A Novel LncRNA MSTRG.310246.1 Promotes Differentiation and Thermogenesis in Goat Brown Adipocytes
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Total RNA Extraction and qPCR
2.3. Bioinformatics Analysis
2.4. Cytoplasmic and Nuclear RNA Extraction
2.5. Primary Goat Brown Adipocytes Culture
2.6. Primary Goat White Adipocytes Culture
2.7. Plasmid Construction and Transfection
2.8. Oil Red O Staining
2.9. Statistical Analysis
3. Results
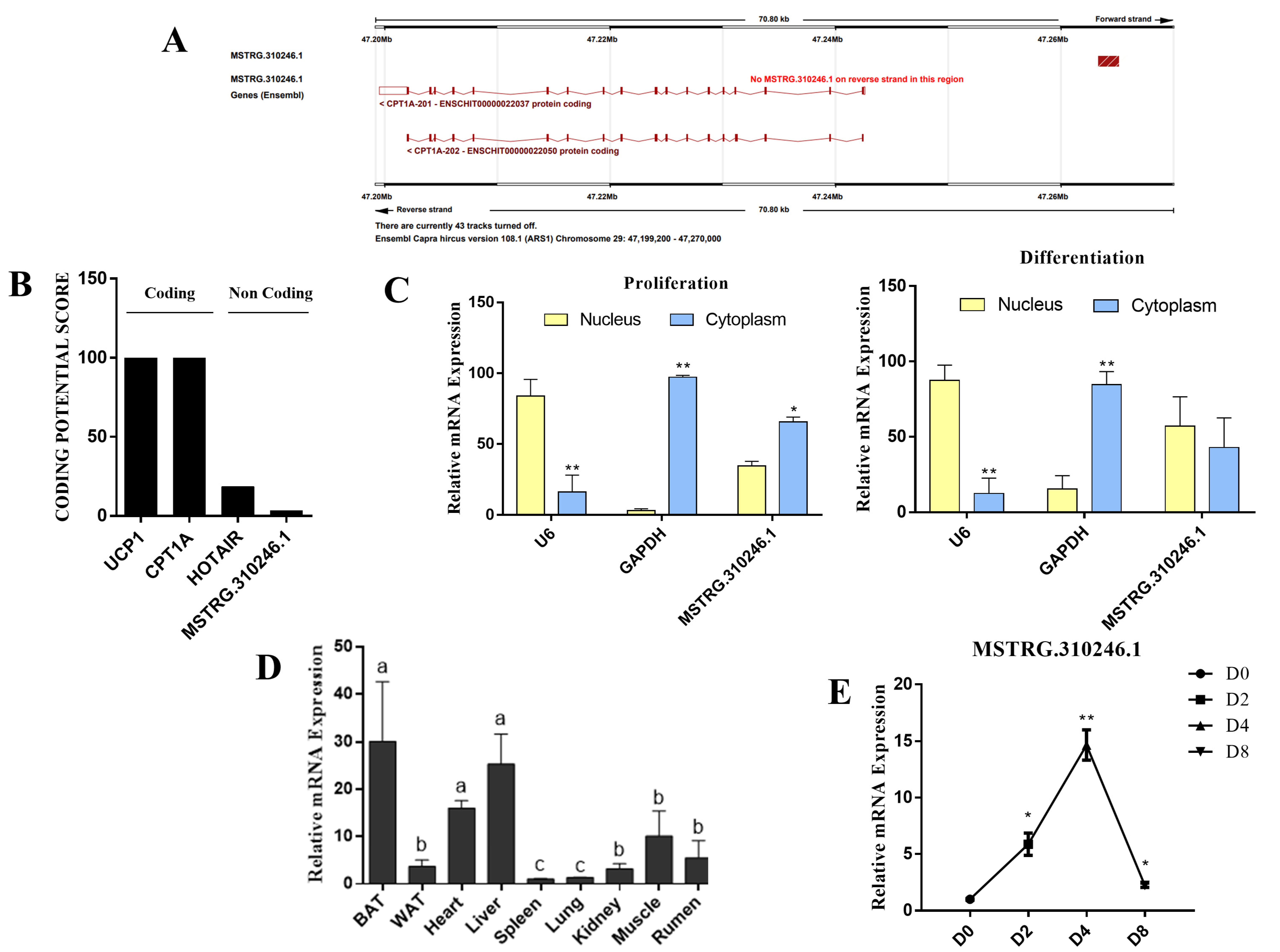
3.1. Identification and Characterization of MSTRG.310246.1 in Goat Brown Adipocytes
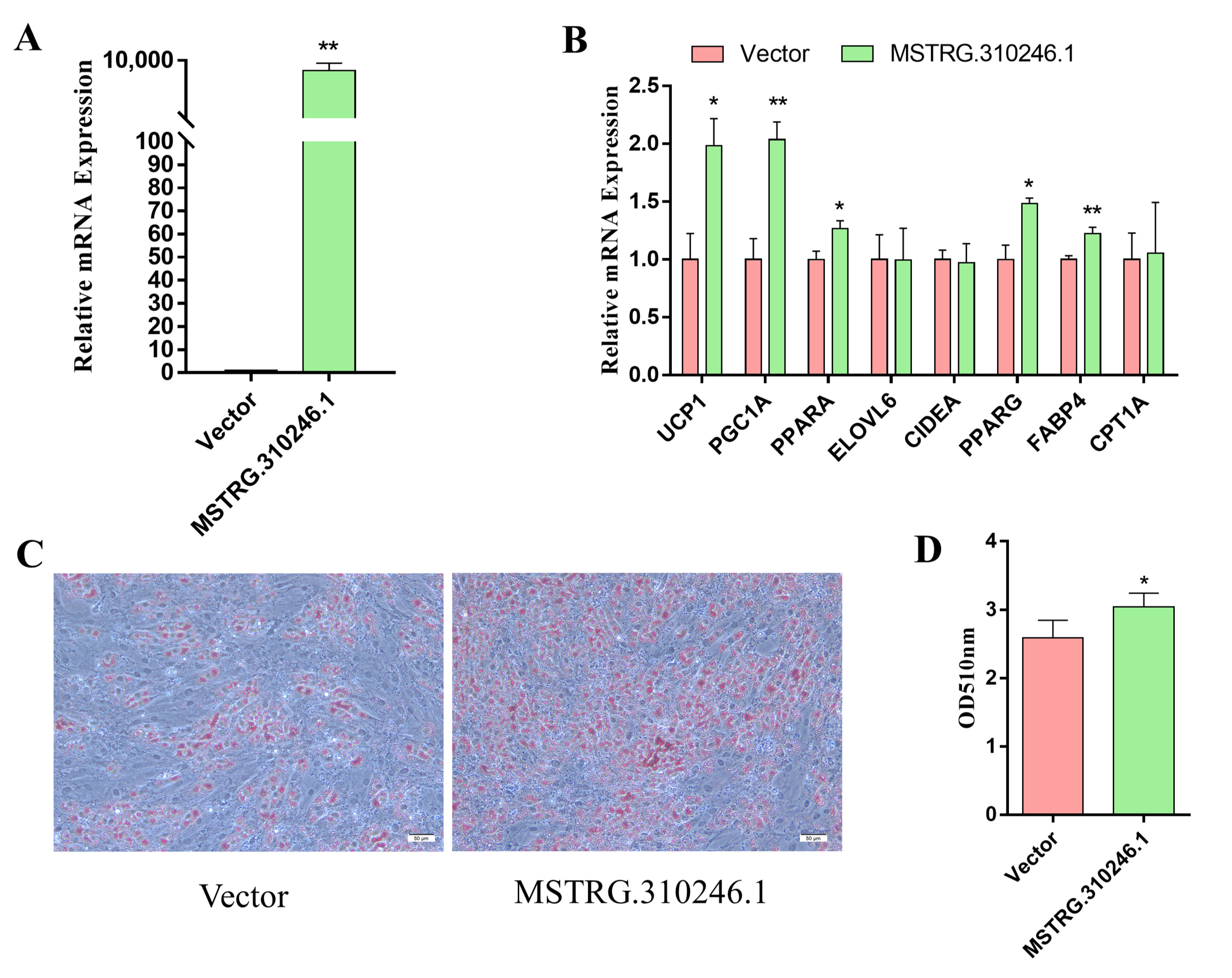
3.2. Overexpression of MSTRG.310246.1 Induced the Differentiation and Thermogenesis of Goat Brown Adipocytes
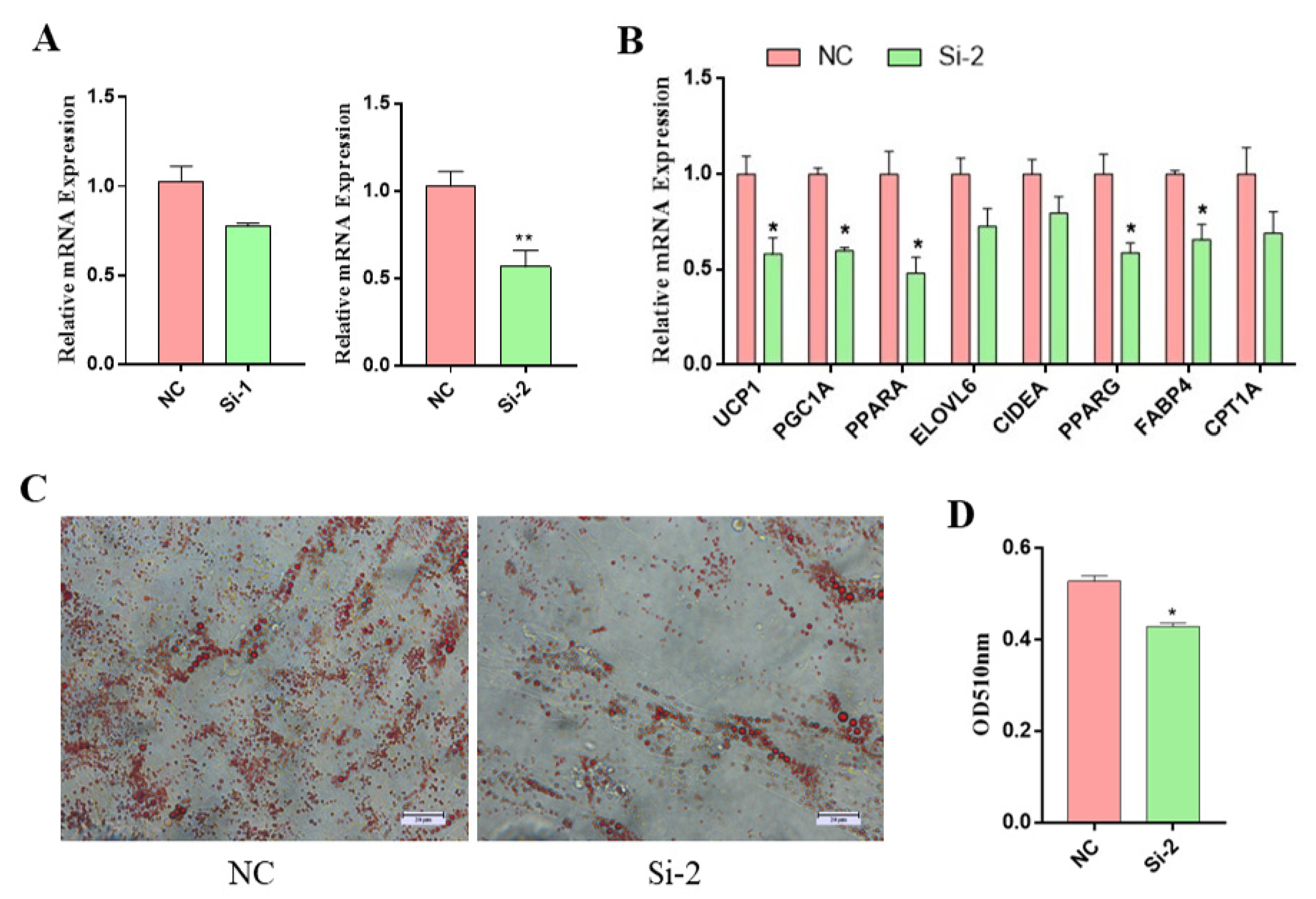
3.3. MSTRG.310246.1 Knockdown Inhibited Brown Adipocyte Differentiation and Thermogenesis
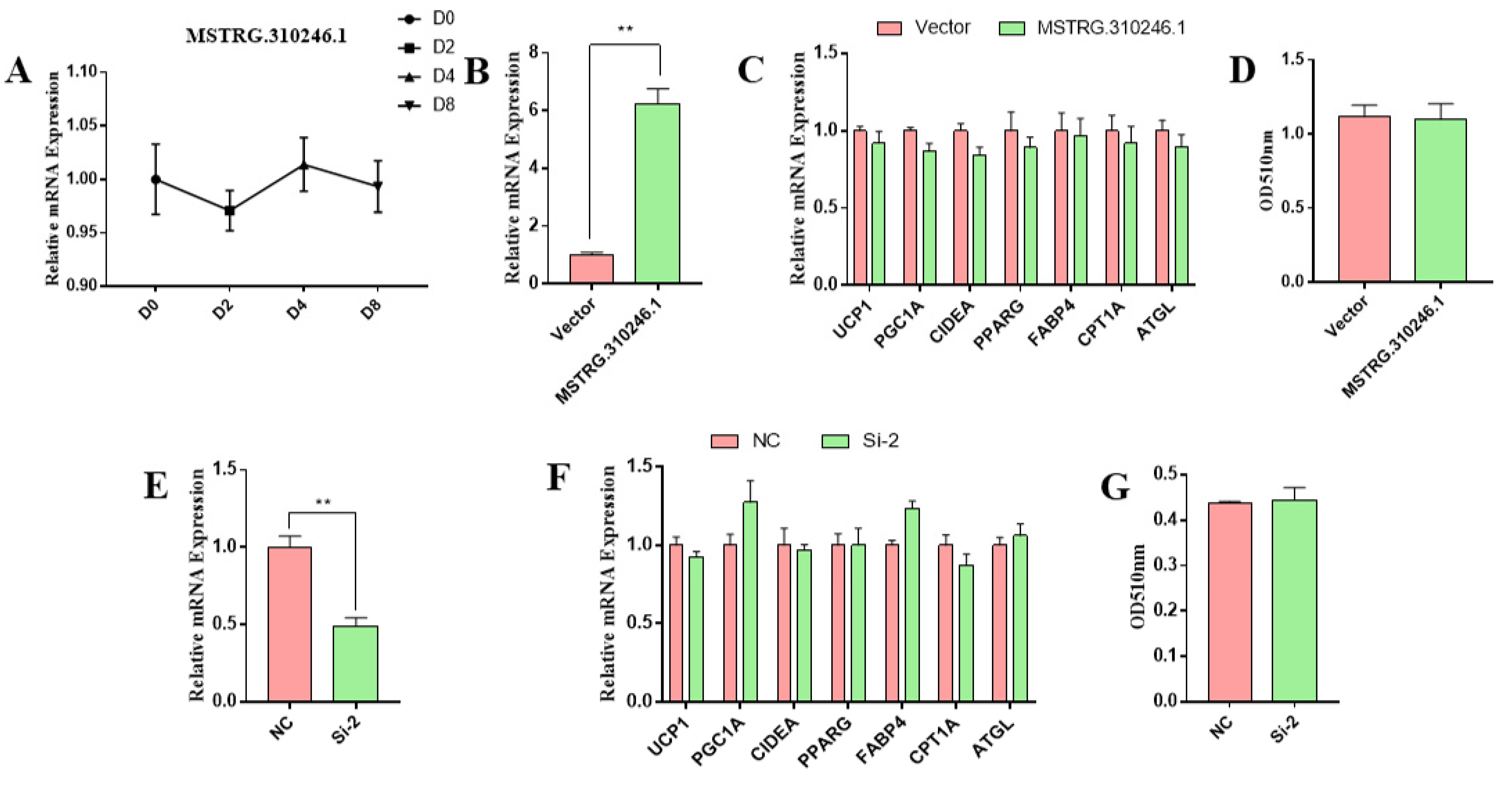
3.4. MSTRG.310246.1 Had No Effect on Goat White Adipocytes Differentiation and Thermogenesis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fenzl, A.; Kiefer, F.W. Brown adipose tissue and thermogenesis. Horm. Mol. Biol. Clin. Investig. 2014, 19, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Frigolet, M.E.; Gutiérrez-Aguilar, R. The colors of adipose tissue. Gac. Med. Mex. 2020, 156, 142–149. [Google Scholar] [CrossRef]
- Lidell, M.E. Brown adipose tissue in human infants. Handb. Exp. Pharmacol. 2019, 251, 107–123. [Google Scholar] [PubMed]
- Lee, Y.H.; Petkova, A.P.; Konkar, A.A.; Granneman, J.G. Cellular origins of cold-induced brown adipocytes in adult mice. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2015, 29, 286–299. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yang, X.; Zhu, Y.; Zhan, S.; Chao, Z.; Zhong, T.; Guo, J.; Wang, Y.; Li, L.; Zhang, H. Genome-wide identification and characterization of long noncoding rnas of brown to white adipose tissue transformation in goats. Cells 2019, 8, 904. [Google Scholar] [CrossRef] [PubMed]
- Nedergaard, J.; Cannon, B. Brown adipose tissue as a heat-producing thermoeffector. Handb. Clin. Neurol. 2018, 156, 137–152. [Google Scholar]
- Gaudry, M.J.; Campbell, K.L.; Jastroch, M. Evolution of ucp1. Handb. Exp. Pharmacol. 2019, 251, 127–141. [Google Scholar]
- Chang, S.H.; Song, N.J.; Choi, J.H.; Yun, U.J.; Park, K.W. Mechanisms underlying ucp1 dependent and independent adipocyte thermogenesis. Obes. Rev. Off. J. Int. Assoc. Study Obes. 2019, 20, 241–251. [Google Scholar] [CrossRef]
- Bouillaud, F.; Alves-Guerra, M.C.; Ricquier, D. Ucps, at the interface between bioenergetics and metabolism. Biochim. Biophys. Acta 2016, 1863, 2443–2456. [Google Scholar] [CrossRef]
- Boström, P.; Wu, J.; Jedrychowski, M.P.; Korde, A.; Ye, L.; Lo, J.C.; Rasbach, K.A.; Boström, E.A.; Choi, J.H.; Long, J.Z.; et al. A pgc1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 2012, 481, 463–468. [Google Scholar] [CrossRef]
- Li, S.; Mi, L.; Yu, L.; Yu, Q.; Liu, T.; Wang, G.X.; Zhao, X.Y.; Wu, J.; Lin, J.D. Zbtb7b engages the long noncoding rna blnc1 to drive brown and beige fat development and thermogenesis. Proc. Natl. Acad. Sci. USA 2017, 114, E7111–E7120. [Google Scholar] [CrossRef]
- Tan, C.Y.; Virtue, S.; Bidault, G.; Dale, M.; Hagen, R.; Griffin, J.L.; Vidal-Puig, A. Brown adipose tissue thermogenic capacity is regulated by elovl6. Cell Rep. 2015, 13, 2039–2047. [Google Scholar] [CrossRef] [PubMed]
- Gu, T.; Xu, G.; Jiang, C.; Hou, L.; Wu, Z.; Wang, C. Prdm16 represses the pig white lipogenesis through promoting lipolysis activity. BioMed Res. Int. 2019, 2019, 1969413. [Google Scholar] [CrossRef]
- Hou, L.; Shi, J.; Cao, L.; Xu, G.; Hu, C.; Wang, C. Pig has no uncoupling protein 1. Biochem. Biophys. Res. Commun. 2017, 487, 795–800. [Google Scholar] [CrossRef]
- Hou, L.; Xie, M.; Cao, L.; Shi, J.; Xu, G.; Hu, C.; Wang, C. Browning of pig white preadipocytes by co-overexpressing pig pgc-1α and mice ucp1. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 48, 556–568. [Google Scholar] [CrossRef]
- Shin, H.; Ma, Y.; Chanturiya, T.; Cao, Q.; Wang, Y.; Kadegowda, A.K.G.; Jackson, R.; Rumore, D.; Xue, B.; Shi, H.; et al. Lipolysis in brown adipocytes is not essential for cold-induced thermogenesis in mice. Cell Metab. 2017, 26, 764–777.e765. [Google Scholar] [CrossRef]
- Picatoste, B.; Yammine, L.; Leahey, R.A.; Soares, D.; Johnson, E.F.; Cohen, P.; McGraw, T.E. Defective insulin-stimulated glut4 translocation in brown adipocytes induces systemic glucose homeostasis dysregulation independent of thermogenesis in female mice. Mol. Metab. 2021, 53, 101305. [Google Scholar] [CrossRef]
- Finlin, B.S.; Memetimin, H.; Zhu, B.; Confides, A.L.; Vekaria, H.J.; El Khouli, R.H.; Johnson, Z.R.; Westgate, P.M.; Chen, J.; Morris, A.J.; et al. The β3-adrenergic receptor agonist mirabegron improves glucose homeostasis in obese humans. J. Clin. Investig. 2020, 130, 2319–2331. [Google Scholar] [CrossRef] [PubMed]
- Cereijo, R.; Gavaldà-Navarro, A.; Cairó, M.; Quesada-López, T.; Villarroya, J.; Morón-Ros, S.; Sánchez-Infantes, D.; Peyrou, M.; Iglesias, R.; Mampel, T.; et al. Cxcl14, a brown adipokine that mediates brown-fat-to-macrophage communication in thermogenic adaptation. Cell Metab. 2018, 28, 750–763.e756. [Google Scholar] [CrossRef]
- Liu, T.; Ju, X.; Zhang, M.; Wei, C.; Wang, D.; Wang, Z.; Lan, X.; Huang, X.X. A 67-bp variable duplication in the promoter region of the adipoq is associated with milk traits in xinjiang brown cattle. Anim. Biotechnol. 2022, 33, 1738–1745. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Davis, M.E.; Chung, H. Effects of genetic variants in the promoter region of the bovine adiponectin (adipoq) gene on marbling of hanwoo beef cattle. Meat Sci. 2015, 105, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Dhanoa, J.K.; Sethi, R.S.; Verma, R.; Arora, J.S.; Mukhopadhyay, C.S. Long non-coding rna: Its evolutionary relics and biological implications in mammals: A review. J. Anim. Sci. Technol. 2018, 60, 25. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding rnas. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wu, S.; He, Y.; Li, X.; Zhu, Y.; Lin, X.; Chen, L.; Zhao, Y.; Niu, L.; Zhang, S.; et al. Lncrna-mediated adipogenesis in different adipocytes. Int. J. Mol. Sci. 2022, 23, 7488. [Google Scholar] [CrossRef]
- Dykes, I.M.; Emanueli, C. Transcriptional and post-transcriptional gene regulation by long non-coding rna. Genom. Proteom. Bioinform. 2017, 15, 177–186. [Google Scholar] [CrossRef]
- Schmitz, S.U.; Grote, P.; Herrmann, B.G. Mechanisms of long noncoding rna function in development and disease. Cell. Mol. Life Sci. CMLS 2016, 73, 2491–2509. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding rnas and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Squillaro, T.; Peluso, G.; Galderisi, U.; Di Bernardo, G. Long non-coding rnas in regulation of adipogenesis and adipose tissue function. eLife 2020, 9, e59053. [Google Scholar] [CrossRef]
- Xu, S.; Chen, P.; Sun, L. Regulatory networks of non-coding rnas in brown/beige adipogenesis. Biosci. Rep. 2015, 35, e00262. [Google Scholar] [CrossRef]
- Yuan, H.; Xu, X.; Feng, X.; Zhu, E.; Zhou, J.; Wang, G.; Tian, L.; Wang, B. A novel long noncoding rna pgc1β-ot1 regulates adipocyte and osteoblast differentiation through antagonizing mir-148a-3p. Cell Death Differ. 2019, 26, 2029–2045. [Google Scholar] [CrossRef]
- Wang, H.; Wei, P.; Zhang, Y.; Li, Y.; Yin, L. Lncrna tcons_00023297 regulates the balance of osteogenic and adipogenic differentiation in bone marrow mesenchymal stem cells and the coupling process of osteogenesis and angiogenesis. Front. Cell Dev. Biol. 2021, 9, 697858. [Google Scholar] [CrossRef] [PubMed]
- Mi, L.; Zhao, X.Y.; Li, S.; Yang, G.; Lin, J.D. Conserved function of the long noncoding rna blnc1 in brown adipocyte differentiation. Mol. Metab. 2017, 6, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Yue, F.; Jia, Z.; Gao, Y.; Jin, W.; Hu, K.; Zhang, Y.; Zhu, D.; Yang, G.; Kuang, S. A novel brown adipocyte-enriched long non-coding rna that is required for brown adipocyte differentiation and sufficient to drive thermogenic gene program in white adipocytes. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hua, S.; Cui, X.; Cao, Y.; Wen, J.; Chi, X.; Ji, C.; Pang, L.; You, L. The effect of foxc2-as1 on white adipocyte browning and the possible regulatory mechanism. Front. Endocrinol. 2020, 11, 565483. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, X.; Song, T.; Zhang, X.; Zhan, S.; Cao, J.; Zhong, T.; Guo, J.; Li, L.; Zhang, H.; et al. Using rna-seq to identify reference genes of the transition from brown to white adipose tissue in goats. Animals 2020, 10, 1626. [Google Scholar] [CrossRef]
- Kang, Y.J.; Yang, D.C.; Kong, L.; Hou, M.; Meng, Y.Q.; Wei, L.; Gao, G. Cpc2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef] [PubMed]
- Ghaben, A.L.; Scherer, P.E. Adipogenesis and metabolic health. Nat. Rev. Mol. Cell Biol. 2019, 20, 242–258. [Google Scholar] [CrossRef]
- Symonds, M.E.; Aldiss, P.; Dellschaft, N.; Law, J.; Fainberg, H.P.; Pope, M.; Sacks, H.; Budge, H. Brown adipose tissue development and function and its impact on reproduction. J. Endocrinol. 2018, 238, R53–R62. [Google Scholar] [CrossRef]
- Basse, A.L.; Dixen, K.; Yadav, R.; Tygesen, M.P.; Qvortrup, K.; Kristiansen, K.; Quistorff, B.; Gupta, R.; Wang, J.; Hansen, J.B. Global gene expression profiling of brown to white adipose tissue transformation in sheep reveals novel transcriptional components linked to adipose remodeling. BMC Genom. 2015, 16, 215. [Google Scholar] [CrossRef]
- Herz, C.T.; Kiefer, F.W. Adipose tissue browning in mice and humans. J. Endocrinol. 2019, 241, R97–R109. [Google Scholar] [CrossRef]
- Liu, X.; Tang, J.; Zhang, R.; Zhan, S.; Zhong, T.; Guo, J.; Wang, Y.; Cao, J.; Li, L.; Zhang, H.; et al. Cold exposure induces lipid dynamics and thermogenesis in brown adipose tissue of goats. BMC Genom. 2022, 23, 528. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, X.; Fan, W.; Zhang, X.; Zhan, S.; Zhong, T.; Guo, J.; Cao, J.; Li, L.; Zhang, H.; et al. Integrated application of metabolomics and rna-seq reveals thermogenic regulation in goat brown adipose tissues. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2021, 35, e21868. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Fan, W.; Zhang, X.; Zhan, S.; Zhong, T.; Guo, J.; Wang, Y.; Cao, J.; Li, L.; Zhang, H.; et al. Maternal l-carnitine supplementation promotes brown adipose tissue thermogenesis of newborn goats after cold exposure. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2022, 36, e22461. [Google Scholar] [CrossRef]
- Sun, L.; Lin, J.D. Function and mechanism of long noncoding rnas in adipocyte biology. Diabetes 2019, 68, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Dominguez, J.R.; Bai, Z.; Xu, D.; Yuan, B.; Lo, K.A.; Yoon, M.J.; Lim, Y.C.; Knoll, M.; Slavov, N.; Chen, S.; et al. De novo reconstruction of adipose tissue transcriptomes reveals long non-coding rna regulators of brown adipocyte development. Cell Metab. 2015, 21, 764–776. [Google Scholar] [CrossRef]
- Cui, X.; You, L.; Li, Y.; Zhu, L.; Zhang, F.; Xie, K.; Cao, Y.; Ji, C.; Guo, X. A transcribed ultraconserved noncoding rna, uc.417, serves as a negative regulator of brown adipose tissue thermogenesis. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2016, 30, 4301–4312. [Google Scholar] [CrossRef]
- Zhang, X.; Zhan, S.; Yang, S.; Zhong, T.; Guo, J.; Cao, J.; Wang, Y.; Li, L.; Zhang, H.; Wang, L. Dynamic expression profiles of circular rnas during brown to white adipose tissue transformation in goats (capra hircus). Animals 2021, 11, 1351. [Google Scholar] [CrossRef]
- Liu, X.; Zhu, Y.; Zhan, S.; Zhong, T.; Guo, J.; Cao, J.; Li, L.; Zhang, H.; Wang, L. Rna-seq reveals mirna role in thermogenic regulation in brown adipose tissues of goats. BMC Genom. 2022, 23, 186. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, J.; Liu, X.; Su, D.; Jiang, T.; Zhan, S.; Zhong, T.; Guo, J.; Cao, J.; Li, L.; Zhang, H.; et al. A Novel LncRNA MSTRG.310246.1 Promotes Differentiation and Thermogenesis in Goat Brown Adipocytes. Genes 2023, 14, 833. https://doi.org/10.3390/genes14040833
Tang J, Liu X, Su D, Jiang T, Zhan S, Zhong T, Guo J, Cao J, Li L, Zhang H, et al. A Novel LncRNA MSTRG.310246.1 Promotes Differentiation and Thermogenesis in Goat Brown Adipocytes. Genes. 2023; 14(4):833. https://doi.org/10.3390/genes14040833
Chicago/Turabian StyleTang, Jing, Xin Liu, Duo Su, Tingting Jiang, Siyuan Zhan, Tao Zhong, Jiazhong Guo, Jiaxue Cao, Li Li, Hongping Zhang, and et al. 2023. "A Novel LncRNA MSTRG.310246.1 Promotes Differentiation and Thermogenesis in Goat Brown Adipocytes" Genes 14, no. 4: 833. https://doi.org/10.3390/genes14040833
APA StyleTang, J., Liu, X., Su, D., Jiang, T., Zhan, S., Zhong, T., Guo, J., Cao, J., Li, L., Zhang, H., & Wang, L. (2023). A Novel LncRNA MSTRG.310246.1 Promotes Differentiation and Thermogenesis in Goat Brown Adipocytes. Genes, 14(4), 833. https://doi.org/10.3390/genes14040833





