Evidence of Association between CTLA-4 Gene Polymorphisms and Colorectal Cancers in Saudi Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. DNA Extraction
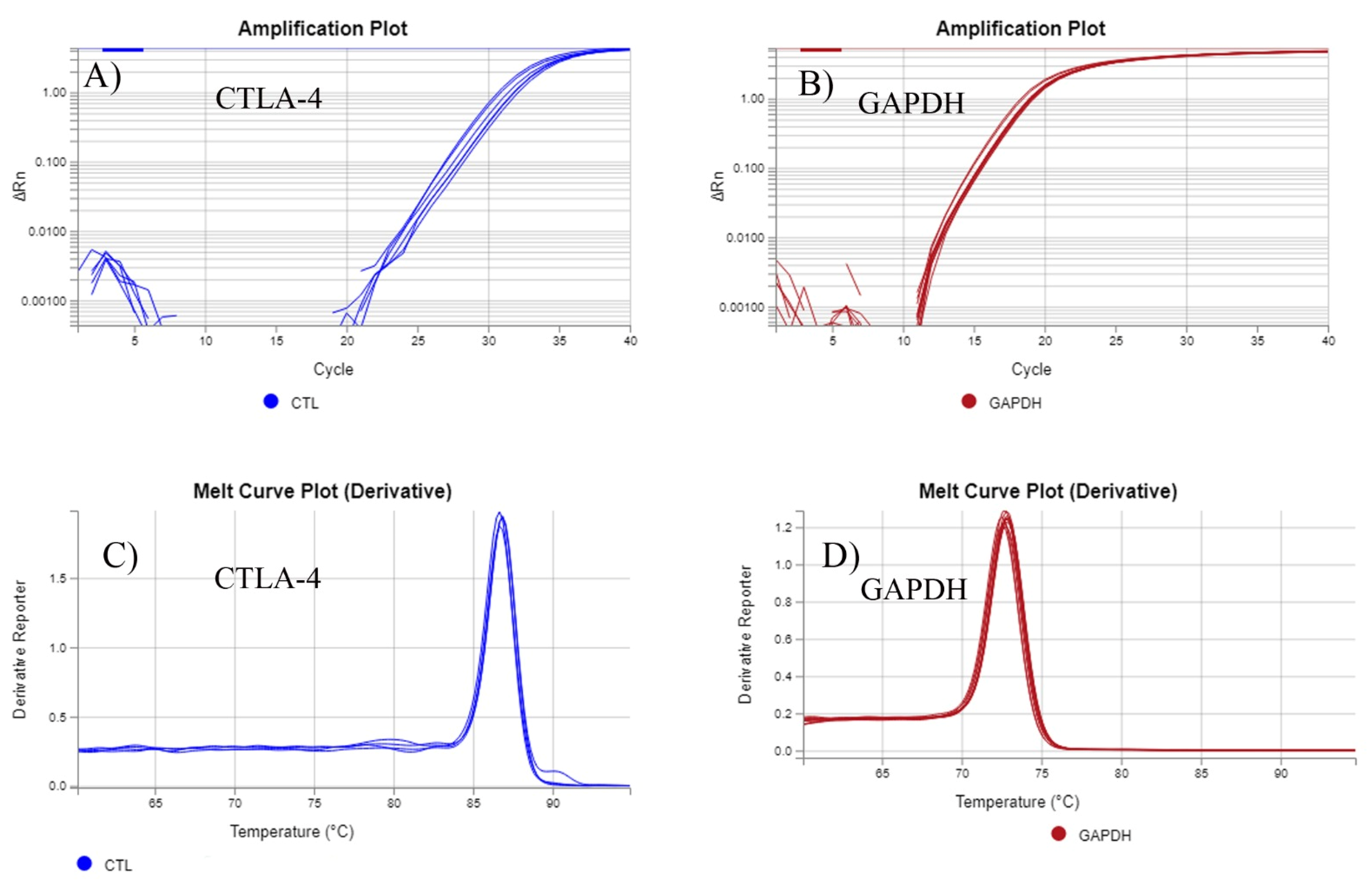
2.3. Preparation of Total RNA and qRT-PCR
2.4. Quantitative Real-Time Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
2.5. SNP Selection and Genotyping
2.6. Construction of Tissue Microarrays (TMAs) and Immunohistochemistry
2.7. Statistical Analysis
3. Results
3.1. Demographic Characteristics of Study Population
3.2. Age and Gender Stratified Analysis
3.3. Haplotype Analysis
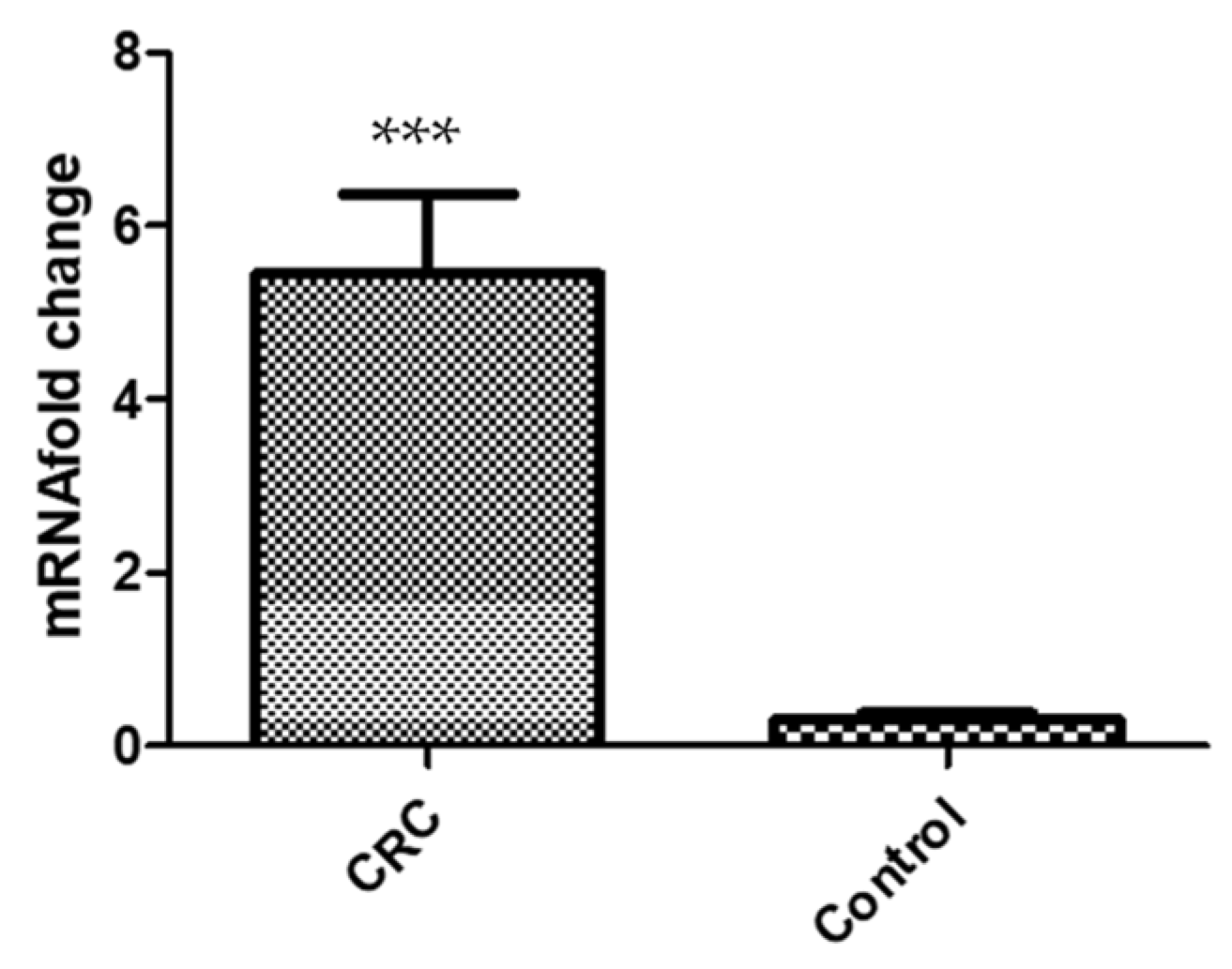
3.4. Gene Expression Analysis
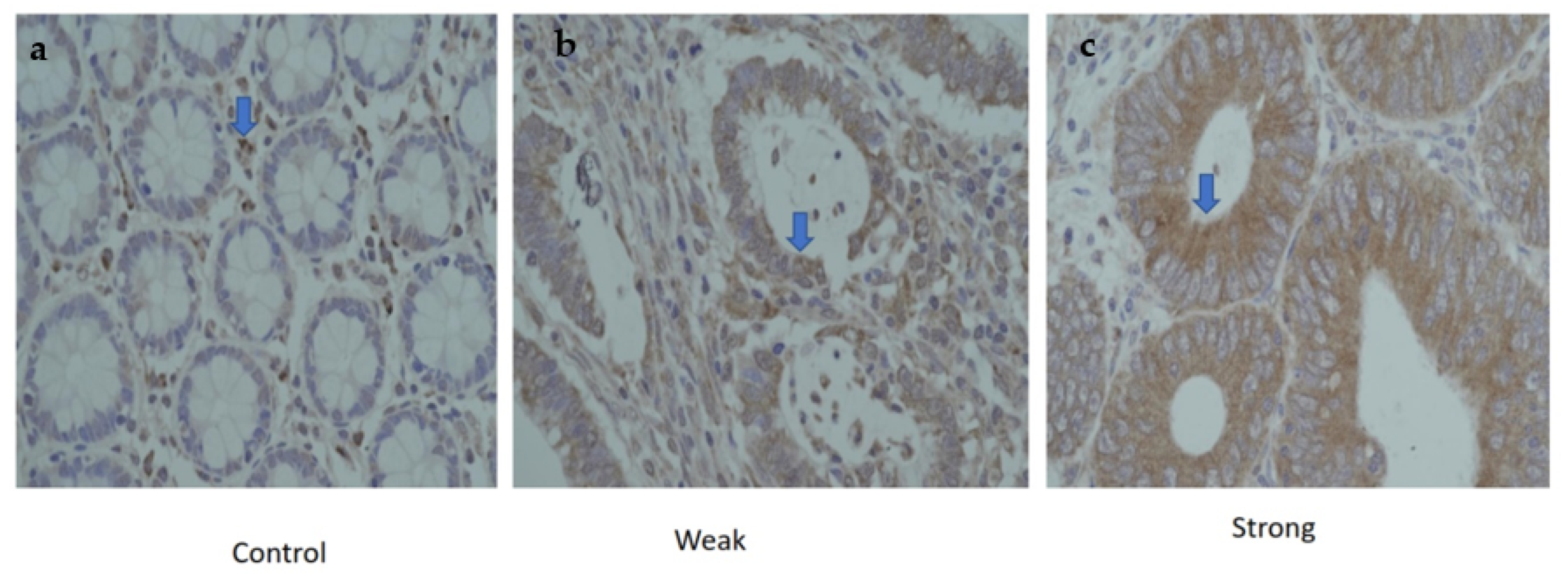
3.5. Protein Expression of CTLA-4 in CRC Patients
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Mojtahedi, Z.; Mohmedi, M.; Rahimifar, S.; Erfani, N.; Hosseini, S.V.; Ghaderi, A. Programmed death-1 gene polymorphism (PD-1.5 C/T) is associated with colon cancer. Gene 2012, 508, 229–232. [Google Scholar] [CrossRef]
- Potter, J.D. Colorectal Cancer: Molecules and Populations. JNCI J. Natl. Cancer Inst. 1999, 91, 916–932. [Google Scholar] [CrossRef] [PubMed]
- Saudi Arabia Source: Globocan Incidence, Mortality and Prevalence by Cancer Site. 2021. Available online: https://gco.iarc.fr/today/data/factsheets/populations/682-saudi-arabia-fact-sheets.pdf (accessed on 26 October 2022).
- Biller, L.H.; Schrag, D. Diagnosis and Treatment of Metastatic Colorectal Cancer. JAMA 2021, 325, 669. [Google Scholar] [CrossRef]
- Qi, P.; Ruan, C.; Wang, H.; Zhou, F.; Xu, X.; Gu, X.; Zhao, Y.; Dou, T.; Gao, C. CTLA-4 +49A > G polymorphism is associated with the risk but not with the progression of colorectal cancer in Chinese. Int. J. Colorectal Dis. 2010, 25, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Wang, H.; Hong, Y.; Ren, A.; Wang, H.; Liu, L.; Zhao, Q. Identification of an at-risk subpopulation with high immune infiltration based on the peroxisome pathway and TIM3 in colorectal cancer. BMC Cancer 2022, 22, 44. [Google Scholar] [CrossRef]
- Zaravinos, A.; Roufas, C.; Nagara, M.; de Lucas Moreno, B.; Oblovatskaya, M.; Efstathiades, C.; Dimopoulos, C.; Ayiomamitis, G.D. Cytolytic activity correlates with the mutational burden and deregulated expression of immune checkpoints in colorectal cancer. J. Exp. Clin. Cancer Res. 2019, 38, 364. [Google Scholar] [CrossRef]
- Hodi, F.S.; O’Day, S.J.; McDermott, D.F.; Weber, R.W.; Sosman, J.A.; Haanen, J.B.; Gonzalez, R.; Robert, C.; Schadendorf, D.; Hassel, J.C.; et al. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. N. Engl. J. Med. 2010, 363, 711–723. [Google Scholar] [CrossRef]
- Khalil, D.N.; Smith, E.L.; Brentjens, R.J.; Wolchok, J.D. The future of cancer treatment: Immunomodulation, CARs and combination immunotherapy. Nat. Rev. Clin. Oncol. 2016, 13, 273–290. [Google Scholar] [CrossRef]
- Ge, J.; Zhu, L.; Zhou, J.; Li, G.; Li, Y.; Li, S.; Wu, Z.; Rong, J.; Yuan, H.; Liu, Y.; et al. Association between co-inhibitory molecule gene tagging single nucleotide polymorphisms and the risk of colorectal cancer in Chinese. J. Cancer Res. Clin. Oncol. 2015, 141, 1533–1544. [Google Scholar] [CrossRef] [PubMed]
- Hung, A.L.; Garzon-Muvdi, T.; Lim, M. Biomarkers and Immunotherapeutic Targets in Glioblastoma. World Neurosurg. 2017, 102, 494–506. [Google Scholar] [CrossRef]
- Jacobs, J.; Smits, E.; Lardon, F.; Pauwels, P.; Deschoolmeester, V. Immune Checkpoint Modulation in Colorectal Cancer: What’s New and What to Expect. J. Immunol. Res. 2015, 2015, 158038. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cai, P.; Li, L.; Shi, L.; Chang, P.; Liang, T.; Yang, Q.; Liu, Y.; Wang, L.; Hu, L. Co-expression of TIM-3 and CEACAM1 promotes T cell exhaustion in colorectal cancer patients. Int. Immunopharmacol. 2017, 43, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Al-Harbi, N.; Vaali-Mohammed, M.-A.; Al-Omar, S.; Zubaidi, A.; Al-Obeed, O.; Abdulla, M.-H.; Mansour, L. Rs10204525 Polymorphism of the Programmed Death (PD-1) Gene Is Associated with Increased Risk in a Saudi Arabian Population with Colorectal Cancer. Medicina 2022, 58, 1439. [Google Scholar] [CrossRef]
- Dehaghani, A.S.; Kashef, M.A.; Ghaemenia, M.; Sarraf, Z.; Khaghanzadeh, N.; Fattahi, M.J.; Ghaderi, A. PDCD1, CTLA-4 and p53 gene polymorphism and susceptibility to gestational trophoblastic diseases. J. Reprod. Med. 2009, 54, 25–31. [Google Scholar] [PubMed]
- Jiang, L.; Luo, R.Y.; Zhang, W.; Wang, L.R.; Wang, F.; Cheng, Y.X. Single nucleotide polymorphisms of CTLA4 gene and their association with human cervical cancer. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2011, 28, 313–317. [Google Scholar]
- Khaghanzadeh, N.; Erfani, N.; Ghayumi, M.A.; Ghaderi, A. CTLA4 gene variations and haplotypes in patients with lung cancer. Cancer Genet. Cytogenet. 2010, 196, 171–174. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Sun, Y.; Wang, C.; Ji, J.; Li, Y.; Ye, Y.; Lv, L.; Guo, Y.; Guo, S.; Li, H.; et al. Genome-Wide Association Study Identifies Two New Susceptibility Loci for Colorectal Cancer at 5q23.3 and 17q12 in Han Chinese. Oncotarget 2015, 6, 40327–40336. [Google Scholar] [CrossRef]
- Le Marchand, L. Genome-Wide Association Studies and Colorectal Cancer. Surg. Oncol. Clin. 2009, 18, 663–668. [Google Scholar] [CrossRef]
- Lu, Y.; Kweon, S.S.; Tanikawa, C.; Jia, W.H.; Xiang, Y.B.; Cai, Q.; Zeng, C.; Schmit, S.L.; Shin, A.; Matsuo, K.; et al. Large-Scale Genome-Wide Association Study of East Asians Identifies Loci Associated With Risk for Colorectal Cancer. Gastroenterology 2019, 156, 1455–1466. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, F.R.; Schmit, S.L.; Jiao, S.; Edlund, C.K.; Wang, H.; Zhang, B.; Hsu, L.; Huang, S.C.; Fischer, C.P.; Harju, J.F.; et al. Genome-wide association study of colorectal cancer identifies six new susceptibility loci. Nat. Commun. 2015, 6, 7138. [Google Scholar] [CrossRef] [PubMed]
- Misra, M.K.; Pandey, S.K.; Kapoor, R.; Sharma, R.K.; Agrawal, S. Cytotoxic T-lymphocyte antigen 4 gene polymorphism influences the incidence of symptomatic human cytomegalovirus infection after renal transplantation. Pharm. Genom. 2015, 25, 19–29. [Google Scholar] [CrossRef]
- Ghaderi, A.; Yeganeh, F.; Kalantari, T.; Talei, A.R.; Mohammad Pezeshki, A.; Doroudchi, M.; Dehaghani, A.S. Cytotoxic T Lymphocyte Antigen-4 Gene in Breast Cancer. Breast Cancer Res. Treat. 2004, 86, 1–7. [Google Scholar] [CrossRef]
- Li Li, D.; Zhang, Q.; Xu, F.; Fu, Z.; Yuan, W.; Li, D.; Pang, D. Association of CTLA-4 gene polymorphisms with sporadic breast cancer risk and clinical features in Han women of Northeast China. Mol. Cell. Biochem. 2012, 364, 283–290. [Google Scholar] [CrossRef]
- Sun, T.; Zhou, Y.; Yang, M.; Hu, Z.; Tan, W.; Han, X.; Shi, Y.; Yao, J.; Guo, Y.; Yu, D.; et al. Functional Genetic Variations in Cytotoxic T-Lymphocyte Antigen 4 and Susceptibility to Multiple Types of Cancer. Cancer Res. 2008, 68, 7025–7034. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, D.; Fu, Z.; Li, H.; Jiang, W.; Li, D. Association of CTLA-4 gene polymorphisms with sporadic breast cancer in Chinese Han population. BMC Cancer 2007, 7, 173. [Google Scholar] [CrossRef] [PubMed]
- Castro, F.A.; Haimila, K.; Sareneva, I.; Schmitt, M.; Lorenzo, J.; Kunkel, N.; Kumar, R.; Försti, A.; Kjellberg, L.; Hallmans, G.; et al. Association of HLA-DRB1, interleukin-6 and cyclin D1 polymorphisms with cervical cancer in the Swedish population-A candidate gene approach. Int. J. Cancer 2009, 125, 1851–1858. [Google Scholar] [CrossRef]
- Gokhale, P.; Kerkar, S.; Tongaonkar, H.; Salvi, V.; Mania-Pramanik, J. CTLA-4 gene polymorphism at position +49A > G in exon 1: A risk factor for cervical cancer in Indian women. Cancer Genet. 2013, 206, 154–161. [Google Scholar] [CrossRef]
- Ivansson, E.L.; Juko-Pecirep, I.; Gyllensten, U.B. Interaction of immunological genes on chromosome 2q33 and IFNG in susceptibility to cervical cancer. Gynecol. Oncol. 2010, 116, 544–548. [Google Scholar] [CrossRef]
- Pawlak, E.; Karabon, L.; Wlodarska-Polinska, I.; Jedynak, A.; Jonkisz, A.; Tomkiewicz, A.; Kornafel, J.; Stepien, M.; Ignatowicz, A.; Lebioda, A.; et al. Influence of CTLA-4/CD28/ICOS gene polymorphisms on the susceptibility to cervical squamous cell carcinoma and stage of differentiation in the Polish population. Hum. Immunol. 2010, 71, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Rahimifar, S.; Erfani, N.; Sarraf, Z.; Ghaderi, A. ctla-4 gene variations may influence cervical cancer susceptibility. Gynecol. Oncol. 2010, 119, 136–139. [Google Scholar] [CrossRef] [PubMed]
- Su, T.H.; Chang, T.Y.; Lee, Y.-J.; Chen, C.K.; Liu, H.F.; Chu, C.C.; Lin, M.; Wang, P.T.; Huang, W.C.; Chen, T.C.; et al. CTLA-4 gene and susceptibility to human papillomavirus-16-associated cervical squamous cell carcinoma in Taiwanese women. Carcinogenesis 2007, 28, 1237–1240. [Google Scholar] [CrossRef]
- Karabon, L.; Pawlak, E.; Tomkiewicz, A.; Jedynak, A.; Passowicz-Muszynska, E.; Zajda, K.; Jonkisz, A.; Jankowska, R.; Krzakowski, M.; Frydecka, I. CTLA-4, CD28, and ICOS gene polymorphism associations with non-small-cell lung cancer. Hum. Immunol. 2011, 72, 947–954. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Liu, Y.; Liu, J.; Song, X.; Wang, Z.; Wang, M.; Zhu, Y.; Han, J. CTLA-4 +49A > G Polymorphism Is Associated with Advanced Non-Small Cell Lung Cancer Prognosis. Respiration 2011, 82, 439–444. [Google Scholar] [CrossRef]
- Kämmerer, P.W.; Toyoshima, T.; Schöder, F.; Kämmerer, P.; Kuhr, K.; Brieger, J.; Al-Nawas, B. Association of T-cell regulatory gene polymorphisms with oral squamous cell carcinoma. Oral Oncol. 2010, 46, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Wong, Y.-K.; Chang, K.-W.; Cheng, C.-Y.; Liu, C.-J. Association of CTLA-4 gene polymorphism with oral squamous cell carcinoma. J. Oral Pathol. Med. 2006, 35, 51–54. [Google Scholar] [CrossRef]
- Cozar, J.M.; Romero, J.M.; Aptsiauri, N.; Vazquez, F.; Vilchez, J.R.; Tallada, M.; Garrido, F.; Ruiz-Cabello, F. High incidence of CTLA-4 AA (CT60) polymorphism in renal cell cancer. Hum. Immunol. 2007, 68, 698–704. [Google Scholar] [CrossRef]
- Jafari, M.; Ghanei, M. Evaluation of plasma, erythrocytes, and brochoalveolar lavage fluid antioxidant defense system in sulfur mustard-injured patients. Clin. Toxicol. 2010, 48, 184–192. [Google Scholar] [CrossRef]
- Jones, G.; Wu, S.; Jang, N.; Fulcher, D.; Hogan, P.; Stewart, G. Polymorphisms within the CTLA4 gene are associated with infant atopic dermatitis. Br. J. Dermatol. 2006, 154, 467–471. [Google Scholar] [CrossRef]
- Ueda, H.; Howson, J.M.M.; Esposito, L.; Heward, J.; Snook; Chamberlain, G.; Rainbow, D.B.; Hunter, K.M.D.; Smith, A.N.; Di Genova, G.; et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 2003, 423, 506–511. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J. Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet. Anal. 1999, 14, 143–149. [Google Scholar] [CrossRef]
- Al Obeed, O.A. Increased expression of tumor necrosis factor-α is associated with advanced colorectal cancer stages. World J. Gastroenterol. 2014, 20, 18390. [Google Scholar] [CrossRef]
- Solé, X.; Guinó, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A web tool for the analysis of association studies. Bioinformatics 2006, 22, 1928–1929. [Google Scholar] [CrossRef]
- El-Hazmi, M.A.; Al-Swailem, A.R.; Warsy, A.S.; Sulaimani, R.; Al-Meshari, A.A. Consanguinity among the Saudi Arabian population. J. Med. Genet. 1995, 32, 623–626. [Google Scholar] [CrossRef] [PubMed]
- Ebrahim, E.; Teklu, T.; Tajebe, F.; Wondmagegn, T.; Akelew, Y.; Fiseha, M. Association of Cytotoxic T-Lymphocyte Antigen-4 Gene Polymorphism with Type 1 Diabetes Mellitus: In silico Analysis of Biological Features of CTLA-4 Protein on Ethiopian Population. Diabetes Metab. Syndr. Obes. 2022, 15, 2733–2751. [Google Scholar] [CrossRef] [PubMed]
- Teft, W.A.; Kirchhof, M.G.; Madrenas, J. A molecular perspective of ctla-4 function. Annu. Rev. Immunol. 2006, 24, 65–97. [Google Scholar] [CrossRef]
- Fang, M.; Huang, W.; Mo, D.; Zhao, W.; Huang, R. Association of Five Snps in Cytotoxic T-Lymphocyte Antigen 4 and Cancer Susceptibility: Evidence from 67 Studies. Cell. Physiol. Biochem. 2018, 47, 414–427. [Google Scholar] [CrossRef]
- Hu, L.; Liu, J.; Chen, X.; Zhang, Y.; Liu, L.; Zhu, J.; Chen, J.; Shen, H.; Qiang, F.; Hu, Z. CTLA-4 gene polymorphism +49A/G contributes to genetic susceptibility to two infection-related cancers—Hepatocellular carcinoma and cervical cancer. Hum. Immunol. 2010, 71, 888–891. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Wang, Y.; Chen, S.; Lin, J.; Chen, B.; Yu, S.; Chen, Y.; Gu, H.; Kang, M. Investigation of Cytotoxic T-lymphocyte antigen 4 Polymorphisms in Gastric Cardia Adenocarcinoma. Scand. J. Immunol. 2016, 83, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; Qiu, H.; Tang, W.; Wang, Y.; Lan, B.; Chen, Y. CTLA4 tagging polymorphisms and risk of colorectal cancer: A case—Control study involving 2306 subjects. OncoTargets Ther. 2018, 11, 4609–4619. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, X.; Zhao, R. The association of CTLA-4 A49G polymorphism with colorectal cancer risk in a Chinese Han population. Int. J. Immunogenet. 2015, 42, 93–99. [Google Scholar] [CrossRef]
- He, L.; Deng, T.; Luo, H.-S. Association between cytotoxic T-lymphocyte antigen-4 +49A/G polymorphism and colorectal cancer risk: A meta-analysis. Int. J. Clin. Exp. Med. 2015, 8, 3752–3760. [Google Scholar] [PubMed]
- Hadinia, A.; Hossieni, S.V.; Erfani, N.; Saberi-Firozi, M.; Fattahi, M.J.; Ghaderi, A. CTLA-4 gene promoter and exon 1 polymorphisms in Iranian patients with gastric and colorectal cancers. J. Gastroenterol. Hepatol. 2007, 22, 2283–2287. [Google Scholar] [CrossRef]
- Arikan, S.; Gümüş, A.; Küçükhüseyin, Ö.; Coşkun, C.; Turan, S.; Cacina, C.; Talu, C.K.; Akyüz, F.; Farooqi, A.A.; Kıran, B.; et al. Mide kanserli hastalarda CTLA-4 ve CD28 gen varyantlarının ve dolaşımdaki düzeylerinin deǧerlendirilmesi. Turk. J. Biochem. 2017, 42, 551–558. [Google Scholar] [CrossRef]
- Geng, R.; Song, F.; Yang, X.; Sun, P.; Hu, J.; Zhu, C.; Zhu, B.; Fan, W. Association between cytotoxic T lymphocyte antigen-4 +49A/G, −1722T/C, and −1661A/G polymorphisms and cancer risk: A meta-analysis. Tumor Biol. 2014, 35, 3627–3639. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, J.; Deng, Y.; Tian, C.; Li, X.; Huang, J.; Fan, H. Polymorphisms in the cytotoxic T-lymphocyte antigen 4 gene and cancer risk. Cancer 2011, 117, 4312–4324. [Google Scholar] [CrossRef] [PubMed]
- Mapara, M.Y.; Sykes, M. Tolerance and Cancer: Mechanisms of Tumor Evasion and Strategies for Breaking Tolerance. J. Clin. Oncol. 2004, 22, 1136–1151. [Google Scholar] [CrossRef]
- Mäurer, S.L.A.M. A polymorphism in the human cytotoxic T-lymphocyte antigen 4 (CTLA4) gene (exon 1 +49) alters T-cell activation. Immunogenetics 2002, 54, 1–8. [Google Scholar] [CrossRef]
- Anjos, S.; Nguyen, A.; Ounissi-Benkalha, H.; Tessier, M.-C.; Polychronakos, C. A Common Autoimmunity Predisposing Signal Peptide Variant of the Cytotoxic T-lymphocyte Antigen 4 Results in Inefficient Glycosylation of the Susceptibility Allele. J. Biol. Chem. 2002, 277, 46478–46486. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Yu, X.; Jiang, L.; Xiao, M.; Bai, B.; Lu, J.; Zhou, Y. Association between the Cytotoxic T-Lymphocyte Antigen 4 +49G > A polymorphism and cancer risk: A meta-analysis. BMC Cancer 2010, 10, 522. [Google Scholar] [CrossRef] [PubMed]
- Pérez-García, A.; De la Cámara, R.; Román-Gómez, J.; Jiménez-Velasco, A.; Encuentra, M.; Nieto, J.B.; de la Rubia, J.; Urbano-Ispizúa, A.; Brunet, S.; Iriondo, A.; et al. CTLA-4 polymorphisms and clinical outcome after allogeneic stem cell transplantation from HLA-identical sibling donors. Blood 2007, 110, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Van Nguyen, S.; Shamoun, L.; Landerholm, K.; Andersson, R.E.; Wagsater, D.; Dimberg, J. Cytotoxic T-lymphocyte Antigen-4 (CTLA-4) Gene Polymorphism (rs3087243) Is Related to Risk and Survival in Patients with Colorectal Cancer. In Vivo 2021, 35, 969–975. [Google Scholar] [CrossRef]
- Aslam, M.M.; Jalil, F.; John, P.; Fan, K.-H.; Bhatti, A.; Feingold, E.; Demirci, F.Y.; Kamboh, M.I. A sequencing study of CTLA4 in Pakistani rheumatoid arthritis cases. PLoS ONE 2020, 15, e0239426. [Google Scholar] [CrossRef]
- Dai, Z.; Tian, T.; Wang, M.; Liu, X.; Lin, S.; Yang, P.; Liu, K.; Zheng, Y.; Xu, P.; Liu, M.; et al. CTLA-4 polymorphisms associate with breast cancer susceptibility in Asians: A meta-analysis. PeerJ 2017, 5, e2815. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, J.; Chen, Y.; Tang, W.; Liu, C.; Sun, Y.; Chen, J. Association of CTLA-4 tagging polymorphisms and haplotypes with hepatocellular carcinoma risk. Medicine 2019, 98, e16266. [Google Scholar] [CrossRef]
- Yan, Q.; Chen, P.; Lu, A.; Zhao, P.; Gu, A. Association between CTLA-4 60G/A and -1661A/G Polymorphisms and the Risk of Cancers: A Meta-Analysis. PLoS ONE 2013, 8, e83710. [Google Scholar] [CrossRef]
- Okada, Y.; Wu, D.; Trynka, G.; Raj, T.; Terao, C.; Ikari, K.; Kochi, Y.; Ohmura, K.; Suzuki, A.; Yoshida, S.; et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 2014, 506, 376–381. [Google Scholar] [CrossRef]
- Stahl, E.A.; Raychaudhuri, S.; Remmers, E.F.; Xie, G.; Eyre, S.; Thomson, B.P.; Li, Y.; Kurreeman, F.A.S.; Zhernakova, A.; Hinks, A.; et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet. 2010, 42, 508–514. [Google Scholar] [CrossRef]
- Torres-Carrillo, N.; Ontiveros-Mercado, H.; Torres-Carrillo, N.M.; Parra-Rojas, I.; Rangel-Villalobos, H.; Ramírez-Dueñas, M.G.; Gutiérrez-Ureña, S.R.; Valle, Y.; Muñoz-Valle, J.F. The −319C/+49G/CT60G Haplotype of CTLA-4 Gene Confers Susceptibility to Rheumatoid Arthritis in Mexican Population. Cell Biochem. Biophys. 2013, 67, 1217–1228. [Google Scholar] [CrossRef] [PubMed]
- Luterek-Puszyńska, K.; Malinowski, D.; Paradowska-Gorycka, A.; Safranow, K.; Pawlik, A. CD28, CTLA-4 and CCL5 gene polymorphisms in patients with rheumatoid arthritis. Clin. Rheumatol. 2017, 36, 1129–1135. [Google Scholar] [CrossRef] [PubMed]
- Frías, J.L.; Gilbert-Barness, E. Human Teratogens: Current Controversies. Adv. Pediatr. 2008, 55, 171–211. [Google Scholar] [CrossRef]
- Kupchella, C.E. Environmental factors in cancer etiology. Semin. Oncol. Nurs. 1986, 2, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Parsa, N. Environmental Factors Inducing Human Cancers. 2012. Available online: http://ijph.tums.ac.ir (accessed on 9 December 2022).
- Cartmel, B.; Loescher, L.J.; Villar-Werstler, P. Professional and Consumer Concerns about the Environment, Lifestyle, and Cancer. Semin. Oncol. Nurs. 1992, 8, 20–29. [Google Scholar] [CrossRef]
- Özyalçin, B.; Sanlier, N. The effect of diet components on cancer with epigenetic mechanisms. In Trends in Food Science and Technology; Elsevier Ltd.: Amsterdam, The Netherlands, 2020; Volume 102, pp. 138–145. [Google Scholar] [CrossRef]
- Bharti, V.; Mohanti, B.K.; Das, S.N. Functional genetic variants of CTLA-4 and risk of tobacco-related oral carcinoma in high-risk North Indian population. Hum. Immunol. 2013, 74, 348–352. [Google Scholar] [CrossRef] [PubMed]
- Saleh, R.; Taha, R.Z.; Toor, S.M.; Sasidharan Nair, V.; Murshed, K.; Khawar, M.; Al-Dhaheri, M.; Petkar, M.A.; Abu Nada, M.; Elkord, E. Expression of immune checkpoints and T cell exhaustion markers in early and advanced stages of colorectal cancer. Cancer Immunol. Immunother. 2020, 69, 1989–1999. [Google Scholar] [CrossRef]
- Yu, H.; Yang, J.; Jiao, S.; Li, Y.; Zhang, W.; Wang, J. Cytotoxic T lymphocyte antigen 4 expression in human breast cancer: Implications for prognosis. Cancer Immunol. Immunother. 2015, 64, 853–860. [Google Scholar] [CrossRef]
- Nistico, L. The CTLA-4 gene region of chromosome 2q33 is linked to, and associated with, type 1 diabetes. Belgian Diabetes Registry. Hum. Mol. Genet. 1996, 5, 1075–1080. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Savost’anov, K.V.; Turakulov, R.I.; Efremov, I.A.; Demurov, L.M. Genetic analysis and functional evaluation of the C/T(−318) and A/G(−1661) polymorphisms of the CTLA-4 gene in patients affected with Graves’ disease. Clin. Immunol. 2006, 118, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Misra, M.K.; Mishra, A.; Phadke, S.R.; Agrawal, S. Association of functional genetic variants of CTLA4 with reduced serum CTLA4 protein levels and increased risk of idiopathic recurrent miscarriages. Fertil. Steril. 2016, 106, 1115–1123.e6. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Hu, Z.; Shen, H.; Lin, D. Genetic Polymorphisms in Cytotoxic T-Lymphocyte Antigen 4 and Cancer: The Dialectical Nature of Subtle Human Immune Dysregulation. Cancer Res. 2009, 69, 6011–6014. [Google Scholar] [CrossRef] [PubMed]
- Cameron, F.; Whiteside, G.; Perry, C. Ipilimumab. Drugs 2011, 71, 1093–1104. [Google Scholar] [CrossRef] [PubMed]
- Waldmann, T.A.; Dubois, S.; Miljkovic, M.D.; Conlon, K.C. IL-15 in the Combination Immunotherapy of Cancer. Front. Immunol. 2020, 11, 868. [Google Scholar] [CrossRef] [PubMed]
| Characteristics | Cancer (100) | Control (100) | |
|---|---|---|---|
| Gender (number) | Male | 64 | 65 |
| Female | 36 | 35 | |
| Age (average ± SD) | 56.33 ± 14.56 | 56.31 ± 14.56 | |
| Localization | Colon | 40 | - |
| Recto-sigmoid | 60 | - | |
| Stage | I | 11 | - |
| II | 57 | - | |
| III | 32 | - | |
| IV | 0 | - | |
| SNP ID/Assay ID | Common Name | Chromosome Position | Nucleotide Change | Region | MAF in Human Populations (1000 Genomes Study) | Present Study | |||
|---|---|---|---|---|---|---|---|---|---|
| Global | European | South Asian | Qatari | ||||||
| rs11571317 | −658C > T | Chr2:203867285 | C/T | promoter | T = 0.024 | T = 0.08 | T = 0.022 | T = 0.078 | T = 0.26 |
| rs231775 | +49A > G | Chr2:203867991 | A/G | Exon 1 | G = 0.42 | G = 0.36 | G = 0.28 | G = 0.208 | G = 0.33 |
| rs3087243 | CT60 G > A | chr2:203874196 | G/A | 3-’UTR | A = 0.36 | A = 0.45 | A = 0.59 | A = 0.567 | A = 0.44 |
| Locus | Model | Genotype | CRC (%) N = 99 | Controls (%) N = 96 | OR (95% CI) | * p-Value | AIC |
|---|---|---|---|---|---|---|---|
| rs11571317 (−658C > T) | Alleles | C | 136 | 143 | Ref | ||
| T | 62 | 49 | 0.75 (0.483–1.170) | 0.20502 | |||
| Codominant | CC | 56 (56.6%) | 61 (63.5%) | 1.00 | |||
| CT | 24 (24.2%) | 21 (21.9%) | 1.24 (0.63–2.48) | 275.2 | |||
| TT | 19 (19.2%) | 14 (14.6%) | 1.48 (0.68–3.22) | 0.57 | |||
| Dominant | CC | 56 (56.6%) | 61 (63.5%) | 1.00 | |||
| CT + TT | 43 (43.4%) | 35 (36.5%) | 1.34 (0.75–2.38) | 0.32 | 273.3 | ||
| Recessive | CC + CT | 80 (80.8%) | 82 (85.4%) | 1.00 | |||
| TT | 19 (19.2%) | 14 (14.6%) | 1.39 (0.65–2.96) | 0.39 | 273.5 | ||
| Overdominant | CC + TT | 75 (75.8%) | 75 (78.1%) | 1.00 | |||
| CT | 24 (24.2%) | 21 (21.9%) | 1.14 (0.59–2.23) | 0.69 | 274.1 | ||
| Log-Additive | 1.22 (0.84–1.770 | 0.29 | 273.2 | ||||
| Locus | Model | Genotype | CRC (%) N = 100 | Controls (%) N = 97 | OR (95% CI) | * p-Value | AIC |
| rs231775 (+49A > G) | Alleles | A | 0.46 | 0.67 | Ref | - | |
| G | 0.54 | 0.33 | 2.337 (1.553–3.516) | <0.0001 | |||
| Codominant | AA | 40 (40%) | 38 (39.2%) | 1.00 | 1 | ||
| AG | 13 (13%) | 54 (55.7%) | 0.23 (0.11–048) | <0.0001 | 212.9 | ||
| GG | 47 (47%) | 5 (5.2%) | 8.93 (3.21–24.84) | ||||
| Dominant | AA | 40 (40%) | 38 (39.2%) | 1.00 | |||
| AG + GG | 60 (60%) | 59 (60.8%) | 0.97 (0.55–1.71) | 0.91 | 277 | ||
| Recessive | AG + AA | 53 (53%) | 92 (94.8%) | 1.00 | |||
| GG | 47 (47%) | 5 (5.2%) | 16.32 (6.11–43.56) | <0.0001 | 227.3 | ||
| Overdominant | AA-GG | 45 (46.4%) | 86 (86%) | 1.00 | 235 | ||
| AG | 13 (13%) | 54 (55.7%) | 0.12 (0.06–0.24) | <0.0001 | |||
| Log-Additive | 1.94 (1.34–2.81) | <0.0001 | 263.9 | ||||
| Locus | Model | Genotype | CRC (%) N = 97 | Controls (%) N = 99 | OR (95% CI) | * p-Value | AIC |
| rs3087243 (CT60 G > A) | Alleles | G | 0.42 | 0.56 | 1 | ||
| A | 0.58 | 0.44 | 1.743 (1.168–2.599) | 0.00631 | |||
| Codominant | GG | 19 (19.6%) | 26 (26.3%) | 1.00 | |||
| GA | 44 (45.4%) | 59 (59.6%) | 1.021 (0.502–2.073) | 0.95521 | 265.8 | ||
| AA | 34 (35%) | 14 (14.1%) | 3.323 (1.408–7.843) | 0.00535 | |||
| Dominant | GG | 19 (19.6%) | 26 (26.3%) | 1.00 | |||
| GA + AA | 78 (80.4%) | 73 (73.7%) | 1.462 (0.747–2.864) | 0.26660 | 263.8 | ||
| Recessive | GG + GA | 63 (65%) | 85(85.9%) | 1.00 | |||
| AA | 34 (35%) | 14 (14.1%) | 0.305 (0.151–0.616) | 0.00067 | 274.5 | ||
| Overdominant | AA + GG | 53 (54.6%) | 40 (40.4%) | 1.00 | |||
| AG | 44 (45.4%) | 59 (59.6%) | 0.56 (0.32–0.99) | 0.046 | 271.7 | ||
| Log-Additive | 1.816 (1.19–2.77) | 0.0050 | 267.7 |
| Locus | Model | Genotype | CRC < 56 (%) N = 41 | CRC > 56 (%) N = 58 | OR (95% CI) | * p-Value | AIC |
|---|---|---|---|---|---|---|---|
| rs11571317 C > T | Allele | C | 0.67 | 0.7 | Ref | ||
| T | 0.33 | 0.3 | 0.880 (0.479–1.616) | 0.68059 | |||
| Codominant | CC | 23 (56.1%) | 33 (56.9%) | 1.00 | |||
| CT | 9 (21.9%) | 15 (25.9%) | 1.22 (0.45–3.29) | 0.79 | 140.7 | ||
| TT | 9 (21.9%) | 10 (17.2%) | 0.79 (0.28–2.26) | ||||
| Dominant | CC | 23 (56.1%) | 33 (56.9%) | 1.00 | 139.2 | ||
| CT + TT | 18 (43.9%) | 25 (43.1%) | 1 (0.44–2.26) | 1 | |||
| Recessive | CC + CT | 32 (78 %) | 48 (82.8%) | 1.00 | |||
| TT | 9 (21.9%) | 10 (17.2%) | 0.74 (0.27–2.05) | 0.57 | 138.8 | ||
| Overdominant | CC + TT | 32 (78%) | 43 (74.1%) | 1.00 | |||
| CT | 9 (21.9%) | 15 (25%) | 1.29 (0.50–3.36) | 0.59 | 138.9 | ||
| Log-Additive | 0.93 (0.56–1.55) | 0.78 | 139.1 | ||||
| Locus | Model | Genotype | CRC < 56 (%) N = 41 | CRC > 56 (%) N = 59 | OR (95% CI) | * p-Value | AIC |
| rs231775 A > G | Allele | A | 0.38 | 0.53 | Ref | ||
| G | 0.62 | 0.47 | 0.549 (0.309–0.975) | 0.03986 | |||
| Codominant | AA | 13 (31.7%) | 27 (45.8%) | 2.02 (0.84–4.86) | |||
| AG | 5 (12.2%) | 8 (13.5%) | 1.50 (0.43–5.31) | 0.28 | 140 | ||
| GG | 23 (56.1%) | 24 (40.7%) | 1.00 | ||||
| Dominant | GG | 13 (31.7%) | 27 (45.8%) | 1.00 | |||
| AG-AA | 28 (68.3%) | 32 (54.2%) | 1.87 (0.83–4.21) | 0.13 | 138.2 | ||
| Recessive | GG-AG | 18 (43.9%) | 35 (59.3%) | 1.00 | |||
| AA | 23 (56.1%) | 24 (40.7%) | 1.85 (0.80–4.28) | 0.15 | 138.4 | ||
| Overdominant | AA-GG | 36 (87.8%) | 51 (87.5%) | 1.00 | |||
| AG | 5 (12.2%) | 8 (13.5%) | 1.10 (0.33–3.67) | 0.87 | 140.4 | ||
| Log-Additive | 1.42(0.92–2.21) | 0.11 | 138 | ||||
| Locus | Model | Genotype | CRC < 56 (%) N = 40 | CRC > 56 (%) N = 57 | OR (95% CI) | * p-Value | AIC |
| rs3087243 G > A | Allele | G | 39 | 43 | Ref | ||
| A | 41 | 71 | 1.571 (0.880–2.803) | 0.12576 | |||
| Codominant | GG | 12 (30%) | 7 (12.3%) | 0.38 (0.12–1.22) | |||
| AG | 15 (37.5%) | 29 (50.9%) | 3.314 (1.080–10.172) | 0.03238 | 134.2 | ||
| AA | 13(32.5%) | 21 (36.8%) | 2.769 (0.867–8.840) | 0.08133 | |||
| Dominant | AA | 12 (30%) | 7 (12.3%) | 1.00 | |||
| AG-GG | 28 (70%) | 50 (87.7%) | 3.061 (1.081–8.666) | 0.03042 | 136.1 | ||
| Recessive | AA-AG | 27 (67.5%) | 36 (63.2%) | 1.00 | |||
| GG | 13(32.5%) | 21 (36.8%) | 0.825 (0.352–1.937) | 0.65906 | 132.3 | ||
| Overdominant | GG-AA | 25 (62.5%) | 28 (49.1%) | 1.00 | |||
| AG | 15 (37.5%) | 29 (50.9%) | 1.65 (0.72–3.79) | 0.24 | 134.9 | ||
| Log-Additive | 0.67 (0.38–1.19) | 0.17 | 134.9 |
| Locus | Model | Genotype | CRC Female (%) N = 34 | CRC Male (%) N = 62 | OR (95% CI) | * p-Value | AIC |
|---|---|---|---|---|---|---|---|
| rs11571317 C > T | Alleles | C | 0.66 | 0.7 | Ref | ||
| T | 0.34 | 0.3 | 0.81 (0.43–1.51) | 0.505 | |||
| Codominant | CC | 18 (51.4%) | 38 (59.4%) | 1.00 | |||
| CT | 10 (28.6%) | 14 (21.9%) | 0.66 (0.25–1.78) | 0.71 | 133.9 | ||
| TT | 7 (20%) | 12 (18.8%) | 0.81 (0.27–2.41) | ||||
| Dominant | CC | 18 (51.4%) | 38 (59.4%) | 1.00 | |||
| CT + TT | 17 (48.6%) | 26 (40.6%) | 0.72 (0.32–1.66) | 0.45 | 132 | ||
| Recessive | CC + CT | 28 (80%) | 52 (81.2%) | 1.00 | |||
| TT | 7 (20%) | 12 (18.8%) | 0.92 (0.32–2.61) | 0.88 | 132.6 | ||
| Overdominant | CC + TT | 25 (71.4%) | 50 (78.1%) | 1.00 | |||
| CT | 10 (28.6%) | 14 (21.9%) | 0.70 (0.27–1.80) | 0.46 | 132.1 | ||
| Log-Additive | 0.86 (0.60–1.25) | 0.58 | 132.3 | ||||
| Locus | Model | Genotype | CRC Female (%) N = 36 | CRC Male (%) N = 64 | OR (95% CI) | * p-Value | AIC |
| rs231775 A > G | Alleles | A | 0.47 | 0.46 | Ref | - | |
| G | 0.53 | 0.54 | 1.05 (0.59–1.87) | 0.88 | |||
| Codominant | AA | 15 (41.7%) | 25 (39.1%) | 0.94 (0.39–2.26) | |||
| AG | 4 (11.1%) | 9 (14.1%) | 1.27 (0.34–4.77) | 0.91 | 136.5 | ||
| GG | 17 (47.2%) | 30 (46.9%) | 1 | ||||
| Dominant | GG | 17 (47.2%) | 30 (46.9%) | 1.00 | |||
| AG + AA | 19 (52.8%) | 34 (53.1%) | 1.01 (0.45–2.30) | 0.97 | 134.7 | ||
| Recessive | AG + GG | 21 (58.3%) | 39 (60.9%) | 1.00 | |||
| AA | 15 (41.7%) | 25 (39.1%) | 0.90 (0.39–2.06) | 0.8 | 134.6 | ||
| Overdominant | AA-GG | 32 (88.9%) | 55 (85.9%) | 1.00 | 0.67 | ||
| AG | 4 (11.1%) | 9 (14.1%) | 1.31 (0.37–4.60) | 134.5 | |||
| Log-Additive | 0.97 (0.63–1.51) | 0.91 | 134.7 | ||||
| Locus | Model | Genotype | CRC Female (%) N = 34 | CRC Male (%) N = 64 | OR (95% CI) | * p-Value | AIC |
| rs3087243 G > A | Alleles | A | 0.53 | 0.6 | Ref | ||
| G | 0.47 | 0.4 | 1.365 (0.756–2.466) | 0.30173 | |||
| Codominant | AA | 10 (28.6%) | 9 (14.5%) | 1 | |||
| AG | 13 (37.1%) | 31 (50%) | 2.650 (0.874–8.034) | 0.08068 | 129.8 | ||
| GG | 12 (34.3%) | 22 (35.5%) | 2.037 (0.650–6.386) | 0.21929 | |||
| Dominant | AA | 10 (28.6%) | 9 (14.5%) | 1 | |||
| AG + GG | 25 (71.4%) | 53 (85.5%) | 2.356 (0.851–6.522) | 0.09392 | 130.8 | ||
| Recessive | AG + AA | 23 (65.7%) | 40 (64.5%) | 1 | |||
| GG | 12 (34.3%) | 22 (35.5%) | 0.949 (0.397–2.265) | 0.90545 | 128.1 | ||
| Overdominant | AA-GG | 22 (62.9%) | 31 (50%) | 1 | |||
| AG | 13 (37.1%) | 31 (50%) | 1.69 (0.73–3.95) | 0.22 | 129.4 | ||
| Log-Additive | 0.75 (0.42–1.33) | 0.32 | 129.9 |
| Rs11571317 | Rs231775 | Rs3087243 | CRC | Control | OR (95% CI) | * p-Value |
|---|---|---|---|---|---|---|
| A | G | G | 0.1971 | 0.2973 | Ref | |
| A | A | G | 0.2066 | 0.2357 | 0.57 (0.26–1.25) | 0.16 |
| G | G | G | 0.1518 | 0.2043 | 1.16 (0.58–2.33) | 0.68 |
| A | G | A | 0.116 | 0.1332 | 1.44 (0.55–3.79) | 0.46 |
| G | A | G | 0.1364 | 0 | 57.66 (6.82–487.84) | 3 × 10−4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Harbi, N.; Abdulla, M.-H.; Vaali-Mohammed, M.-A.; Bin Traiki, T.; Alswayyed, M.; Al-Obeed, O.; Abid, I.; Al-Omar, S.; Mansour, L. Evidence of Association between CTLA-4 Gene Polymorphisms and Colorectal Cancers in Saudi Patients. Genes 2023, 14, 874. https://doi.org/10.3390/genes14040874
Al-Harbi N, Abdulla M-H, Vaali-Mohammed M-A, Bin Traiki T, Alswayyed M, Al-Obeed O, Abid I, Al-Omar S, Mansour L. Evidence of Association between CTLA-4 Gene Polymorphisms and Colorectal Cancers in Saudi Patients. Genes. 2023; 14(4):874. https://doi.org/10.3390/genes14040874
Chicago/Turabian StyleAl-Harbi, Nouf, Maha-Hamadien Abdulla, Mansoor-Ali Vaali-Mohammed, Thamer Bin Traiki, Mohammed Alswayyed, Omar Al-Obeed, Islem Abid, Suliman Al-Omar, and Lamjed Mansour. 2023. "Evidence of Association between CTLA-4 Gene Polymorphisms and Colorectal Cancers in Saudi Patients" Genes 14, no. 4: 874. https://doi.org/10.3390/genes14040874
APA StyleAl-Harbi, N., Abdulla, M.-H., Vaali-Mohammed, M.-A., Bin Traiki, T., Alswayyed, M., Al-Obeed, O., Abid, I., Al-Omar, S., & Mansour, L. (2023). Evidence of Association between CTLA-4 Gene Polymorphisms and Colorectal Cancers in Saudi Patients. Genes, 14(4), 874. https://doi.org/10.3390/genes14040874







