Association between HLA Class II Alleles/Haplotypes and Genomic Ancestry in Brazilian Patients with Type 1 Diabetes: A Nationwide Exploratory Study
Abstract
1. Introduction
2. Methods
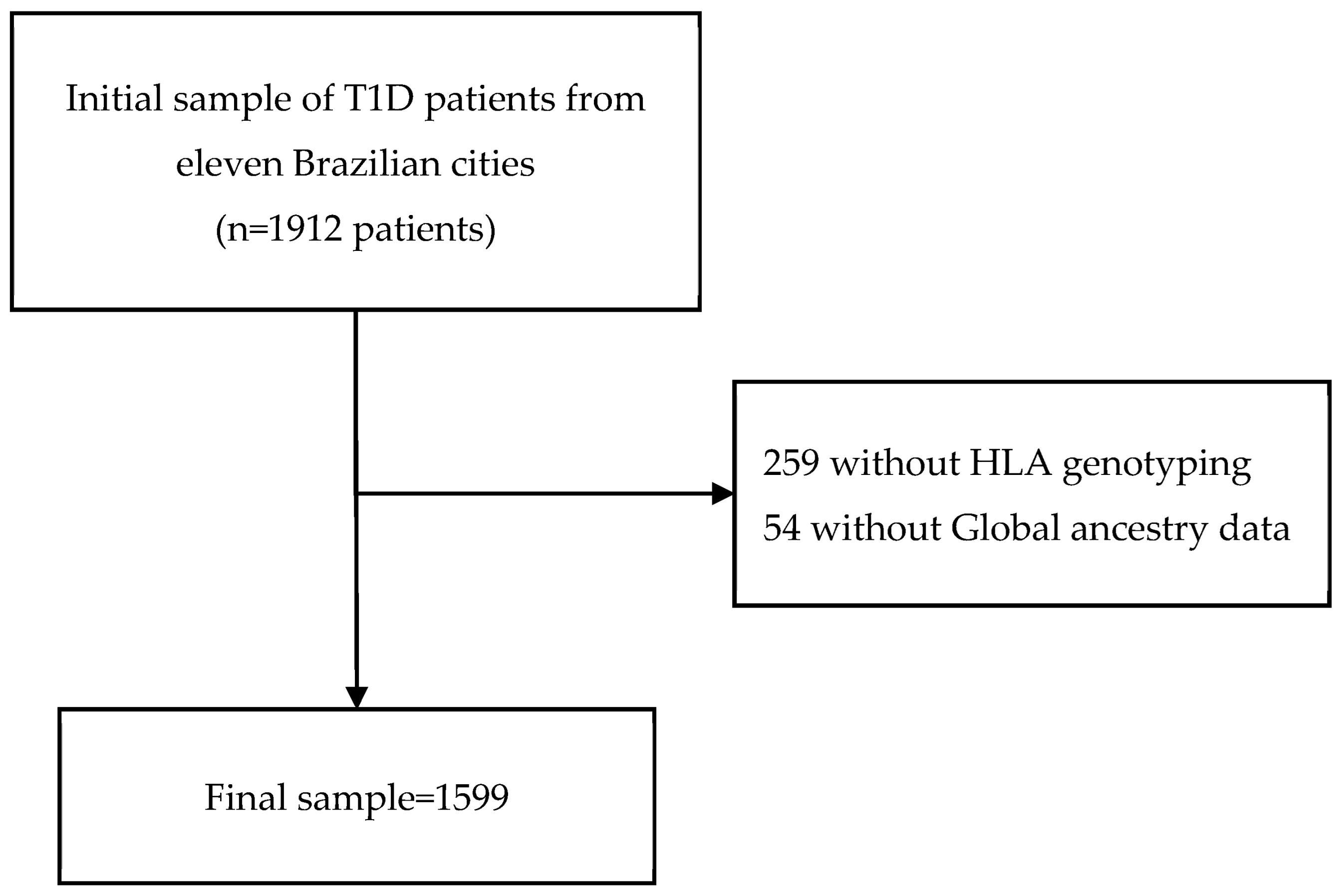
2.1. Study Design and Sample
2.2. Data Collection
2.3. DNA Extraction and AIM-Indel Genotyping
2.4. HLA Genotyping
2.5. Statistical Analysis
3. Results
3.1. Description of the Study Sample
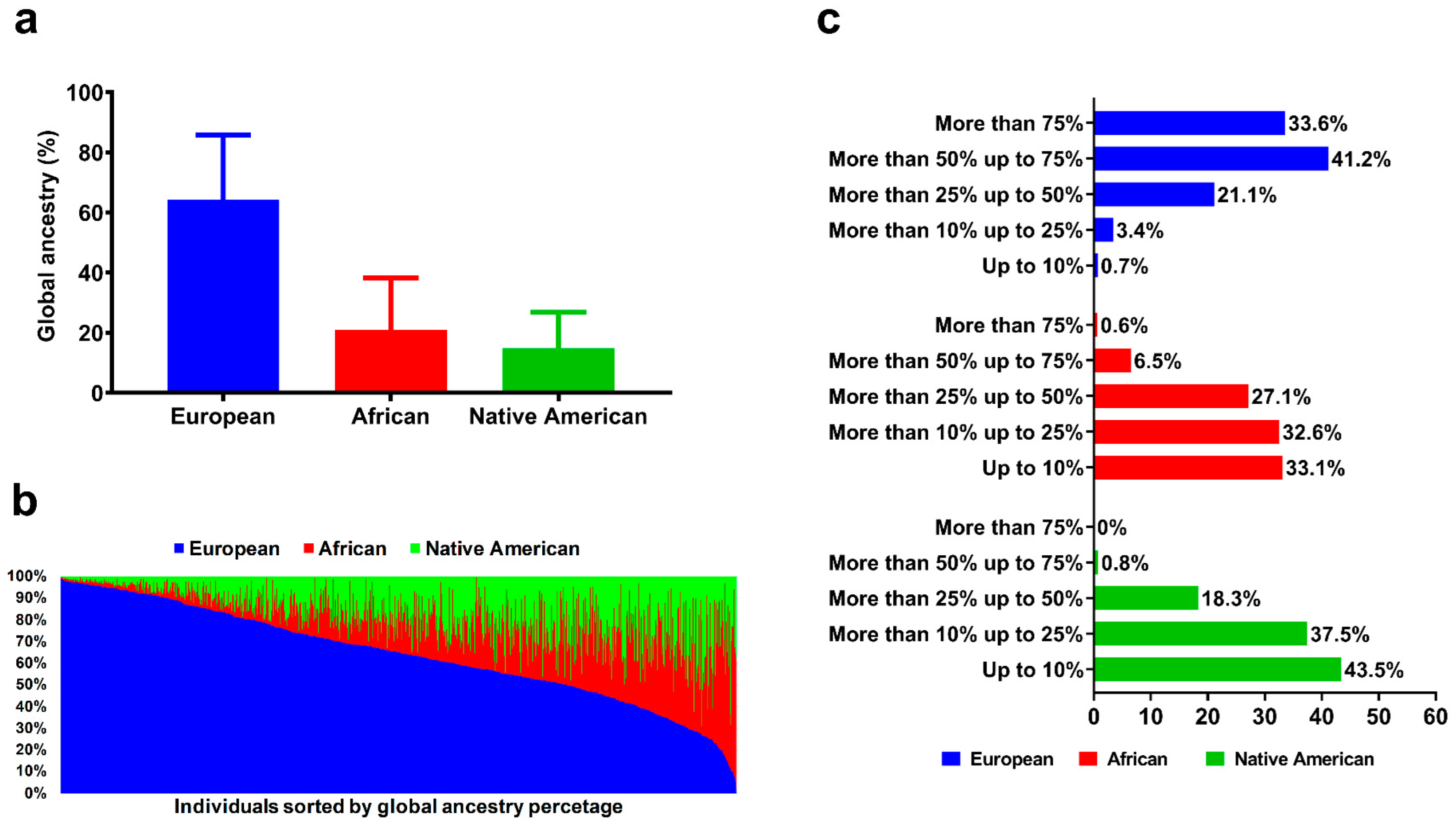
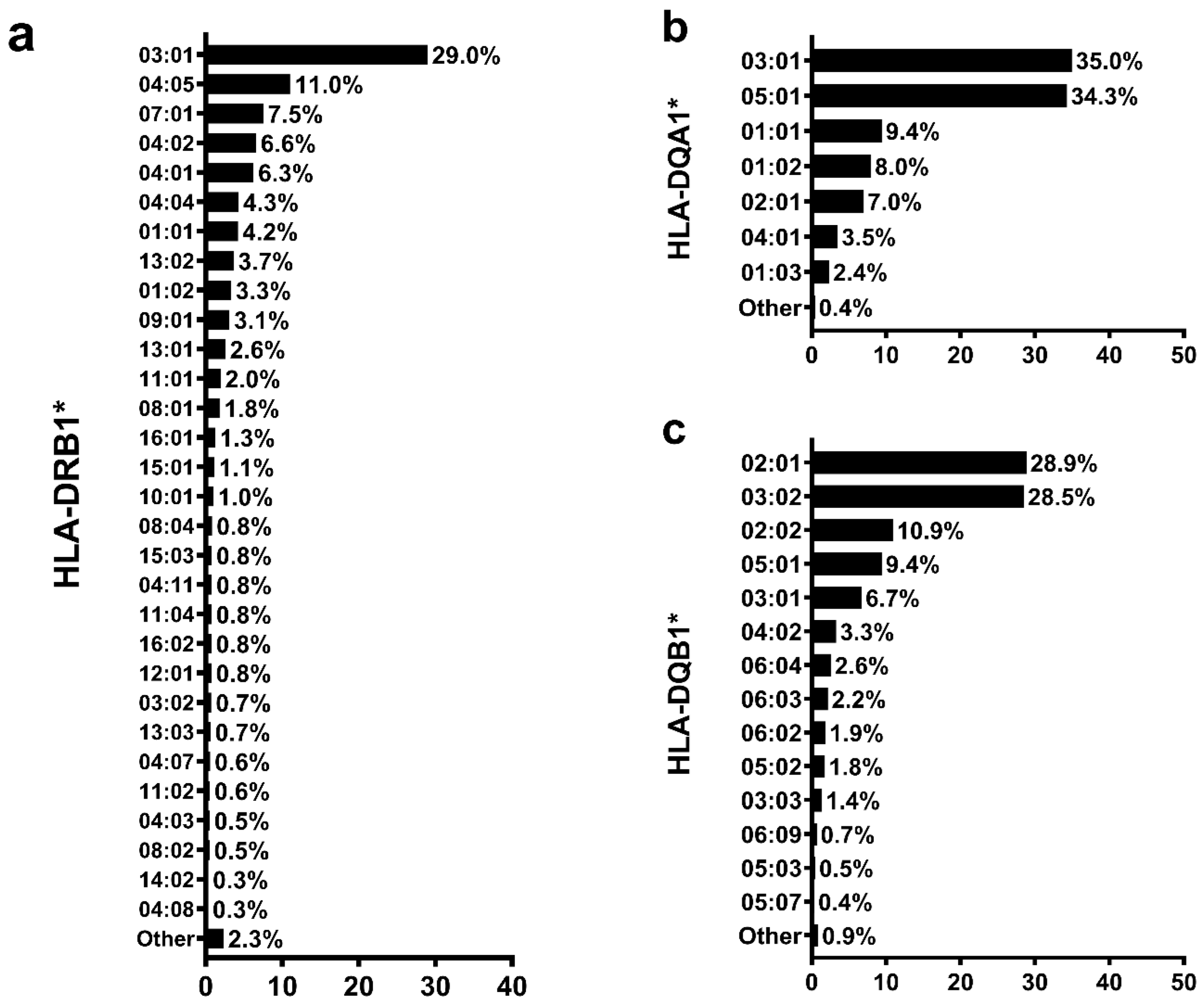
3.2. Global Genomic Ancestry Percentage and HLA Allele Distribution
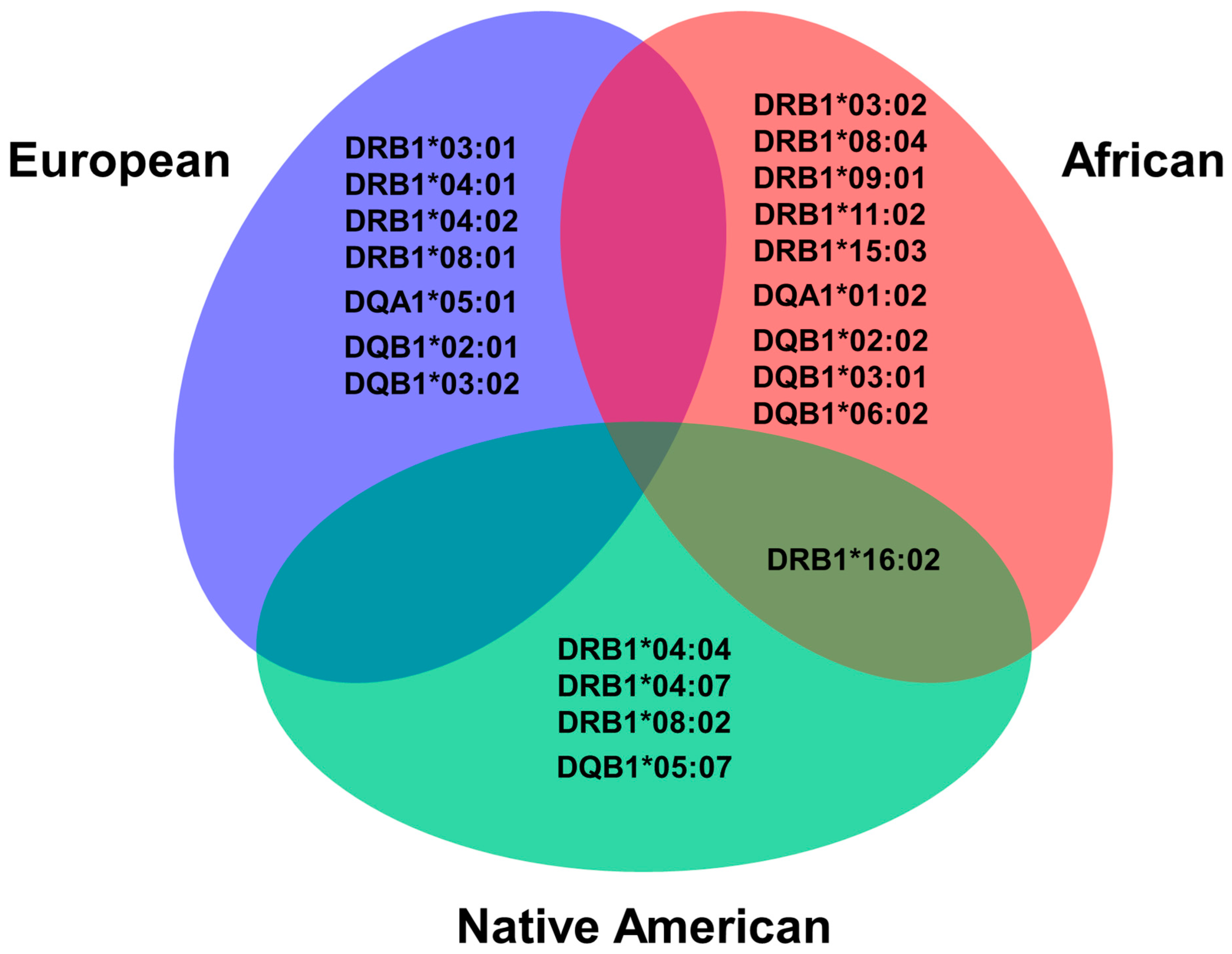
3.3. HLA Alleles Related to Global Ancestry Percentage
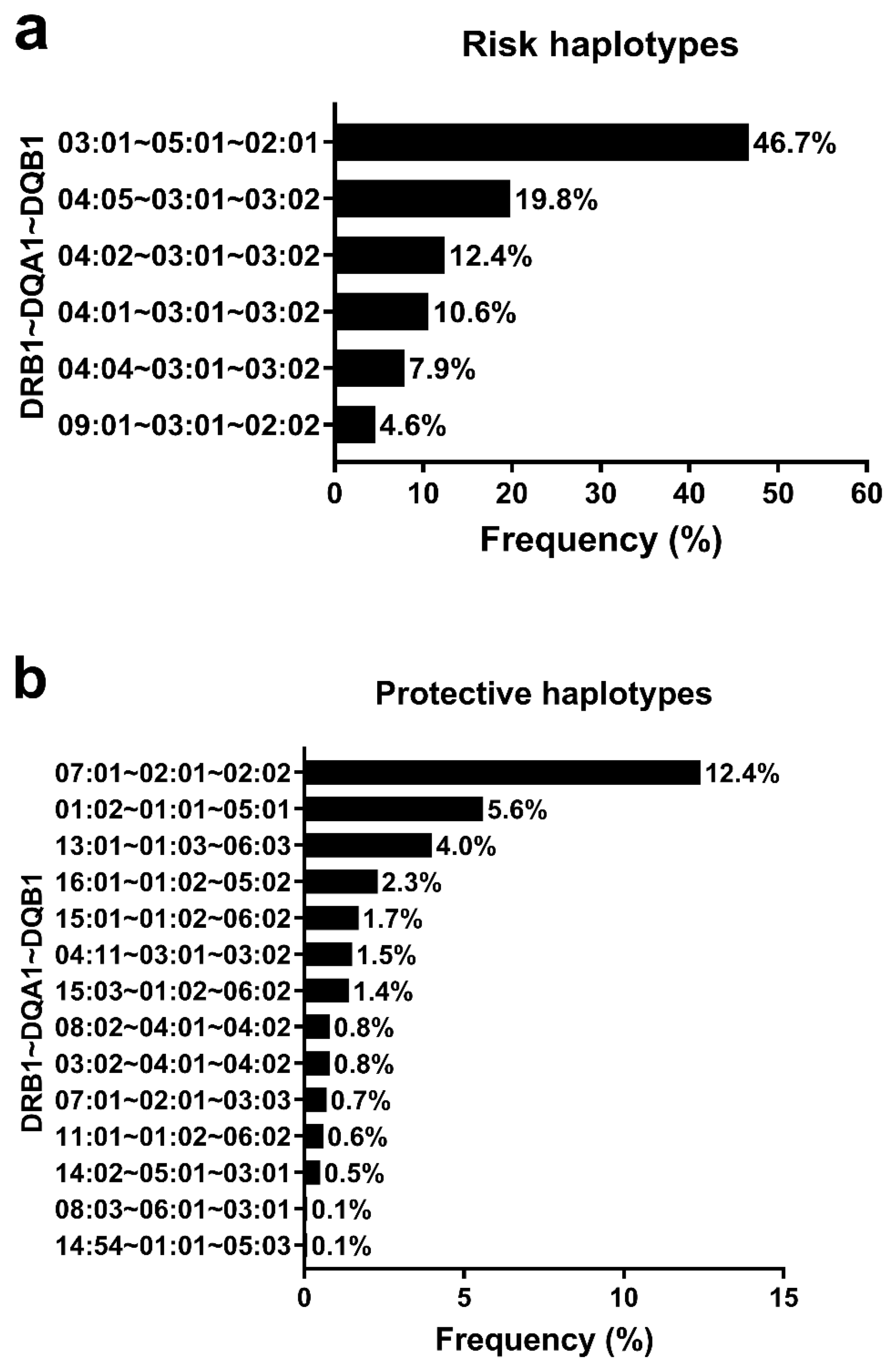
3.4. HLA Haplotypes Associated with T1D and Global Genomic Ancestry Percentage
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Ethical Statement
Appendix A. Brazilian Type 1 Diabetes Study Group (BrazDiab1SG) Participants That Are Listed Below
| Name | Institution | |
|---|---|---|
| Marilia Brito Gomes * | Rio de Janeiro State University | mariliabgomes@gmail.com |
| Laura Nunes Melo | Rio de Janeiro State University | lauragnmelo@gmail.com |
| Roberta Cobas | Rio de Janeiro State University | robertacobas@gmail.com |
| Lucianne Righeti Monteiro Tannus | Rio de Janeiro State University | luciannetannus@ig.com.br |
| Melanie Rodacki * | Federal University Hospital of Rio de Janeiro | mrodacki2001@yahoo.com.br |
| Lenita Zajdenverg | Federal University Hospital of Rio de Janeiro | lenitazaj@gmail.com |
| Joana Rodrigues Dantas | Federal University Hospital of Rio de Janeiro | joanardantasp@ig.com.br |
| Maria Lúcia Cardillo Corrêa-Giannella * | University Hospital of São Paulo | malugia@lim25fm.usp.br |
| Sharon Nina Admoni | University Hospital of São Paulo | sharonadmoni@gmail.com |
| Daniele Pereira dos Santos | University Hospital of São Paulo | dps.daniele@hotmail.com |
| Carlos Antonio Negrato * | Bauru’s Diabetics Association | carlosnegrato@uol.com.br |
| Maria de Fatima Guedes | Bauru’s Diabetics Association | tatiguedeses@hotmail.com |
| Sergio Atala Dib * | Federal University of São Paulo State | sergio.dib@unifesp.br |
| Celso Ferreira de Camargo Sallum Filho | Federal University of São Paulo State | celsosallum@superig.com.br |
| Elisabeth João Pavin * | University of Campinas | ejpavin@fcm.unicamp.br |
| Caroline Takano | University of Campinas | caroline.takano@gmail.com |
| Rosângela Roginski Rea * | Federal University of Paraná | rosangelarea@uol.com.br |
| Nicole Balster Romanzini | Federal University of Paraná | nikbr@hotmail.com |
| Mirela Azevedo * | Clinical Hospital of Porto Alegre | mirelajobimazevedo@gmail.com |
| Luis Henrique Canani | Clinical Hospital of Porto Alegre | luishenriquecanani@gmail.com |
| Hermelinda Cordeiro Pedrosa * | Regional Hospital of Taguatinga | pedrosa.hc@globo.com |
| Monica Tolentino | Regional Hospital of Taguatinga | monicatolentino@uol.com.br |
| Cejana Hamu Aguiar | Regional Hospital of Taguatinga | cejanahamu@yahoo.com.br |
| Reine Marie Chaves Fonseca * | Diabetes and Endocrinology Center of Bahia | reinemar@terra.com.br |
| Ludmila Chaves Fonseca | Diabetes and Endocrinology Center of Bahia | ludchaves@yahoo.com.br |
| Raffaele Kasprowicz | Diabetes and Endocrinology Center of Bahia | raffaellebarros@hotmail.com |
| Adriana Costa e Forti * | Diabetes and Hypertension Center of Ceará | adrianaforti@uol.com.br |
| Angela Delmira Nunes Mendes | Diabetes and Hypertension Center of Ceará | angeladelmira@terra.com.br |
| Renan Montenegro Junior * | Federal University of Ceará | renanjr@ufc.br |
| Virgínia Oliveira Fernandes | Federal University of Ceará | virginiafernande@hotmail.com |
| João Soares Felício * | Federal University Hospital of Pará | felicio.bel@terra.com.br |
| Flavia Marques Santos | Federal University Hospital of Pará | drafms@bol.com.br |
References
- Cudworth, A.G.; Woodrow, J.C. HL-A antigens and diabetes mellitus. Lancet 1974, 304, 1153. [Google Scholar] [CrossRef]
- Noble, J.A. Immunogenetics of type 1 diabetes: A comprehensive review. J. Autoimmun. 2015, 64, 101–112. [Google Scholar] [CrossRef]
- Ogle, G.D.; James, S.; Dabelea, D.; Pihoker, C.; Svennson, J.; Maniam, J.; Klatman, E.L.; Patterson, C.C. Global estimates of incidence of type 1 diabetes in children and adolescents: Results from the International Diabetes Federation Atlas, 10th edition. Diabetes Res. Clin. Pract. 2022, 183, 109083. [Google Scholar] [CrossRef]
- Negrato, C.A.; Lauris, J.R.P.; Saggioro, I.B.; Corradini, M.C.M.; Borges, P.R.; Crês, M.C.; Junior, A.L.; Guedes, M.F.S.; Gomes, M.B. Increasing incidence of type 1 diabetes between 1986 and 2015 in Bauru, Brazil. Diabetes Res. Clin. Pract. 2017, 127, 198–204. [Google Scholar] [CrossRef]
- Redondo, M.J.; Steck, A.K.; Pugliese, A. Genetics of type 1 diabetes. Pediatr. Diabetes 2018, 19, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Noble, J.A.; Erlich, H.A. Genetics of Type 1 Diabetes. Cold Spring Harb. Perspect. Med. 2012, 2, a007732. [Google Scholar] [CrossRef] [PubMed]
- Santos, D.C.; Porto, L.C.; Oliveira, R.V.; Secco, D.; Hanhoerderster, L.; Pizarro, M.H.; Barros, B.S.V.; Mello, L.G.N.; Muniz, L.H.; Silva, D.A.; et al. HLA class II genotyping of admixed Brazilian patients with type 1 diabetes according to self-reported color/race in a nationwide study. Sci. Rep. 2020, 10, 6628. [Google Scholar] [CrossRef] [PubMed]
- Noble, J.A.; Johnson, J.; Lane, J.A.; Valdes, A.M. HLA Class II Genotyping of African American Type 1 Diabetic Patients Reveals Associations Unique to African Haplotypes. Diabetes 2013, 62, 3292–3299. [Google Scholar] [CrossRef]
- Halagan, M.; Oliveira, D.C.; Maiers, M.; Fabreti-Oliveira, R.A.; Moraes, M.E.H.; Visentainer, J.E.L.; Pereira, N.F.; Romero, M.; Cardoso, J.F.; Porto, L.C. The distribution of HLA haplotypes in the ethnic groups that make up the Brazilian Bone Marrow Volunteer Donor Registry (REDOME). Immunogenetics 2018, 70, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Suarez-Kurtz, G.; Pena, S.D.J.; Struchiner, C.J.; Hutz, M.H. Pharmacogenomic Diversity among Brazilians: Influence of Ancestry, Self-Reported Color, and Geographical Origin. Front. Pharmacol. 2012, 3, 191. [Google Scholar] [CrossRef]
- Gomes, M.B.; Gabrielli, A.B.; Santos, D.C.; Pizarro, M.H.; Barros, B.S.V.; Negrato, C.A.; Dib, S.A.; Porto, L.C.; Silva, D.A. Self-reported color-race and genomic ancestry in an admixed population: A contribution of a nationwide survey in patients with type 1 diabetes in Brazil. Diabetes Res. Clin. Pract. 2018, 140, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Gomes, K.F.B.; Santos, A.S.; Semzezem, C.; Correia, M.R.; Brito, L.A.; Ruiz, M.O.; Fukui, R.T.; Matioli, S.R.; Passos-Bueno, M.R.; Silva, M.E.R.D. The influence of population stratification on genetic markers associated with type 1 diabetes. Sci. Rep. 2017, 7, 43513. [Google Scholar] [CrossRef]
- Sharp, S.A.; Rich, S.S.; Wood, A.R.; Jones, S.E.; Beaumont, R.N.; Harrison, J.W.; Schneider, D.A.; Locke, J.M.; Tyrrell, J.; Weedon, M.N.; et al. Development and Standardization of an Improved Type 1 Diabetes Genetic Risk Score for Use in Newborn Screening and Incident Diagnosis. Diabetes Care 2019, 42, 200–207. [Google Scholar] [CrossRef]
- Harrison, J.W.; Tallapragada, D.S.P.; Baptist, A.; Sharp, S.A.; Bhaskar, S.; Jog, K.S.; Patel, K.A.; Weedon, M.N.; Chandak, G.R.; Yajnik, C.S.; et al. Type 1 diabetes genetic risk score is discriminative of diabetes in non-Europeans: Evidence from a study in India. Sci. Rep. 2020, 10, 9450. [Google Scholar] [CrossRef] [PubMed]
- Onengut-Gumuscu, S.; Chen, W.M.; Robertson, C.C.; Bonnie, J.K.; Farber, E.; Zhu, Z.; Oksenberg, J.R.; Brant, S.R.; Louis Bridges, S.; Edberg, J.C.; et al. Type 1 diabetes risk in African-ancestry participants and utility of an ancestry-specific genetic risk score. Diabetes Care 2019, 42, 406–415. [Google Scholar] [CrossRef] [PubMed]
- Pena, S.D.J.; Santos, F.R.; Tarazona-Santos, E. Genetic admixture in Brazil. Am. J. Med. Genet. Part C Semin. Med. Genet. 2020, 184, 928–938. [Google Scholar] [CrossRef]
- Saloum de Neves Manta, F.; Pereira, R.; Vianna, R.; Beuttenmüller de Araújo, A.R.; Leite Góes Gitaí, D.; Aparecida da Silva, D.; de Vargas Wolfgramm, E.; da Mota Pontes, I.; Ivan Aguiar, J.; Ozório Moraes, M.; et al. Revisiting the Genetic Ancestry of Brazilians Using Autosomal AIM-Indels. PLoS ONE 2013, 8, e75145. [Google Scholar] [CrossRef]
- Torres, L.; da Silva Bouzas, L.F.; Almada, A.; de Sobrino Porto, L.C.M.; Abdelhay, E. Distribution of HLA-A, -B and -DRB1 antigenic groups and haplotypes from the Brazilian bone marrow donor registry (REDOME). Hum. Immunol. 2017, 78, 602–609. [Google Scholar] [CrossRef]
- Boquett, J.A.; Bisso-Machado, R.; Zagonel-Oliveira, M.; Schüler-Faccini, L.; Fagundes, N.J.R. HLA diversity in Brazil. HLA 2020, 95, 3–14. [Google Scholar] [CrossRef]
- Gomes, M.B.; Negrato, C.A. Adherence to insulin therapeutic regimens in patients with type 1 diabetes. A nationwide survey in Brazil. Diabetes Res. Clin. Pract. 2016, 120, 47–55. [Google Scholar] [CrossRef]
- Instituto Brasileiro de Geografia e Estatística (IBGE). Censo 2000. 2000. Available online: http:/www.ibge.gov.br/censo (accessed on 15 May 2022).
- Pereira, R.; Phillips, C.; Pinto, N.; Santos, C.; dos Santos, S.E.B.; Amorim, A.; Carracedo, Á.; Gusmão, L. Straightforward inference of ancestry and admixture proportions through ancestry-informative insertion deletion multiplexing. PLoS ONE 2012, 7, e29684. [Google Scholar] [CrossRef] [PubMed]
- Hurley, C.K.; Kempenich, J.; Wadsworth, K.; Sauter, J.; Hofmann, J.A.; Schefzyk, D.; Schmidt, A.H.; Galarza, P.; Cardozo, M.B.; Dudkiewicz, M.; et al. Common, intermediate and well-documented HLA alleles in world populations: CIWD version 3.0.0. HLA 2020, 95, 516–531. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Schisterman, E.F.; Perkins, N.J.; Liu, A.; Bondell, H. Optimal Cut-point and Its Corresponding Youden Index to Discriminate Individuals Using Pooled Blood Samples. Epidemiology 2005, 16, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Mazas, A.; Fernandez-Viña, M.; Middleton, D.; Hollenbach, J.A.; Buhler, S.; Di, D.; Rajalingam, R.; Dugoujon, J.-M.; Mack, S.J.; Thorsby, E. Immunogenetics as a tool in anthropological studies. Immunology 2011, 133, 143–164. [Google Scholar] [CrossRef]
- Levy, M.S.F. O papel da migração internacional na evolução da população brasileira (1872 a 1972). Rev. Saude Publica 1974, 8, 49–90. [Google Scholar] [CrossRef]
- Sanchez-Mazas, A.; Nunes, J.M.; Middleton, D.; Sauter, J.; Buhler, S.; McCabe, A.; Hofmann, J.; Baier, D.M.; Schmidt, A.H.; Nicoloso, G.; et al. Common and well-documented HLA alleles over all of Europe and within European sub-regions: A catalogue from the European Federation for Immunogenetics. HLA 2017, 89, 104–113. [Google Scholar] [CrossRef]
- Besançon, S.; Govender, D.; Sidibé, A.T.; Noble, J.A.; Togo, A.; Lane, J.A.; Mack, S.J.; Atkinson, M.A.; Wasserfall, C.H.; Kakkat, F.; et al. Clinical features, biochemistry, and HLA - DRB1 status in youth-onset type 1 diabetes in Mali. Pediatr. Diabetes 2022, 23, 1552–1559. [Google Scholar] [CrossRef]
- Salzano, F.M.; Sans, M. Interethnic admixture and the evolution of Latin American populations. Genet. Mol. Biol. 2014, 37 (Suppl. S1), 151–170. [Google Scholar] [CrossRef]
- Redondo, M.J.; Gignoux, C.R.; Dabelea, D.; Hagopian, W.A.; Onengut-Gumuscu, S.; Oram, R.A.; Rich, S.S. Type 1 diabetes in diverse ancestries and the use of genetic risk scores. Lancet Diabetes Endocrinol. 2022, 10, 597–608. [Google Scholar] [CrossRef]
- Probst, C.M.; Hutz, M.H.; Salzano, F.M.; Zago, M.A.; Hill, K.; Hurtado, A.M. HLA class II diversity in seven Amerindian populations. Clues about the origins of the Ache. Tissue Antigens 2003, 62, 512–526. [Google Scholar] [CrossRef]
- Parolín, M.L.; Carnese, F.R. HLA-DRB1 alleles in four Amerindian populations from Argentina and Paraguay. Genet. Mol. Biol. 2009, 32, 212–219. [Google Scholar] [CrossRef]
- Jinam, T.A.; Hosomichi, K.; Nakaoka, H.; Phipps, M.E.; Saitou, N.; Inoue, I. Allelic and haplotypic HLA diversity in indigenous Malaysian populations explored using Next Generation Sequencing. Hum. Immunol. 2022, 83, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Tracey, M.C.; Carter, J.M. Class II HLA allele polymorphism: DRB1, DQB1 and DPB1 alleles and haplotypes in the New Zealand Maori population. Tissue Antigens 2006, 68, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Uinuk-ool, T.S.; Takezaki, N.; Sukernik, R.I.; Nagl, S.; Klein, J. Origin and affinities of indigenous Siberian populations as revealed by HLA class II gene frequencies. Hum. Genet. 2002, 110, 209–226. [Google Scholar] [CrossRef] [PubMed]
- Instituto Brasileiro de Geografia e Estatística (IBGE). Brasil: 500 Anos de Povoamento; IBGE: Rio de Janeiro, Brazil, 2007; pp. 35–99. [Google Scholar]
- Gomes, M.B.; Porto, L.C.; Silva, D.A.; Negrato, C.A.; Pavin, E.J.; Montenegro Junior, R.; Dib, S.A.; Felício, J.S.; Santos, D.C.; Muniz, L.H.; et al. HLA Genotypes and Type 1 Diabetes and Its Relationship to Reported Race/Skin Color in Their Relatives: A Brazilian Multicenter Study. Genes 2022, 13, 972. [Google Scholar] [CrossRef] [PubMed]
| Variables | n | (%) |
|---|---|---|
| Sex | ||
| Male | 738 | (46.2) |
| Female | 861 | (53.8) |
| Age | ||
| <20 years old | 360 | (22.5) |
| ≥20 years old | 1239 | (77.5) |
| Age at type 1 diabetes diagnosis (mean ± SD) | 14.8 ± 8.9 | |
| Age group at type 1 diabetes onset | ||
| 0–4 years | 145 | (9.1) |
| 5–9 years | 328 | (20.5) |
| 10–14 years | 442 | (27.6) |
| 15–19 years | 281 | (17.6) |
| 20–24 years | 158 | (9.9) |
| 25–29 years | 134 | (8.4) |
| ≥30 years | 111 | (6.9) |
| Brazilian region of birth | ||
| Southeast | 680 | (42.5) |
| Northeast | 539 | (33.7) |
| South | 208 | (13.0) |
| Midwest | 127 | (7.9) |
| North | 45 | (2.8) |
| Self-reported skin-color/race | ||
| White | 824 | (51.5) |
| Brown | 616 | (38.5) |
| Black | 130 | (8.1) |
| Yellow | 16 | (1.0) |
| Indigenous | 13 | (0.8) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomes, M.B.; Rodrigues, V.; Santos, D.C.; Bôas, P.R.V.; Silva, D.A.; de Sousa Azulay, R.S.; Dib, S.A.; Pavin, E.J.; Fernandes, V.O.; Montenegro Junior, R.M.; et al. Association between HLA Class II Alleles/Haplotypes and Genomic Ancestry in Brazilian Patients with Type 1 Diabetes: A Nationwide Exploratory Study. Genes 2023, 14, 991. https://doi.org/10.3390/genes14050991
Gomes MB, Rodrigues V, Santos DC, Bôas PRV, Silva DA, de Sousa Azulay RS, Dib SA, Pavin EJ, Fernandes VO, Montenegro Junior RM, et al. Association between HLA Class II Alleles/Haplotypes and Genomic Ancestry in Brazilian Patients with Type 1 Diabetes: A Nationwide Exploratory Study. Genes. 2023; 14(5):991. https://doi.org/10.3390/genes14050991
Chicago/Turabian StyleGomes, Marília Brito, Vandilson Rodrigues, Deborah Conte Santos, Paulo Ricardo Villas Bôas, Dayse A. Silva, Rossana Santiago de Sousa Azulay, Sergio Atala Dib, Elizabeth João Pavin, Virgínia Oliveira Fernandes, Renan Magalhães Montenegro Junior, and et al. 2023. "Association between HLA Class II Alleles/Haplotypes and Genomic Ancestry in Brazilian Patients with Type 1 Diabetes: A Nationwide Exploratory Study" Genes 14, no. 5: 991. https://doi.org/10.3390/genes14050991
APA StyleGomes, M. B., Rodrigues, V., Santos, D. C., Bôas, P. R. V., Silva, D. A., de Sousa Azulay, R. S., Dib, S. A., Pavin, E. J., Fernandes, V. O., Montenegro Junior, R. M., Felicio, J. S., Réa, R., Negrato, C. A., & Porto, L. C. (2023). Association between HLA Class II Alleles/Haplotypes and Genomic Ancestry in Brazilian Patients with Type 1 Diabetes: A Nationwide Exploratory Study. Genes, 14(5), 991. https://doi.org/10.3390/genes14050991







