Transcriptome Profiling of Different Developmental Stages on Longissimus Dorsi to Identify Genes Underlying Intramuscular Fat Content in Wannanhua Pigs
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement and Collection of Tissue Samples
2.2. RNA Extraction, Library Construction, and Illumina Sequencing
2.3. Time Series Expression Pattern Clustering
2.4. Functional Annotation
2.5. Reliability Verification of RNA-seq Data
2.6. Data Availability
3. Results
3.1. Body Weight and IMF Content
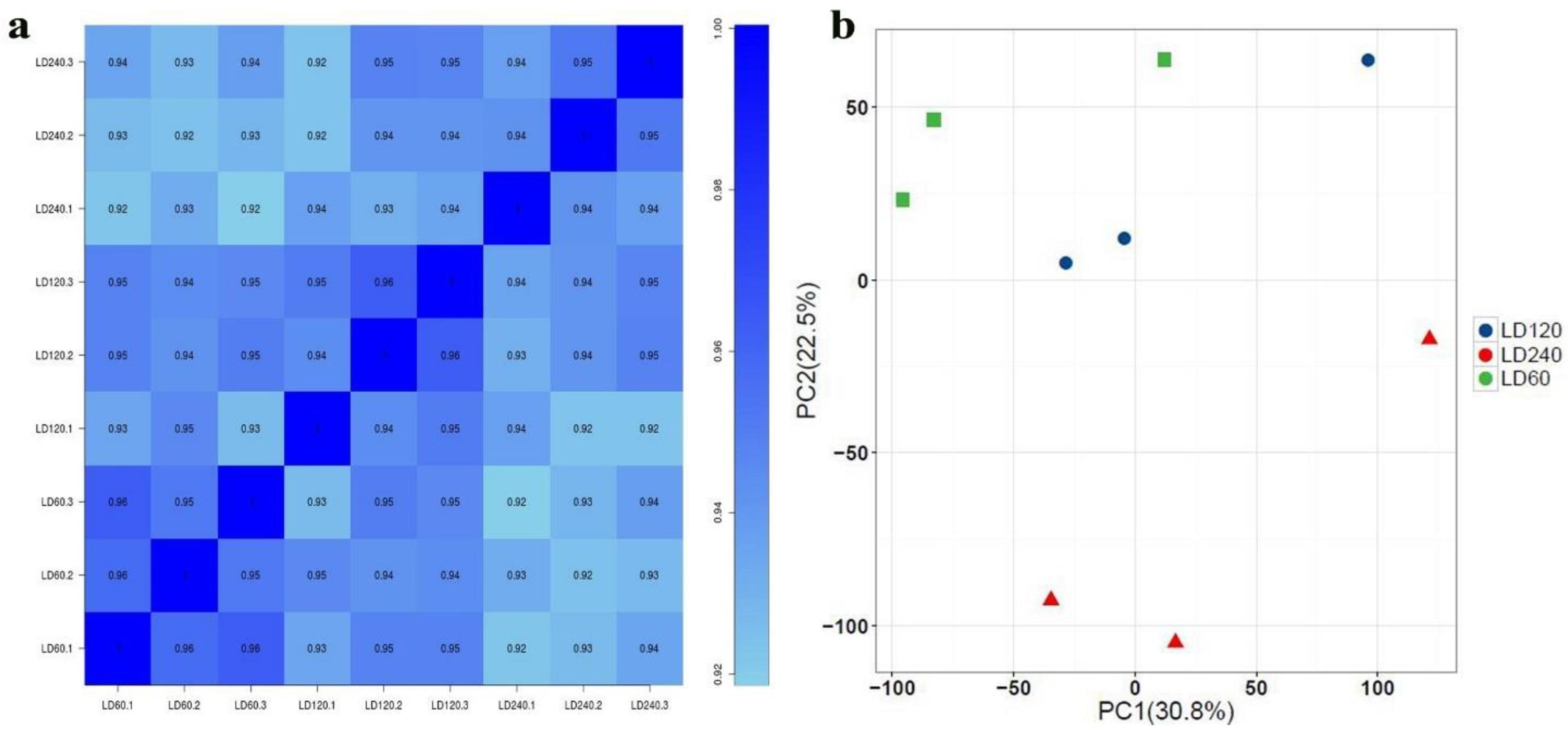
3.2. Summary of RNA-seq Results
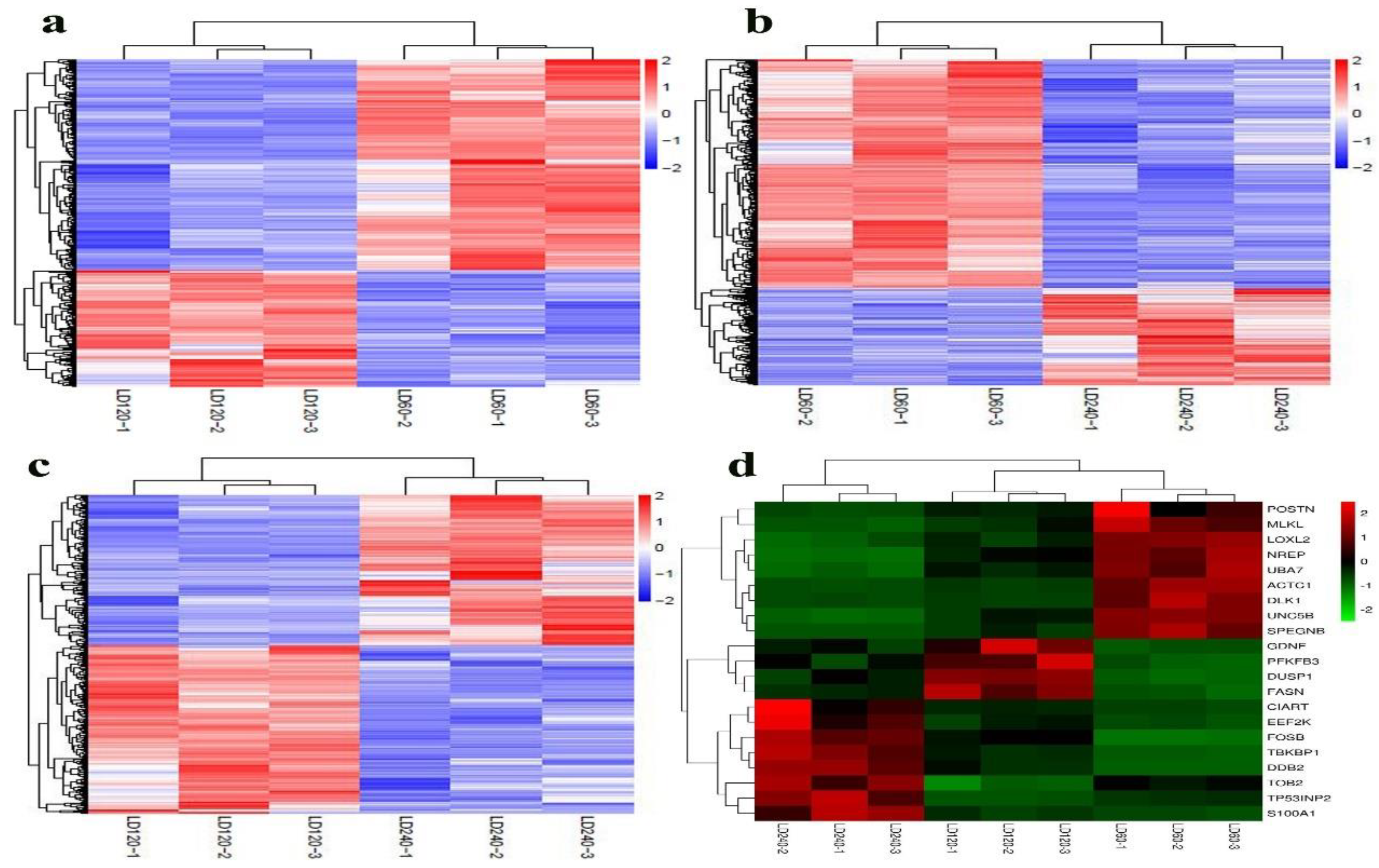
3.3. Differential Expression Analysis
3.4. STEM Analysis
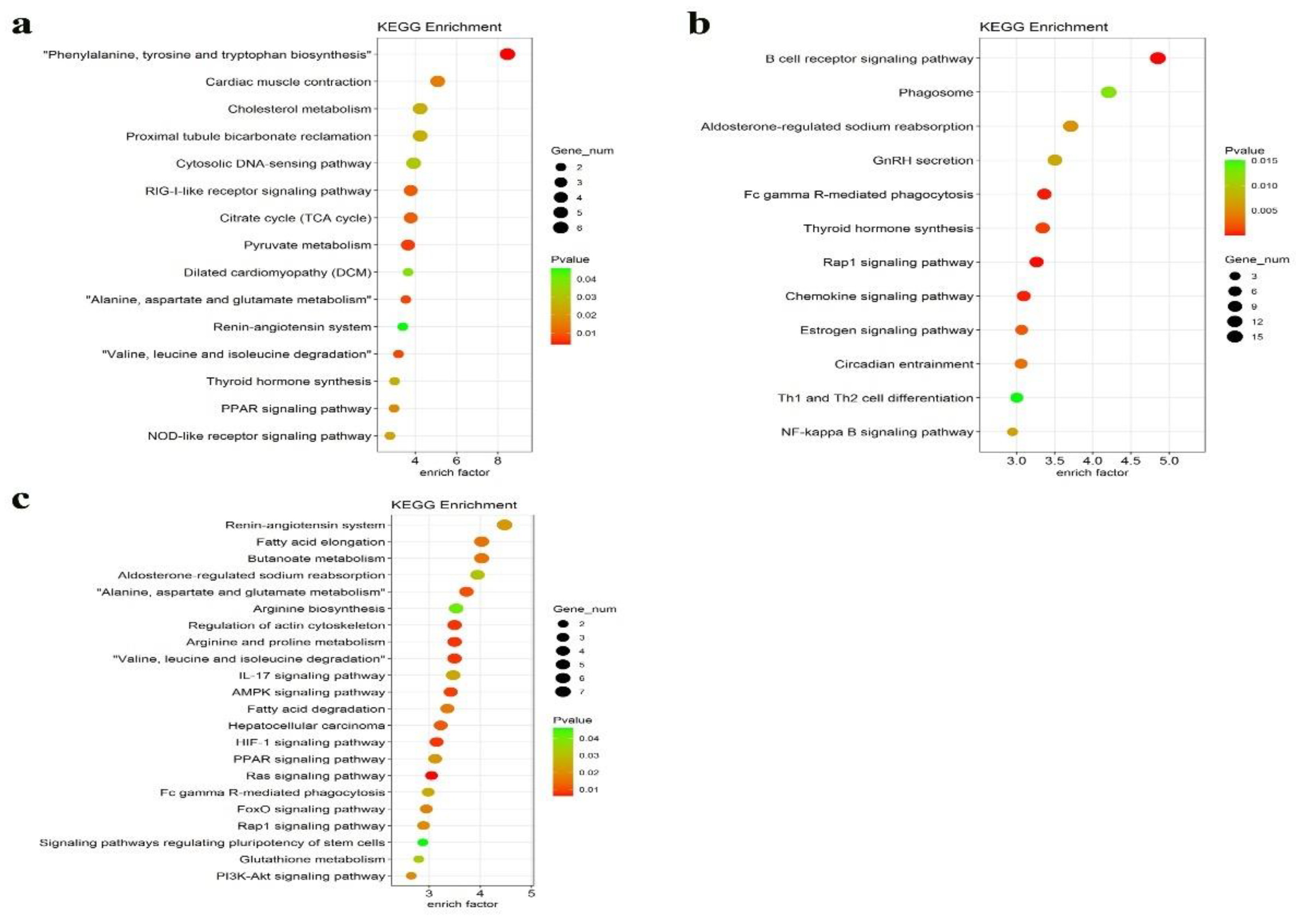
3.5. Functional Annotation and Enrichment Analysis of DEGs
3.6. Quantitative Validation of RNA-seq Data
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Sá, P.M.; Richard, A.J.; Hang, H.; Stephens, J.M. Transcriptional regulation of adipogenesis. Compr. Physiol. 2011, 7, 635–674. [Google Scholar]
- Campos, C.F.; Duarte, M.S.; Guimarães, S.E.F.; Verardo, L.L.; Wei, S.; Du, M.; Jiang, Z.; Bergen, W.G.; Hausman, G.J.; Fernyhough-Culver, M.; et al. Review: Animal model and the current understanding of molecule dynamics of adipogenesis. Animal 2016, 10, 927–932. [Google Scholar] [CrossRef]
- Hocquette, J.F.; Gondret, F.; Baéza, E.; Médale, F.; Jurie, C.; Pethick, D.W. Intramuscular fat content in meat-producing animals: Development, genetic and nutritional control, and identification of putative markers. Animal 2010, 4, 303–319. [Google Scholar] [CrossRef]
- Du, M.; Tong, J.; Zhao, J.; Underwood, K.R.; Zhu, M.; Ford, S.P.; Nathanielsz, P.W. Fetal programming of skeletal muscle development in ruminant animals. J. Anim. Sci. 2010, 88 (Suppl. 13), E51–E60. [Google Scholar] [CrossRef]
- Tang, Q.Q.; Otto, T.C.; Lane, M.D. Commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proc. Natl. Acad. Sci. USA 2004, 101, 9607–9611. [Google Scholar] [CrossRef]
- Ayuso, M.; Fernandez, A.; Nunez, Y.; Benitez, R.; Isabel, B.; Fernandez, A.I.; Rey, A.I.; Gonzalez-Bulnes, A.; Medrano, J.F.; Canovas, A.; et al. Developmental stage, muscle and genetic type modify muscle transcriptome in pigs: Effects on gene expression and regulatory factors involved in growth and metabolism. PLoS ONE 2016, 11, e0167858. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, Y.; Zhang, B.; Zhong, H.; Lu, Y.; Zhang, H. Candidate gene screening for lipid deposition using combined transcriptomic and proteomic data from Nanyang black pigs. BMC Genom. 2021, 22, 441. [Google Scholar] [CrossRef]
- Qiao, R.; Li, X.; Han, X.; Wang, K.; Lv, G.; Ren, G.; Li, X. Population structure and genetic diversity of four Henan pig populations. Anim. Genet. 2019, 50, 262–265. [Google Scholar] [CrossRef]
- Li, X.J.; Zhou, J.; Liu, L.Q.; Qian, K.; Wang, C.L. Identification of genes in longissimus dorsi muscle differentially expressed between Wannanhua and Yorkshire pigs using RNA-sequencing. Anim. Genet. 2016, 47, 324–333. [Google Scholar] [CrossRef]
- Picard, B.; Lefaucheur, L.; Berri, C.; Duclos, M.J. Muscle fibre ontogenesis in farm animal species. Reprod. Nutr. Dev. 2002, 42, 415–431. [Google Scholar] [CrossRef]
- Bérard, J.; Kalbe, C.; Lösel, D.; Tuchscherer, A.; Rehfeldt, C. Potential sources of early-postnatal increase in myofibre number in pig skeletal muscle. Histochem. Cell. Biol. 2011, 136, 217–225. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, J.; Liu, H.; Xi, Y.; Xue, M.; Liu, W.; Zhuang, Z.; Lei, M. Dynamic transcriptome profiles of skeletal muscle tissue across 11 developmental stages for both Tongcheng and Yorkshire pigs. BMC Genom. 2015, 16, 377. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Ernst, J.; Bar-Joseph, Z. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinform. 2006, 7, 191. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Zhang, J. Effects of Adiponection on Skeletal Muscle Growth and Types of Muscle Fibers of Wannanhua Pigs; Anhui Agricultural University: Hefei, China, 2010. (In Chinese) [Google Scholar]
- Mai, M.; Jin, L.; Tian, S.; Liu, R.; Huang, W.; Tang, Q.; Ma, J.; Wang, X.; Hu, Y.; Wang, D.; et al. Deciphering the microRNA transcriptome of skeletal muscle during porcine development. PeerJ 2016, 4, e1504. [Google Scholar] [CrossRef]
- Völkers, M.; Rohde, D.; Goodman, C.; Most, P. 100A1: A regulator of striated muscle sarcoplasmic reticulum Ca2+ handling, sarcomeric, and mitochondrial function. J. Biomed. Biotechnol. 2010, 2010, 178614. [Google Scholar] [CrossRef]
- Cortez-Toledo, O.; Schnair, C.; Sangngern, P.; Metzger, D.; Chao, L.C. Nur77 deletion impairs muscle growth during developmental myogenesis and muscle regeneration in mice. PLoS ONE 2017, 12, e0171268. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Bai, L.; Qin, S.; Li, Q. Expression of TGFBI gene in different periods and tissues of Landrace pigs. Heilongjiang Anim. Husb. Vet. Sci. 2016, 9, 111–112. (In Chinese) [Google Scholar]
- Pan, Y.; Yu, C.; Huang, J.; Rong, Y.; Chen, J.; Chen, M. Bioinformatics analysis of vascular RNA-seq data revealed hub genes and pathways in a novel Tibetan minipig atherosclerosis model induced by a high fat/cholesterol diet. Lipids Health Dis. 2020, 19, 54. [Google Scholar] [CrossRef]
- Rehfeldt, C.; Kuhn, G. Consequences of birth weight for postnatal growth performance and carcass quality in pigs as related to myogenesis. J. Anim. Sci. 2006, 84 (Suppl. 13), E113–E123. [Google Scholar] [CrossRef]
- Bee, G.; Calderini, M.; Biolley, C.; Guex, G.; Herzog, W.; Lindemann, M.D. Changes in the histochemical properties and meat quality traits of porcine muscles during the growing-finishing period as affected by feed restriction, slaughter age, or slaughter weight. J. Anim. Sci. 2007, 85, 1030–1045. [Google Scholar] [CrossRef]
- Peters, E.L.; Van der Linde, S.M.; Vogel, I.S.; Haroon, M.; Offringa, C.; De Wit, G.M.; Koolwijk, P.; Van der Laarse, W.J.; Jaspers, R.T. IGF-1 attenuates hypoxia-induced atrophy but inhibits myoglobin expression in C2C12 skeletal muscle myotubes. Int. J. Mol. Sci. 2017, 18, 1889. [Google Scholar] [CrossRef]
- Piccinin, E.; Cariello, M.; De Santis, S.; Ducheix, S.; Sabbà, C.; Ntambi, J.M.; Moschetta, A. Role of oleic acid in the gut-liver axis: From diet to the regulation of its synthesis via stearoyl-CoA desaturase 1 (SCD1). Nutrients 2019, 11, 2283. [Google Scholar] [CrossRef]
- Bessa, R.J.B.; Hughes, R.A.; Jeronimo, E.; Moreira, O.C.; Prates, J.A.M.; Doran, O. Effect of pig breed and dietary protein level on selected fatty acids and stearoyl-coenzyme A desaturase protein expression in longissimus muscle and subcutaneous fat. J. Anim. Sci. 2013, 91, 4540–4546. [Google Scholar] [CrossRef]
- Agarwal, A.K.; Arioglu, E.; De Almeida, S.; Akkoc, N.; Taylor, S.I.; Bowcock, A.M.; Barnes, R.I.; Garg, A. AGPAT2 is mutated in congenital generalized lipodystrophy linked to chromosome 9q34. Nat. Genet. 2002, 31, 21–23. [Google Scholar] [CrossRef]
- Verschuure, P.; Tatard, C.; Boelens, W.C.; Grongnet, J.F.; David, J.C. Expression of small heat shock proteins HspB2, HspB8, Hsp20 and cvHsp in different tissues of the perinatal developing pig. Eur. J. Cell Biol. 2003, 82, 523–530. [Google Scholar] [CrossRef]
- Zambonelli, P.; Gaffo, E.; Zappaterra, M.; Bortoluzzi, S.; Davoli, R. Transcriptional profiling of subcutaneous adipose tissue in Italian Large White pigs divergent for backfat thickness. Anim. Genet. 2016, 47, 306–323. [Google Scholar] [CrossRef]
- Gozalo-Marcilla, M.; Buntjer, J.; Johnsson, M.; Batista, L.; Diez, F.; Werner, C.R.; Chen, C.Y.; Gorjanc, G.; Mellanby, R.J.; Hickey, J.M.; et al. Genetic architecture and major genes for backfat thickness in pig lines of diverse genetic backgrounds. Genet. Sel. Evol. 2021, 53, 76. [Google Scholar] [CrossRef] [PubMed]
- Hong, F.; Haldeman, B.D.; Jackson, D.; Carter, M.; Baker, J.E.; Cremo, C.R. Biochemistry of smooth muscle myosin light chain kinase. Arch. Biochem. Biophys. 2011, 510, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, Y.; Pan, J.; Liu, X.; Chen, H.; Zhou, X.; Yuan, Z.; Wang, X.; Mo, D. iTRAQ-based quantitative proteomic analysis reveals the distinct early embryo myofiber type characteristics involved in landrace and miniature pig. BMC Genom. 2016, 17, 137. [Google Scholar] [CrossRef]
- Wang, Y.; Sul, H.S. Ectodomain shedding of preadipocyte factor 1 (Pref-1) by tumor necrosis factor α converting enzyme (TACE) and inhibition of adipocyte differentiation. Mol. Cell. Biol. 2006, 26, 5421–5435. [Google Scholar] [CrossRef]
- Fu, Y.; Hao, X.; Shang, P.; Chamba, Y.; Zhang, B.; Zhang, H. Functional Identification of Porcine DLK1 during Muscle Development. Animals 2022, 12, 1523. [Google Scholar] [CrossRef]
- Wang, Y.; Ma, C.; Sun, Y.; Li, Y.; Kang, L.; Jiang, Y. Dynamic transcriptome and DNA methylome analyses on longissimus dorsi to identify genes underlying intramuscular fat content in pigs. BMC Genom. 2017, 18, 780. [Google Scholar] [CrossRef]
- Lu, Y.; Mao, J.; Han, X.; Zhang, W.; Li, Y.; Liu, Y.; Li, Q. Downregulated hypoxia-inducible factor 1α improves myoblast differentiation under hypoxic condition in mouse genioglossus. Mol. Cell. Biochem. 2021, 476, 1351–1364. [Google Scholar] [CrossRef]
- Ke, Q.; Costa, M. Hypoxia-inducible factor-1 (HIF-1). Mol. Pharmacol. 2006, 70, 1469–1480. [Google Scholar] [CrossRef] [PubMed]
- Dehne, N.; Kerkweg, U.; Flohé, S.B.; Brüne, B.; Fandrey, J. Activation of hypoxia-inducible factor 1 in skeletal muscle cells after exposure to damaged muscle cell debris. Shock 2011, 35, 632–638. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.J.; Wang, L.Y.; Chodosh, L.A.; Keith, B.; Simon, M.C. Differential roles of hypoxia-inducible factor 1α (HIF-1α) and HIF-2α in hypoxic gene regulation. Mol. Cell. Biol. 2003, 23, 9361–9374. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.; Long, H.; Yuan, Y.; Zhang, H.; Peng, Y.; Zhou, D.; Liu, C.; Xiang, B.; Huang, Y.; Zhao, Y.; et al. Identification of Body Size Determination Related Candidate Genes in Domestic Pig Using Genome-Wide Selection Signal Analysis. Animals 2022, 12, 1839. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.J.; Gao, T.; Li, J.L.; Zhang, L.; Gao, F.; Zhou, G.H. Effects of dietary starch types on early postmortem muscle energy metabolism in finishing pigs. Meat Sci. 2017, 133, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Chen, Y.; Zhu, Y. The molecular basis of targeting PFKFB3 as a therapeutic strategy against cancer. Oncotarget 2017, 8, 62793. [Google Scholar] [CrossRef] [PubMed]
- Velez-Irizarry, D.; Casiro, S.; Daza, K.R.; Bates, R.O.; Raney, N.E.; Steibel, J.P.; Ernst, C.W. Genetic control of longissimus dorsi muscle gene expression variation and joint analysis with phenotypic quantitative trait loci in pigs. BMC Genom. 2019, 20, 3. [Google Scholar] [CrossRef]





| Genes | Sequence (5′-to-3′) | Product Size (bp) | TM (°C) |
|---|---|---|---|
| PCK2 | F: CCAACCAGAGGGCATCCAC R: AGTTACAATCACCGTCTTGCT | 175 | 59.4 |
| ACTC1 | F: TCTGCTGGCATCCATGAAACGA R: GGCAGTGATTTCCTTTTGCAT | 147 | 59.2 |
| PHGDH | F: GAAATCGCCATCCAGTTCGT R: AGAGCTTCAGCCAGACCAA | 125 | 56.3 |
| POSTN | F: TCCCCATAACTGTCTACACC R: CCTTCCGTTTTGATAATAGGC | 100 | 53.4 |
| HOXD1 | F: AGATAGCCAACTCATTGCAG R: GCAAAAGCCCTTCTCGTTC | 99 | 54.5 |
| TBKBP1 | F: AACGGCTCAACCAATTCCA R: TCACTCTTCTCCACGCAGA | 106 | 55.6 |
| TP53INP2 | F: CTGGCTCATCATTGACCTG R: TGCTGCCAGTAACATAAACTGA | 119 | 53.7 |
| UBE2L6 | F: TGCCCATCATCAGCAACGA R: GTCTATTCACCAGCACGTTGA | 84 | 52.6 |
| S100A1 | F: GAACCTGCTCCCAATACACC R: GAGTCTCCATCGCTGTCTC | 76 | 56.0 |
| SPEGNB | F: CATCAACATAACCAACCCCTT R: TTAATCTTCGCCGGGACTTCCA | 75 | 58.5 |
| NREP | F: CCAAAGGAAGTGAACCGCAAG R: TCATTTTCATTGCTGCCGAGT | 77 | 59.0 |
| CIART | F: GGAGCCAAGTCTAAATATCCAC R: TCCTCTTTCAAATCGACCCAT | 139 | 56.8 |
| β-actin a | F: GTCATCACCATCGGCAATGAG R: AATGCCGCAGGATTCCATG | 84 | 56.4 |
| Samples ID | Raw Reads | Clean Reads | Mapped Reads | Clean Ratio | Unique Reads | Mapping Ratio |
|---|---|---|---|---|---|---|
| LD60-1 | 50,054,498 | 45,849,605 | 42,757,962 | 91.60% | 42,502,777 | 96.28% |
| LD60-2 | 54,562,360 | 50,025,837 | 46,411,138 | 91.69% | 46,130,037 | 96.24% |
| LD60-3 | 51,280,966 | 46,622,900 | 43,592,197 | 90.92% | 43,333,563 | 96.46% |
| LD120-1 | 50,000,566 | 46,569,727 | 43,889,677 | 93.14% | 43,674,962 | 96.89% |
| LD120-2 | 53,709,810 | 48,261,833 | 44,860,139 | 89.86% | 44,592,694 | 95.87% |
| LD120-3 | 53,573,860 | 49,887,575 | 46,710,172 | 93.12% | 46,427,981 | 96.05% |
| LD240-1 | 48,876,746 | 43,937,385 | 41,037,844 | 89.89% | 40,835,618 | 96.72% |
| LD240-2 | 53,312,294 | 49,045,957 | 45,672,971 | 92.00% | 45,385,253 | 96.02% |
| LD240-3 | 53,267,442 | 48,786,293 | 45,298,674 | 91.59% | 45,048,382 | 95.84% |
| Gene Name | Description | LD60 vs. LD120 | LD60 vs. LD240 | LD120 vs. LD240 | |||
|---|---|---|---|---|---|---|---|
| log2FC | p-Value | log2FC | p-Value | log2FC | p-Value | ||
| ACTC1 | actin α cardiac muscle 1 | 2.92 | 9.29E-16 | 4.17 | 5.69E-29 | 1.25 | 9.40E-04 |
| TGFBI | transforming growth factor β induced | 0.21 | 8.74E-01 | 1.28 | 1.26E-04 | 1.08 | 1.73E-03 |
| UCP2 | uncoupling protein 2 | 0.25 | 9.92E-01 | 2.16 | 1.22E-04 | 1.92 | 8.37E-04 |
| TNNT2 | Sus scrofa troponin T2, cardiac type (TNNT2). | 1.63 | 7.00E-03 | 3.77 | 2.89E-07 | 2.14 | 1.32E-02 |
| FAM184B | family with sequence similarity 184 member B | 0.59 | 2.29E-01 | 1.45 | 7.47E-04 | 0.85 | 3.16E-02 |
| FADS1 | fatty acid desaturase 1 | 0.47 | 4.59E-01 | 1.59 | 2.46E-03 | 1.11 | 2.52E-02 |
| WWTR1 | WW domain containing transcription regulator 1 | 0.58 | 2.56E-01 | 1.19 | 2.52E-03 | 0.61 | 7.04E-02 |
| MYL9 | myosin light chain 9 | 1.24 | 2.01E-03 | 1.59 | 4.38E-03 | 0.34 | 5.50E-01 |
| SH3PXD2B | SH3 and PX domains 2B | 0.63 | 2.94E-01 | 1.63 | 3.14E-03 | 1.00 | 5.45E-02 |
| NCF1 | neutrophil cytosolic factor 1 | 0.40 | 8.08E-01 | 4.13 | 2.58E-04 | 3.73 | 8.47E-04 |
| ANXA1 | annexin A1 | 1.14 | 2.37E-02 | 1.72 | 6.31E-04 | 0.58 | 1.72E-01 |
| FGFR4 | fibroblast growth factor receptor 4 | −0.10 | 3.00E-01 | 1.52 | 3.62E-04 | 1.62 | 7.17E-05 |
| PHGDH | phosphoglycerate dehydrogenase | 2.07 | 2.08E-05 | 5.38 | 1.72E-22 | 3.31 | 1.32E-07 |
| POSTN | periostin | 1.79 | 1.69E-03 | 4.54 | 5.81E-11 | 2.75 | 1.24E-05 |
| TP53INP2 | tumor protein p53 inducible nuclear protein 2 | 1.30 | 2.58E-05 | −2.05 | 2.22E-09 | −3.35 | 1.49E-16 |
| NREP | neuronal regeneration related protein | 1.24 | 4.18E-05 | 4.22 | 5.26E-27 | 2.99 | 3.53E-14 |
| DLK1 | delta like non-canonical Notch ligand 1 | 4.07 | 3.64E-19 | 7.52 | 1.12E-27 | 3.45 | 1.06E-03 |
| HSPB8 | heat shock protein family B (small) member 8 | −0.32 | 5.09E-02 | −1.06 | 3.45E-04 | −0.74 | 4.32E-02 |
| SETD7 | SET domain containing 7, histone lysine methyltransferase | −0.49 | 3.70E-02 | −1.39 | 1.39E-05 | −0.91 | 3.13E-02 |
| S100A1 | S100 calcium binding protein A1 | −1.09 | 9.64E-04 | −3.76 | 5.85E-17 | −2.67 | 5.19E-09 |
| SCD | stearoyl-CoA desaturase | −0.37 | 2.52E-01 | −2.21 | 1.61E-03 | −1.84 | 7.84E-03 |
| LEPR | leptin receptor | −1.30 | 3.96E-01 | −4.32 | 2.94E-04 | −3.03 | 1.21E-02 |
| IRX3 | iroquois homeobox 3 | −0.69 | 3.22E-02 | −2.41 | 1.21E-05 | −1.72 | 3.19E-03 |
| AGPAT2 | 1-acylglycerol-3-phosphate O-acyltransferase 2 | −0.33 | 5.44E-02 | −1.04 | 6.77E-04 | −0.71 | 9.00E-02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Yang, Y.; Li, L.; Ren, M.; Zhou, M.; Li, S. Transcriptome Profiling of Different Developmental Stages on Longissimus Dorsi to Identify Genes Underlying Intramuscular Fat Content in Wannanhua Pigs. Genes 2023, 14, 903. https://doi.org/10.3390/genes14040903
Li X, Yang Y, Li L, Ren M, Zhou M, Li S. Transcriptome Profiling of Different Developmental Stages on Longissimus Dorsi to Identify Genes Underlying Intramuscular Fat Content in Wannanhua Pigs. Genes. 2023; 14(4):903. https://doi.org/10.3390/genes14040903
Chicago/Turabian StyleLi, Xiaojin, Yanan Yang, Lei Li, Man Ren, Mei Zhou, and Shenghe Li. 2023. "Transcriptome Profiling of Different Developmental Stages on Longissimus Dorsi to Identify Genes Underlying Intramuscular Fat Content in Wannanhua Pigs" Genes 14, no. 4: 903. https://doi.org/10.3390/genes14040903
APA StyleLi, X., Yang, Y., Li, L., Ren, M., Zhou, M., & Li, S. (2023). Transcriptome Profiling of Different Developmental Stages on Longissimus Dorsi to Identify Genes Underlying Intramuscular Fat Content in Wannanhua Pigs. Genes, 14(4), 903. https://doi.org/10.3390/genes14040903





