4818 variants and haplotype combinations with mostly lower severity of several individual PANSS symptoms in patients with schizophrenia. Longitudinal studies are needed to establish whether those correlations remain significant during treatment and if different COMT genotypes and haplotypes have predictive value on trajectories of treatment.
Author Contributions
Conceptualization, M.S. and N.P.; methodology, N.P., G.N.E., M.N.P., M.K., S.U., O.K., N.M., Z.M., M.Z. and M.S.; validation, M.S. and N.P.; formal analysis, G.N.E., M.N.P. and M.K.; investigation, M.S. and N.P.; resources, N.P.; data curation, G.N.E. and M.N.P.; writing—original draft preparation, N.P., L.T. and M.S.; writing—review and editing, N.P., D.S.S. and M.S.; visualization, L.T.; supervision, N.P.; project administration, N.P. and M.S.; funding acquisition, N.P. and M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partly supported by CRO-USA collaborative project Pivac 1463002/1463002U, and by the project “Predictors of treatment response in schizophrenia”, sponsored by University of Zagreb, project code: BM106.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committees of the University Hospital Center Zagreb and University Psychiatric Hospital Vrapce, Zagreb, Croatia.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bradley, A.J.; Dinan, T.G. A systematic review of hypothalamic-pituitary-adrenal axis function in schizophrenia: Implications for mortality. J. Psychopharmacol. 2010, 24 (Suppl. S4), 91–118. [Google Scholar] [CrossRef]
- Mizuno, Y.; Hofer, A.; Suzuki, T.; Frajo-Apor, B.; Wartelsteiner, F.; Kemmler, G.; Saruta, J.; Tsukinoki, K.; Mimura, M.; Fleischhacker, W.W.; et al. Clinical and biological correlates of resilience in patients with schizophrenia and bipolar disorder: A cross-sectional study. Schizophr. Res. 2016, 175, 148–153. [Google Scholar] [CrossRef]
- Chuang, S.P.; Wu, J.Y.W.; Wang, C.S. Resilience and Quality of Life in People with Mental Illness: A Systematic Review and Meta-Analysis. Neuropsychiatr. Dis. Treat. 2023, 4, 507–514. [Google Scholar] [CrossRef]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; American Psychiatric Association: Washington, DC, USA, 2013. [Google Scholar]
- Alkan, E.; Davies, G.; Evans, S.L. Cognitive impairment in schizophrenia: Relationships with cortical thickness in fronto-temporal regions, and dissociability from symptom severity. NPJ Schizophr. 2021, 7, 20. [Google Scholar] [CrossRef] [PubMed]
- Harvey, P.D.; Bosia, M.; Cavallaro, R.; Howes, O.D.; Kahn, R.S.; Leucht, S.; Müller, D.R.; Penadés, R.; Vita, A. Cognitive dysfunction in schizophrenia: An expert group paper on the current state of the art. Schizophr. Res. Cogn. 2022, 29, 100249. [Google Scholar] [CrossRef] [PubMed]
- Rekhi, G.; Saw, Y.E.; Lim, K.; Keefe, R.S.E.; Lee, J. Impact of Cognitive Impairments on Health-Related Quality of Life in Schizophrenia. Brain Sci. 2023, 13, 215. [Google Scholar] [CrossRef]
- Etkin, A.; Gyurak, A.; O’Hara, R. A neurobiological approach to the cognitive deficits of psychiatric disorders. Dialogues Clin. Neurosci. 2013, 15, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Li, S.C. Neuromodulation and developmental contextual influences on neural and cognitive plasticity across the lifespan. Neurosci. Biobehav. Rev. 2013, 37 Pt B, 2201–2208. [Google Scholar] [CrossRef]
- Goldberg, T.E.; Weinberger, D.R. Genes and the parsing of cognitive processes. Trends Cogn. Sci. 2004, 8, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Nikolac Perkovic, M.; Nedic Erjavec, G.; Svob Strac, D.; Uzun, S.; Kozumplik, O.; Pivac, N. Theranostic biomarkers for schizophrenia. Int. J. Mol. Sci. 2017, 18, 733. [Google Scholar] [CrossRef]
- Brisch, R.; Saniotis, A.; Wolf, R.; Bielau, H.; Bernstein, H.G.; Steiner, J.; Bogerts, B.; Braun, K.; Jankowski, Z.; Kumaratilake, J.; et al. The role of dopamine in schizophrenia from a neurobiological and evolutionary perspective: Old fashioned, but still in vogue. Front. Psychiatry 2014, 19, 47. [Google Scholar]
- Westbrook, A.; Braver, T.S. Dopamine does double duty in motivating cognitive effort. Neuron 2016, 89, 695–710. [Google Scholar] [CrossRef] [PubMed]
- Jatana, N.; Sharma, A.; Latha, N. Pharmacophore modeling and virtual screening studies to design potential COMT inhibitors as new leads. J. Mol. Graph. Model. 2013, 39, 145–164. [Google Scholar] [CrossRef] [PubMed]
- Zubieta, C.; Kota, P.; Ferrer, J.L.; Dixon, R.A.; Noel, J.P. Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase. Plant Cell 2002, 14, 1265–1277. [Google Scholar] [CrossRef]
- Craddock, N.; Owen, M.J.; O’Donovan, M.C. The catechol-O-methyl transferase (COMT) gene as a candidate for psychiatric phenotypes: Evidence and lessons. Mol. Psychiatry 2006, 11, 446–458. [Google Scholar] [CrossRef]
- Tunbridge, E.M.; Narajos, M.; Harrison, C.H.; Beresford, C.; Cipriani, A.; Harrison, P.J. Which dopamine polymorphisms are functional? Systematic review and meta-analysis of COMT, DAT, DBH, DDC, DRD1-5, MAOA, MAOB, TH, VMAT1, and VMAT2. Biol. Psychiatry 2019, 86, 608–620. [Google Scholar] [CrossRef]
- Lachman, H.M.; Papolos, D.F.; Saito, T.; Yu, Y.M.; Szumlanski, C.L.; Weinshilboum, R.M. Human catechol-O-methyltransferase pharmacogenetics: Description of a functional polymorphism and its potential application to neuropsychiatric disorders. Pharmacogenetics 1996, 6, 243–250. [Google Scholar] [CrossRef]
- Roussos, P.; Giakoumaki, S.G.; Pavlakis, S.; Bitsios, P. Planning, decision-making and the COMT rs4818 polymorphism in healthy males. Neuropsychologia 2008, 46, 757–763. [Google Scholar] [CrossRef]
- Nackley, A.G.; Shabalina, S.A.; Tchivileva, I.E.; Satterfield, K.; Korchynskyi, O.; Makarov, S.S.; Maixner, W.; Diatchenko, L. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science 2006, 314, 1930–1933. [Google Scholar] [CrossRef]
- González-Castro, T.B.; Hernández-Díaz, Y.; Juárez-Rojop, I.E.; López-Narváez, M.L.; Tovilla-Zárate, C.A.; Fresan, A. The role of a Catechol-O-Methyltransferase (COMT) Val158Met genetic polymorphism in schizophrenia: A Systematic review and updated meta-analysis on 32,816 subjects. Neuromolecular. Med. 2016, 18, 216–231. [Google Scholar] [CrossRef]
- Gong, J.; Zhang, T.; Zhou, L.; Mo, Y.; Yu, F.; Liu, M.; Yang, L.; Liu, J. Gender divergent effect of COMT gene rs4680 polymorphism on an association between executive dysfunction and psychotic-like experiences. Behav. Brain Res. 2023, 439, 114215. [Google Scholar] [CrossRef] [PubMed]
- Shifman, S.; Bronstein, M.; Sternfeld, M.; Pisanté-Shalom, A.; Lev-Lehman, E.; Weizman, A.; Reznik, I.; Spivak, B.; Grisaru, N.; Karp, L.; et al. A highly significant association between the COMT haplotype and schizophrenia. Am. J. Hum. Genet. 2002, 71, 1296–1302. [Google Scholar] [CrossRef] [PubMed]
- Dennison, C.A.; Legge, S.E.; Pardiñas, A.F.; Walters, J.T.R. Genome-wide association studies in schizophrenia: Recent advances, challenges and future perspective. Schizophr. Res. 2020, 217, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Nikolac, M.; Sagud, M.; Nedic, G.; Nenadic Sviglin, K.; Mihaljevic Peles, A.; Uzun, S.; Vuskan Cusa, B.; Kozumplik, O.; Zivkovic, M.; Mustapic, M.; et al. The lack of association between catechol-O-methyl-transferase Val108/158Met polymorphism and smoking in schizophrenia and alcohol dependence. Psychiatry Res. 2013, 205, 179–180. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Lu, R.B.; Yeh, Y.W.; Shih, M.C.; Huang, S.Y. Association study of catechol-O-methyltransferase gene polymorphisms with schizophrenia and psychopathological symptoms in Han Chinese. Genes Brain Behav. 2011, 10, 316–324. [Google Scholar] [CrossRef]
- Li, W.J.; Kou, C.G.; Yu, Y.; Sun, S.; Zhang, X.; Kosten, T.R.; Zhang, X.Y. Association of catechol-O-methyltransferase gene polymorphisms with schizophrenia and negative symptoms in a Chinese population. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2012, 159, 370–375. [Google Scholar] [CrossRef]
- Madzarac, Z.; Tudor, L.; Sagud, M.; Nedic Erjavec, G.; Mihaljevic Peles, A.; Pivac, N. The Associations between COMT and MAO-B genetic variants with negative symptoms in patients with schizophrenia. Curr. Issues Mol. Biol. 2021, 43, 618–636. [Google Scholar] [CrossRef]
- Sagud, M.; Tudor, L.; Uzun, S.; Nikolac Perkovic, M.; Zivkovic, M.; Konjevod, M.; Kozumplik, O.; Vuksan Cusa, B.; Svob Strac, D.; Rados, I.; et al. Haplotypic and genotypic association of Catechol-O-Methyltransferase rs4680 and rs4818 polymorphisms and treatment resistance in schizophrenia. Front. Pharmacol. 2018, 9, 705. [Google Scholar] [CrossRef]
- Nikolac Perkovic, M.; Sagud, M.; Zivkovic, M.; Uzun, S.; Nedic Erjavec, G.; Kozumplik, O.; Svob Strac, D.; Mimica, N.; Mihaljevic Peles, A.; Pivac, N. Catechol-O-methyltransferase rs4680 and rs4818 haplotype association with treatment response to olanzapine in patients with schizophrenia. Sci. Rep. 2020, 10, 10049. [Google Scholar] [CrossRef]
- Tunbridge, E.M.; Harrison, P.J. Importance of the COMT gene for sex differences in brain function and predisposition to psychiatric disorders. Curr. Top. Behav. Neurosci. 2011, 8, 119–140. [Google Scholar]
- Kay, S.R.; Fiszbein, A.; Opler, L.A. The positive and negative syndrome scale (PANSS) for schizophrenia. Schizophr. Bull. 1987, 13, 261–276. [Google Scholar] [CrossRef] [PubMed]
- Lindström, E.; Tuninger, E.; Levander, S. PECC-factor structure and findings in three longitudinal cohorts of patients with schizophrenia. Nord. J. Psychiatry 2012, 66, 33–39. [Google Scholar] [CrossRef]
- Bagney, A.; Dompablo, M.; Santabárbara, J.; Moreno-Ortega, M.; Lobo, A.; Jimenez-Arriero, M.A.; Palomo, T.; Rodriguez-Jimenez, R. Are negative symptoms really related to cognition in schizophrenia? Psychiatry Res. 2015, 230, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.; Peh, O.H.; Yang, Z.; Rekhi, G.; Rapisarda, A.; See, Y.M.; Rashid, N.A.A.; Ang, M.S.; Lee, S.A.; Sim, K.; et al. Large-scale evaluation of the Positive and Negative Syndrome Scale (PANSS) symptom architecture in schizophrenia. Asian J. Psychiatr. 2021, 62, 102732. [Google Scholar] [CrossRef]
- First, M.B.; Spitzer, R.L.; Williams, J.B.W.; Gibbons, M. Structured Clinical Interview for DSM-IV Patient Edition (SCID-P); American Psychiatric Press: Washington, DC, USA, 1995. [Google Scholar]
- Montoya, A.; Valladares, A.; Lizán, L.; San, L.; Escobar, R.; Paz, S. Validation of the Excited Component of the Positive and Negative Syndrome Scale (PANSS-EC) in a naturalistic sample of 278 patients with acute psychosis and agitation in a psychiatric emergency room. Health Qual. Life Outcomes 2011, 9, 18. [Google Scholar] [CrossRef]
- Miller, S.A.; Dykes, D.D.; Polesky, H.F. A Simple Salting out Procedure for Extracting DNA from Human Nucleated Cells. Nucleic Acids Res. 1988, 16, 1215. [Google Scholar] [CrossRef]
- Unwin, A. Discovering Statistics Using R by Andy Field, Jeremy Miles, Zoë Field. Int. Stat. Rev. 2013, 81, 169–170. [Google Scholar] [CrossRef]
- Rodriguez, S.; Gaunt, T.R.; Day, I.N. Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am. J. Epidemiol. 2009, 169, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and Visualization of LD and Haplotype Maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Faul, F.; Erdfelder, E.; Lang, A.G.; Buchner, A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 2007, 39, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Shafer, A.; Dazzi, F. Meta-analysis of the positive and Negative Syndrome Scale (PANSS) factor structure. J. Psychiatr. Res. 2019, 115, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Mihaljević-Peleš, A.; Bajs Janović, M.; Šagud, M.; Živković, M.; Janović, Š.; Jevtović, S. Cognitive deficit in schizophrenia: An overview. Psychiatr. Danub. 2019, 31 (Suppl. S2), 139–142. [Google Scholar]
- Rosen, C.; Harrow, M.; Tong, L.; Jobe, T.H.; Harrow, H. A word is worth a thousand pictures: A 20-year comparative analysis of aberrant abstraction in schizophrenia, affective psychosis, and non-psychotic depression. Schizophr. Res. 2021, 238, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Suetani, S.; Baker, A.; Garner, K.; Cosgrove, P.; Mackay-Sim, M.; Siskind, D.; Murray, G.K.; Scott, J.G.; Kesby, J.P. Impairments in goal-directed action and reversal learning in a proportion of individuals with psychosis. Cogn. Affect. Behav. Neurosci. 2022, 22, 1390–1403. [Google Scholar] [CrossRef]
- Ricarte, J.J.; Del Rey, F.; Ros, L.; Latorre, J.M.; Berna, F. Abstract and experiential thinking differentially account for anomalous perception of reality in people with or without schizophrenia. Schizophr. Res. 2018, 193, 43–50. [Google Scholar] [CrossRef]
- Vignapiano, A.; Koenig, T.; Mucci, A.; Giordano, G.M.; Amodio, A.; Altamura, M.; Bellomo, A.; Brugnoli, R.; Corrivetti, G.; Di Lorenzo, G.; et al. Italian Network for Research on Psychoses. Disorganization and cognitive impairment in schizophrenia: New insights from electrophysiological findings. Int. J. Psychophysiol. 2019, 145, 99–108. [Google Scholar] [CrossRef]
- Ortiz, B.B.; Higuchi, C.H.; Noto, C.; Joyce, D.W.; Correll, C.U.; Bressan, R.A.; Gadelha, A. A symptom combination predicting treatment-resistant schizophrenia—A strategy for real-world clinical practice. Schizophr. Res. 2020, 218, 195–200. [Google Scholar] [CrossRef]
- Farreny, A.; Usall, J.; Cuevas-Esteban, J.; Ochoa, S.; Brébion, G. Amendment of traditional assessment measures for the negative symptoms of schizophrenia. Eur. Psychiatry 2018, 49, 50–55. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, Y.; Lu, Z.; Yan, H.; Guo, L.; Liao, Y.; Lu, T.; Wang, L.; Li, J.; Li, W.; et al. Longitudinal network analysis reveals interactive change of schizophrenia symptoms during acute antipsychotic treatment. Schizophr. Bull. 2023, 49, 208–217. [Google Scholar] [CrossRef]
- Cheng, Z.; Yuan, Y.; Han, X.; Yang, L.; Zeng, X.; Yang, F.; Lu, Z.; Wang, C.; Deng, H.; Zhao, J.; et al. Which subgroup of first-episode schizophrenia patients can remit during the first year of antipsychotic treatment? Front. Psychiatry 2020, 11, 566. [Google Scholar] [CrossRef] [PubMed]
- Hoprekstad, G.E.; Kjelby, E.; Gjestad, R.; Fathian, F.; Larsen, T.K.; Reitan, S.K.; Rettenbacher, M.; Torsvik, A.; Skrede, S.; Johnsen, E.; et al. Depression trajectories and cytokines in schizophrenia spectrum disorders—A longitudinal observational study. Schizophr. Res. 2023, 252, 77–87. [Google Scholar] [CrossRef]
- Morozova, A.; Zorkina, Y.; Pavlov, K.; Pavlova, O.; Storozheva, Z.; Zubkov, E.; Zakharova, N.; Karpenko, O.; Reznik, A.; Chekhonin, V.; et al. Association of rs4680 COMT, rs6280 DRD3, and rs7322347 5HT2A With Clinical Features of Youth-Onset Schizophrenia. Front. Psychiatry 2019, 10, 830. [Google Scholar] [CrossRef] [PubMed]
- Rajji, T.K.; Ismail, Z.; Mulsant, B.H. Age at onset and cognition in schizophrenia: Meta-analysis. Br. J. Psychiatry 2009, 195, 286–293. [Google Scholar] [CrossRef]
- Twamley, E.W.; Hua, J.P.; Burton, C.Z.; Vella, L.; Chinh, K.; Bilder, R.M.; Kelsoe, J.R. Effects of COMT genotype on cognitive ability and functional capacity in individuals with schizophrenia. Schizophr. Res. 2014, 159, 114–117. [Google Scholar] [CrossRef] [PubMed]
- Rolls, E.T.; Lu, W.; Wan, L.; Yan, H.; Wang, C.; Yang, F.; Tan, Y.; Li, L.; Chinese Schizophrenia Collaboration Group; Yu, H.; et al. Individual differences in schizophrenia. BJPsych Open 2017, 3, 265–273. [Google Scholar] [CrossRef]
- Parkin, G.M.; Udawela, M.; Gibbons, A.; Scarr, E.; Dean, B. Catechol-O-methyltransferase (COMT) genotypes are associated with varying soluble, but not membrane-bound COMT protein in the human prefrontal cortex. J. Hum. Genet. 2018, 63, 1251–1258. [Google Scholar] [CrossRef]
- Oh, J.; Chun, J.W.; Joon, J.H.; Kim, E.; Park, H.J.; Lee, B.; Kim, J.J. The neural basis of a deficit in abstract thinking in patients with schizophrenia. Psychiatry Res. 2015, 234, 66–73. [Google Scholar] [CrossRef]
- Martens, M.; McConnell, F.K.; Filippini, N.; Mackay, C.E.; Harrison, P.J.; Tunbridge, E.M. Dopaminergic modulation of regional cerebral blood flow: An arterial spin labelling study of genetic and pharmacological manipulation of COMT activity. Neuroimage 2021, 234, 117999. [Google Scholar] [CrossRef]
- Marder, S.R.; Davis, J.M.; Chouinard, G. The effects of risperidone on the five dimensions of schizophrenia derived by factor analysis: Combined results of the North American trials. J. Clin. Psychiatry 1997, 58, 538–546. [Google Scholar] [CrossRef]
- Peitl, V.; Štefanović, M.; Karlović, D. Depressive symptoms in schizophrenia and dopamine and serotonin gene polymorphisms. Prog. Neuropsychopharmacol. Biol. Psychiatry 2017, 77, 209–215. [Google Scholar] [CrossRef]
- Tosato, S.; Bonetto, C.; de Santi, K.; Lasalvia, A.; Gennarelli, M.; Cristofalo, D.; Bertani, M.; Ruggeri, M. on behalf of the Picos-Veneto group. COMT but Not 5HTTLPR gene is associated with depression in first-episode psychosis: The role of stressful life events. Genes 2023, 14, 350. [Google Scholar] [CrossRef] [PubMed]
- Nour El Huda, A.R.; Norsidah, K.Z.; Nabil Fikri, M.R.; Hanisah, M.N.; Kartini, A.; Norlelawati, A.T. DNA methylation of membrane-bound catechol-O-methyltransferase in Malaysian schizophrenia patients. Psychiatry Clin. Neurosci. 2018, 72, 266–279. [Google Scholar] [CrossRef]
- Pirio Richardson, S.; Triggiani, A.I.; Matsuhashi, M.; Voon, V.; Peckham, E.; Nahab, F.; Mari, Z.; Hallett, M. Timing of the sense of volition in patients with schizophrenia. Front. Neurosci. 2020, 14, 574472. [Google Scholar] [CrossRef]
- Cesková, E.; Prikryl, R.; Kaspárek, T.; Ondrusová, M. Psychopathology and treatment responsiveness of patients with first-episode schizophrenia. Neuropsychiatr. Dis. Treat. 2005, 1, 179–185. [Google Scholar] [CrossRef]
- Mellem, M.S.; Kollada, M.; Tiller, J.; Lauritzen, T. Explainable AI enables clinical trial patient selection to retrospectively improve treatment effects in schizophrenia. BMC Med. Inform. Decis. Mak. 2021, 21, 162. [Google Scholar] [CrossRef]
- Ogyu, K.; Noda, Y.; Yoshida, K.; Kurose, S.; Masuda, F.; Mimura, Y.; Nishida, H.; Plitman, E.; Tarumi, R.; Tsugawa, S.; et al. Early improvements of individual symptoms as a predictor of treatment response to asenapine in patients with schizophrenia. Neuropsychopharmacol. Rep. 2020, 40, 138–149. [Google Scholar] [CrossRef]
- Laatikainen, L.M.; Sharp, T.; Harrison, P.J.; Tunbridge, E.M. Sexually dimorphic effects of catechol-O-methyltransferase (COMT) inhibition on dopamine metabolism in multiple brain regions. PLoS ONE 2013, 8, e61839. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Bai, W.; Wang, W.; Jiang, H.; Jin, B.; Liu, Y.; Liu, S.; Wang, K.; Jia, J.; Qin, L. Mechanisms underlying alterations in norepinephrine levels in the locus coeruleus of ovariectomized rats: Modulation by estradiol valerate and black cohosh. Neuroscience 2017, 354, 110–121. [Google Scholar] [CrossRef]
- Louis, C.C.; Jacobs, E.; D’Esposito, M.; Moser, J. Estradiol and the Catechol-O-methyltransferase Gene Interact to Predict Working Memory Performance: A Replication and Extension. J. Cogn. Neurosci. 2023, 35 (Suppl. S7), 1144–1153. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.K.; Yang, M.C.; Chen, C.S.; Wang, I.C.; Kao, W.T.; Shi, M.D. A gender-specific COMT haplotype contributes to risk modulation rather than disease severity of major depressive disorder in a Chinese population. J. Affect. Disord. 2019, 246, 376–386. [Google Scholar] [CrossRef]
- Sagud, M.; Mück-Seler, D.; Mihaljević-Peles, A.; Vuksan-Cusa, B.; Zivković, M.; Jakovljević, M.; Pivac, N. Catechol-O-methyl transferase and schizophrenia. Psychiat. Danub. 2010, 22, 270–274. [Google Scholar]
- Correll, C.U.; Schooler, N.R. Negative symptoms in schizophrenia: A review and clinical guide for recognition, assessment, and treatment. Neuropsychiatr. Dis. Treat. 2020, 16, 519–534. [Google Scholar] [CrossRef] [PubMed]
- Vidal, P.M.; Pacheco, R. The cross-talk between the dopaminergic and the immune system involved in schizophrenia. Front. Pharmacol. 2020, 11, 394. [Google Scholar] [CrossRef] [PubMed]
- Cumming, P.; Abi-Dargham, A.; Gründer, G. Molecular imaging of schizophrenia: Neurochemical findings in a heterogeneous and evolving disorder. Behav. Brain Res. 2021, 398, 113004. [Google Scholar] [CrossRef] [PubMed]
- Werner, M.C.F.; Wirgenes, K.V.; Haram, M.; Bettella, F.; Lunding, S.H.; Rødevand, L.; Hjell, G.; Agartz, I.; Djurovic, S.; Melle, I.; et al. Indicated association between polygenic risk score and treatment-resistance in a naturalistic sample of patients with schizophrenia spectrum disorders. Schizophr. Res. 2020, 218, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Wimberley, T.; Gasse, C.; Meier, S.M.; Agerbo, E.; MacCabe, J.H.; Horsdal, H.T. Polygenic Risk Score for Schizophrenia and Treatment-Resistant Schizophrenia. Schizophr. Bull. 2017, 43, 1064–1069. [Google Scholar] [CrossRef]
- Zhang, J.P.; Robinson, D.; Yu, J.; Gallego, J.; Fleischhacker, W.W.; Kahn, R.S.; Crespo-Facorro, B.; Vazquez-Bourgon, J.; Kane, J.M.; Malhotra, A.K.; et al. Schizophrenia Polygenic Risk Score as a Predictor of Antipsychotic Efficacy in First-Episode Psychosis. Am. J. Psychiatry 2019, 176, 21–28. [Google Scholar] [CrossRef]
- Richards, A.L.; Pardinas, A.F.; Frizzati, A.; Tansey, K.E.; Lynham, A.J.; Holmans, P.; Legge, S.E.; Savage, J.E.; Agartz, I.; Andreassen, O.A.; et al. The Relationship Between Polygenic Risk Scores and Cognition in Schizophrenia. Schizophr. Bull. 2020, 46, 336–344. [Google Scholar] [CrossRef]
- Habtewold, T.D.; Liemburg, E.J.; Islam, M.A.; de Zwarte, S.M.C.; Boezen, H.M.; GROUP Investigators; Bruggeman, R.; Alizadeh, B.Z. Association of schizophrenia polygenic risk score with data-driven cognitive subtypes: A six-year longitudinal study in patients, siblings and controls. Schizophr. Res. 2020, 223, 135–147. [Google Scholar] [CrossRef]
- Rantala, M.J.; Luoto, S.; Borráz-León, J.I.; Krams, I. Schizophrenia: The new etiological synthesis. Neurosci. Biobehav. Rev. 2022, 142, 104894. [Google Scholar] [CrossRef] [PubMed]
- Harrison, P.J.; Tunbridge, E.M. Catechol-O-methyltransferase (COMT): A gene contributing to sex differences in brain function, and to sexual dimorphism in the predisposition to psychiatric disorders. Neuropsychopharmacology 2008, 33, 3037–3045. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Shang, S.; Su, Y. Genetic influences on insight problem solving: The role of catechol-O-methyltransferase (COMT) gene polymorphisms. Front. Psychol. 2015, 6, 1569. [Google Scholar] [CrossRef] [PubMed]
- Piffer, D. Correlation of the COMT Val158Met polymorphism with latitude and hunter-gather lifestyle suggests culture–gene coevolution and selective pressure on cognition genes due to climate. Anthropol. Sci. 2013, 121, 161–171. [Google Scholar] [CrossRef]
- Käenmäki, M.; Tammimäki, A.; Myöhänen, T.; Pakarinen, K.; Amberg, C.; Karayiorgou, M.; Gogos, J.A.; Männistö, P.T. Quantitative role of COMT in dopamine clearance in the prefrontal cortex of freely moving mice. J. Neurochem. 2010, 114, 1745–1755. [Google Scholar] [CrossRef]
- Tunbridge, E.; Burnet, P.W.; Sodhi, M.S.; Harrison, P.J. Catechol-O-methyltransferase (COMT) and proline dehydrogenase (PRODH) mRNAs in the dorsolateral prefrontal cortex in schizophrenia, bipolar disorder, and major depression. Synapse 2004, 51, 112–118. [Google Scholar] [CrossRef]
- Aggarwal, N.K.; Tao, H.; Xu, K.; Stefanovics, E.; Liu, Z.; Rosenheck, R.A. Comparing the PANSS in Chinese and American inpatients: Cross-cultural psychiatric analyses of instrument translation and implementation. Schizophr. Res. 2011, 132, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Bray, N.J.; Buckland, P.R.; Williams, N.M.; Williams, H.J.; Norton, N.; Owen, M.J.; O’Donovan, M.C. A haplotype implicated in schizophrenia susceptibility is associated with reduced COMT expression in human brain. Am. J. Hum. Genet. 2003, 73, 152–161. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).