Identification of Key Functional Genes and LncRNAs Influencing Muscle Growth and Development in Leizhou Black Goats
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. RNA Isolation, Library Preparation, and Sequencing
2.3. Data Analysis
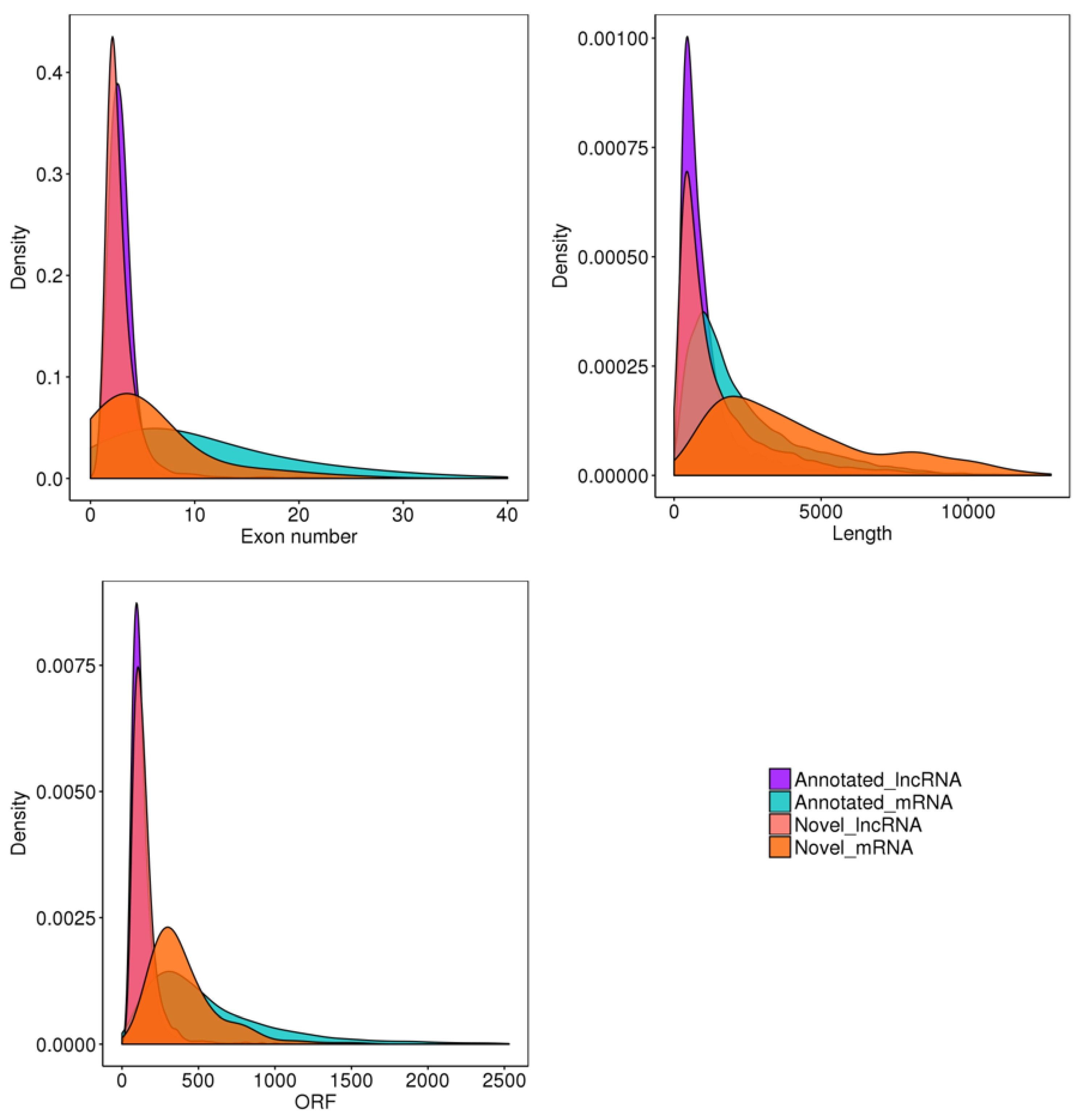
2.4. LncRNA Identification
2.5. Expression Analysis
2.6. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analyses
2.7. Target Gene Prediction
2.8. qRT-PCR Verification
3. Results
3.1. Sequencing Data Quality Control
3.2. Differential Expression of mRNAs and LncRNAs
3.3. GO Analyses of DE-mRNAs and Target Genes of DE-lncRNAs
3.4. KEGG Analyses of DE-mRNAs and Target Genes of DE-lncRNAs
3.5. Predicted Target Genes of DE-lncRNAs
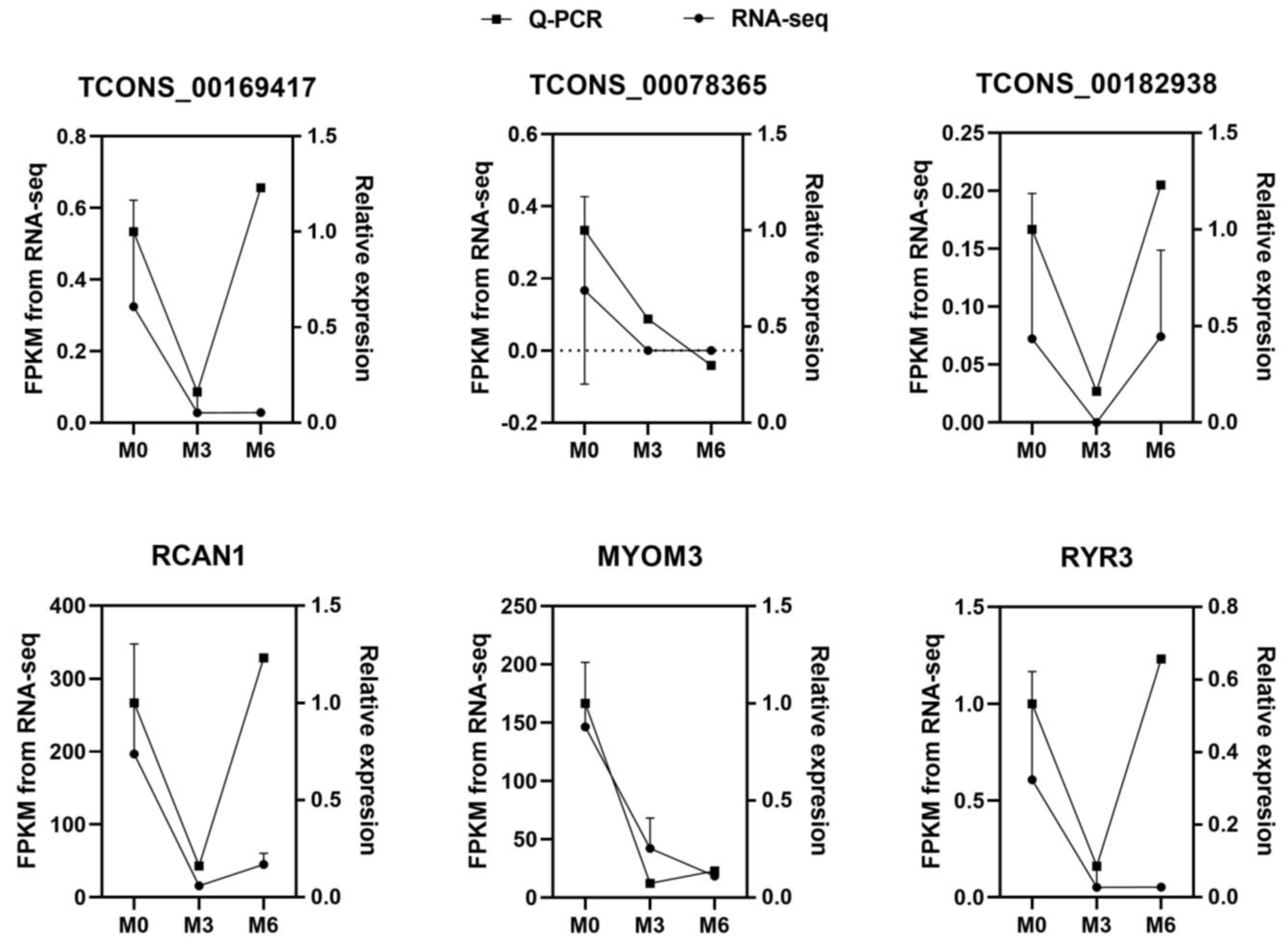
3.6. Validation of RNA-Seq Data
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Braun, T.; Gautel, M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat. Rev. Mol. Cell Biol. 2011, 12, 349–361. [Google Scholar] [CrossRef]
- Buckingham, M.; Vincent, S.D. Distinct and dynamic myogenic populations in the vertebrate embryo. Curr. Opin. Genet. Dev. 2009, 19, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Quiat, D.; Voelker, K.A.; Pei, J.; Grishin, N.V.; Grange, R.W.; Bassel-Duby, R.; Olson, E.N. Concerted regulation of myofiber-specific gene expression and muscle performance by the transcriptional repressor Sox6. Proc. Natl. Acad. Sci. USA 2011, 108, 10196–10201. [Google Scholar] [CrossRef] [PubMed]
- Valencia, P.; Dias, A.P.; Reed, R. Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc. Natl. Acad. Sci. USA 2008, 105, 3386–3391. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Donaghey, J.; Carey, B.W.; Garber, M.; Grenier, J.K.; Munson, G.; Young, G.; Lucas, A.B.; Ach, R.; Bruhn, L.; et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011, 477, 295–300. [Google Scholar] [CrossRef]
- Kretz, M.; Siprashvili, Z.; Chu, C.; Webster, D.E.; Zehnder, A.; Qu, K.; Lee, C.S.; Flockhart, R.J.; Groff, A.F.; Chow, J.; et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 2013, 493, 231–235. [Google Scholar] [CrossRef]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef]
- Prensner, J.R.; Zhao, S.; Erho, N.; Schipper, M.; Iyer, M.K.; Dhanasekaran, S.M.; Magi-Galluzzi, C.; Mehra, R.; Sahu, A.; Siddiqui, J.; et al. RNA biomarkers associated with metastatic progression in prostate cancer: A multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol. 2014, 15, 1469–1480. [Google Scholar] [CrossRef]
- Peng, W.X.; Koirala, P.; Mo, Y.Y. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017, 36, 5661–5667. [Google Scholar] [CrossRef]
- Watts, R.; Johnsen, V.L.; Shearer, J.; Hittel, D.S. Myostatin-induced inhibition of the long noncoding RNA Malat1 is associated with decreased myogenesis. Am. J. Physiol. Cell Physiol. 2013, 304, C995–C1001. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.H.; Zhang, Y.; Li, T.T.; Ma, Z.; Jia, H.X.; Chen, Q.; Zhao, Y.X.; Zhai, L.L.; Zhong, R.; Li, C.Y.; et al. Long non-coding RNA Linc-RAM enhances myogenic differentiation by interacting with MyoD. Nat. Commun. 2017, 8, 14016. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.S.; Gong, C.G.; Maquat, L.E. Control of myogenesis by rodent SINE-containing lncRNAs. Genes Dev. 2013, 27, 793–804. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.Q.; Wang, Y.; Xiong, Y.; Chen, X.C.; Ma, M.L.; Cai, R.; Gao, Y.; Sun, Y.M.; Yang, G.S.; Pang, W.J. Sirt1 AS lncRNA interacts with its mRNA to inhibit muscle formation by attenuating function of miR-34a. Sci. Rep. 2016, 6, 21865. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.S.; Zhang, X.H.; Gu, L.H.; Zhou, H.L.; Rong, G.; Sun, W.P. Identification and characterization of genes related to the development of skeletal muscle in the Hainan black goat. Biosci. Biotechnol. Biochem. 2012, 76, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Zhan, S.; Dong, Y.; Zhao, W.; Guo, J.; Zhong, T.; Wang, L.; Li, L.; Zhang, H. Genome-wide identification and characterization of long non-coding RNAs in developmental skeletal muscle of fetal goat. BMC Genom. 2016, 17, 666. [Google Scholar] [CrossRef]
- Ling, Y.; Zheng, Q.; Sui, M.; Zhu, L.; Xu, L.; Zhang, Y.; Liu, Y.; Fang, F.; Chu, M.; Ma, Y.; et al. Comprehensive Analysis of LncRNA Reveals the Temporal-Specific Module of Goat Skeletal Muscle Development. Int. J. Mol. Sci. 2019, 20, 3950. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- van Diepen, J.A.; Jansen, P.A.; Ballak, D.B.; Hijmans, A.; Hooiveld, G.J.; Rommelaere, S.; Galland, F.; Naquet, P.; Rutjes, F.P.; Mensink, R.P.; et al. PPAR-Alpha dependent regulation of vanin-1 mediates hepatic lipid metabolism. J. Hepatol. 2014, 61, 366–372. [Google Scholar] [CrossRef]
- Li, Y.; Wang, X.; Yu, J.; Shao, F.; Zhang, Y.; Lu, X.; Gu, Z. MiR-122 targets the vanin 1 gene to regulate its expression in chickens. Poult. Sci. 2016, 95, 1145–1150. [Google Scholar] [CrossRef]
- Uemura, T.; Goto, T.; Kang, M.S.; Mizoguchi, N.; Hirai, S.; Lee, J.Y.; Nakano, Y.; Shono, J.; Hoshino, S.; Taketani, K.; et al. Diosgenin, the main aglycon of fenugreek, inhibits LXRα activity in HepG2 cells and decreases plasma and hepatic triglycerides in obese diabetic mice. J. Nutr. 2011, 141, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, T.; Treuter, E.; Gustafsson, J.; Steffensen, K.R. Liver X receptor biology and pharmacology: New pathways, challenges and opportunities. Trends Pharm. Sci. 2012, 33, 394–404. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kong, L.; Deng, M.; Lian, Z.; Han, Y.; Sun, B.; Guo, Y.; Liu, G.; Liu, D. Heat Stress-Responsive Transcriptome Analysis in the Liver Tissue of Hu Sheep. Genes 2019, 10, 395. [Google Scholar] [CrossRef]
- Young, M.D.; Wakeeld, M.J.; Smyth, G.K.; Oshlack, A. goseq: Gene Ontology testing for RNA-seq datasets. R Bioconduct. 2012, 8, 1–25. [Google Scholar]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Bonasio, R.; Shiekhattar, R. Regulation of transcription by long noncoding RNAs. Annu. Rev. Genet. 2014, 48, 433–455. [Google Scholar] [CrossRef]
- Singh, M. Dysregulated A to I RNA editing and non-coding RNAs in neurodegeneration. Front. Genet. 2012, 3, 326. [Google Scholar] [CrossRef]
- Ermak, G.; Cheadle, C.; Becker, K.G.; Harris, C.D.; Davies, K.J. DSCR1(Adapt78) modulates expression of SOD1. FASEB J. 2004, 18, 62–69. [Google Scholar] [CrossRef]
- Kim, Y.S.; Cho, K.O.; Lee, H.J.; Kim, S.Y.; Sato, Y.; Cho, Y.J. Down syndrome candidate region 1 increases the stability of the IkappaBalpha protein: Implications for its anti-inflammatory effects. J. Biol. Chem. 2006, 281, 39051–39061. [Google Scholar] [CrossRef]
- Feingold, E.A.; Penny, L.A.; Nienhuis, A.W.; Forget, B.G. An olfactory receptor gene is located in the extended human beta-globin gene cluster and is expressed in erythroid cells. Genomics 1999, 61, 15–23. [Google Scholar] [CrossRef]
- Spehr, M.; Schwane, K.; Riffell, J.A.; Zimmer, R.K.; Hatt, H. Odorant receptors and olfactory-like signaling mechanisms in mammalian sperm. Mol. Cell. Endocrinol. 2006, 250, 128–136. [Google Scholar] [CrossRef]
- Yuan, T.T.; Toy, P.; McClary, J.A.; Lin, R.J.; Miyamoto, N.G.; Kretschmer, P.J. Cloning and genetic characterization of an evolutionarily conserved human olfactory receptor that is differentially expressed across species. Gene 2001, 278, 41–51. [Google Scholar] [CrossRef]
- Griffin, C.A.; Kafadar, K.A.; Pavlath, G.K. MOR23 promotes muscle regeneration and regulates cell adhesion and migration. Dev. Cell 2009, 17, 649–661. [Google Scholar] [CrossRef] [PubMed]
- Pichavant, C.; Burkholder, T.J.; Pavlath, G.K. Decrease of myofiber branching via muscle-specific expression of the olfactory receptor mOR23 in dystrophic muscle leads to protection against mechanical stress. Skelet. Muscle 2016, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Thach, T.T.; Wu, C.; Hwang, K.Y.; Lee, S.J. Azelaic Acid Induces Mitochondrial Biogenesis in Skeletal Muscle by Activation of Olfactory Receptor 544. Front. Physiol. 2020, 11, 329. [Google Scholar] [CrossRef]
- Wen, Y.; Alimov, A.P.; McCarthy, J.J. Ribosome Biogenesis is Necessary for Skeletal Muscle Hypertrophy. Exerc. Sport Sci. Rev. 2016, 44, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, C.K.; Kristensen, N.B. Nitrogen recycling through the gut and the nitrogen economy of ruminants: An asynchronous symbiosis. J. Anim. Sci. 2008, 86, E293–E305. [Google Scholar] [CrossRef]
- Stewart, G.S.; Graham, C.; Cattell, S.; Smith, T.P.; Simmons, N.L.; Smith, C.P. UT-B is expressed in bovine rumen: Potential role in ruminal urea transport. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005, 289, R605–R612. [Google Scholar] [CrossRef]
- Reese, A.T.; Pereira, F.C.; Schintlmeister, A.; Berry, D.; Wagner, M.; Hale, L.P.; Wu, A.; Jiang, S.; Durand, H.K.; Zhou, X.; et al. Microbial nitrogen limitation in the mammalian large intestine. Nat. Microbiol. 2018, 3, 1441–1450. [Google Scholar] [CrossRef]
- Holmes, A.J.; Chew, Y.V.; Colakoglu, F.; Cliff, J.B.; Klaassens, E.; Read, M.N.; Solon-Biet, S.M.; McMahon, A.C.; Cogger, V.C.; Ruohonen, K.; et al. Diet-Microbiome Interactions in Health Are Controlled by Intestinal Nitrogen Source Constraints. Cell Metab. 2017, 25, 140–151. [Google Scholar] [CrossRef] [PubMed]
- Kernstock, S.; Davydova, E.; Jakobsson, M.; Moen, A.; Pettersen, S.; Mælandsmo, G.M.; Egge-Jacobsen, W.; Falnes, P. Lysine methylation of VCP by a member of a novel human protein methyltransferase family. Nat. Commun. 2012, 3, 1038. [Google Scholar] [CrossRef] [PubMed]
- Wiederstein, J.L.; Nolte, H.; Günther, S.; Piller, T.; Baraldo, M.; Kostin, S.; Bloch, W.; Schindler, N.; Sandri, M.; Blaauw, B.; et al. Skeletal Muscle-Specific Methyltransferase METTL21C Trimethylates p97 and Regulates Autophagy-Associated Protein Breakdown. Cell Rep. 2018, 23, 1342–1356. [Google Scholar] [CrossRef] [PubMed]
- Zoabi, M.; Zhang, L.; Li, T.M.; Elias, J.E.; Carlson, S.M.; Gozani, O. Methyltransferase-like 21C (METTL21C) methylates alanine tRNA synthetase at Lys-943 in muscle tissue. J. Biol. Chem. 2020, 295, 11822–11832. [Google Scholar] [CrossRef]
- Olszewski, P.K.; Rozman, J.; Jacobsson, J.A.; Rathkolb, B.; Strömberg, S.; Hans, W.; Klockars, A.; Alsiö, J.; Risérus, U.; Becker, L.; et al. Neurobeachin, a regulator of synaptic protein targeting, is associated with body fat mass and feeding behavior in mice and body-mass index in humans. PLoS Genet. 2012, 8, e1002568. [Google Scholar] [CrossRef]
- Li, Y.-D.; Bai, X.; Liu, X.; Wang, W.-J.; Li, Z.-W.; Wang, N.; Xiao, F.; Gao, H.-H.; Guo, H.-S.; Li, H.; et al. Integration of genome-wide association study and selection signatures reveals genetic determinants for skeletal muscle production traits in an F2 chicken population. J. Integr. Agric. 2022, 21, 2065–2075. [Google Scholar] [CrossRef]
| Gene | Primer Sequence 5′→3′ | Product Length (bp) |
|---|---|---|
| TCONS_00169417 | CCCGATTCCCCAGATAGCGA | 117 |
| ACCGGATAAAGATCGGCTCG | ||
| TCONS_00078365 | AGGCAGGAATTGCGGTGTAT | 288 |
| CCCAGGATCAGTCAACACCA | ||
| TCONS_00182938 | TTTCTTCACTGCCATCCTCCC | 250 |
| AAGTTCTGTTGCCTTCCCCG | ||
| RCAN1 | GTTTGTATGTAGAGTTGCG | 437 |
| TTGATGTATTAGTGGGGGT | ||
| MYOM3 | CACATTCTTCTCCCGGTCCC | 124 |
| TGCGAGAGCAAAAACAGAAGC | ||
| RYR3 | TGGTCATCAACACGCCATCT | 280 |
| ATGGTTGTGTACCAGGCGAG |
| Sample Name | Raw Reads | Clean Reads | Q20 | Q30 | Total Mapped | Multiple Mapped | Uniquely Mapped |
|---|---|---|---|---|---|---|---|
| M0_1 | 91,451,366 | 90,296,570 | 97.0% | 92.1% | 81,031,701 (94.2%) | 11,681,824 (12.9%) | 71,341,877 (81.2%) |
| M0_2 | 99,145,950 | 97,674,536 | 97.3% | 92.6% | 91,661,001 (95.9%) | 1,491,508 (9.7%) | 81,171,493 (86.2%) |
| M0_3 | 97,522,592 | 96,325,710 | 97.0% | 91.9% | 91,851,053 (96.4%) | 1,741,407 (7.0%) | 81,111,646 (89.4%) |
| M3_1 | 93,972,708 | 92,817,732 | 96.8% | 91.5% | 81,171,540 (92.8%) | 11,311,379 (11.1%) | 71,861,161 (81.7%) |
| M3_2 | 115,506,982 | 114,030,402 | 97.5% | 93.4% | 101,861,223 (91.1%) | 11,141,916 (10.6%) | 91,721,307 (80.4%) |
| M3_3 | 109,977,218 | 108,672,712 | 97.4% | 93.1% | 101,661,879 (92.6%) | 1,921,360 (9.1%) | 91,731,519 (83.5%) |
| M6_1 | 91,389,622 | 90,146,316 | 97.5% | 93.4% | 81,141,246 (94.4%) | 1,011,263 (7.8%) | 71,131,983 (86.7%) |
| M6_2 | 109,277,192 | 107,969,018 | 97.4% | 93.1% | 101,741,280 (93.3%) | 11,131,444 (11.2%) | 81,601,836 (82.1%) |
| M6_3 | 115,475,130 | 114,090,040 | 97.5% | 93.4% | 101,911,408 (93.7%) | 11,511,743 (11.8%) | 91,401,665 (81.9%) |
| Upregulated | Downregulated | Uniquely Expressed | Total | ||
|---|---|---|---|---|---|
| DE-mRNAs | M0 vs. M3 | 103 | 89 | 59 | 192 |
| M0 vs. M6 | 167 | 121 | 134 | 288 | |
| M6 vs. M3 | 65 | 94 | 58 | 159 | |
| DE-lncRNAs | M0 vs. M3 | 23 | 32 | 28 | 55 |
| M0 vs. M6 | 45 | 38 | 59 | 83 | |
| M6 vs. M3 | 10 | 23 | 18 | 33 |
| GO Accession | Description | Term Type | p-Value | Gene Count |
|---|---|---|---|---|
| M0 vs. M3 | ||||
| GO:0070469 | respiratory chain | CC | 3.23 × 10−3 | 6 |
| GO:0006091 | generation of precursor metabolites and energy | BP | 7.48 × 10−3 | 10 |
| M0 vs. M6 | ||||
| GO:0005840 | ribosome | CC | 4.65 × 10−27 | 59 |
| GO:0003735 | structural constituent of ribosome | MF | 1.08 × 10−26 | 58 |
| GO:0030529 | ribonucleoprotein complex | CC | 1.86 × 10−23 | 64 |
| GO:0006412 | translation | BP | 9.49 × 10−22 | 67 |
| GO:0005198 | structural molecule activity | MF | 4.10 × 10−20 | 73 |
| GO:0043228 | non-membrane-bound organelle | CC | 1.54 × 10−15 | 106 |
| GO:0043232 | intracellular non-membrane-bound organelle | CC | 1.54 × 10−15 | 106 |
| GO:0044444 | cytoplasmic part | CC | 5.13 × 10−10 | 114 |
| GO:0044391 | ribosomal subunit | CC | 5.25 × 10−10 | 16 |
| GO:0044267 | cellular protein metabolic process | BP | 2.51 × 10−8 | 147 |
| GO:0005737 | cytoplasm | CC | 4.47 × 10−8 | 138 |
| GO:0044724 | single-organism carbohydrate catabolic process | BP | 4.96 × 10−8 | 13 |
| GO:0006096 | glycolysis | BP | 6.23 × 10−8 | 11 |
| GO:0016052 | carbohydrate catabolic process | BP | 8.55 × 10−8 | 13 |
| GO:0015935 | small ribosomal subunit | CC | 1.40 × 10−7 | 9 |
| GO:0019538 | protein metabolic process | BP | 2.35 × 10−7 | 171 |
| GO:0006091 | generation of precursor metabolites and energy | BP | 3.55 × 10−7 | 27 |
| GO:0006006 | glucose metabolic process | BP | 6.83 × 10−7 | 13 |
| GO:0006007 | glucose catabolic process | BP | 9.27 × 10−7 | 11 |
| GO:0019320 | hexose catabolic process | BP | 1.17 × 10−6 | 11 |
| M6 vs. M3 | ||||
| GO:0015204 | urea transmembrane transporter activity | MF | 0.017515 | 2 |
| GO:0015840 | urea transport | BP | 0.017515 | 2 |
| GO:0019755 | one-carbon compound transport | BP | 0.017515 | 2 |
| GO:0071918 | urea transmembrane transport | BP | 0.017515 | 2 |
| GO Accession | Description | Term Type | p-Value | Gene Count |
|---|---|---|---|---|
| GO:0015204 | urea transmembrane transporter activity | MF | 0.047565 | 2 |
| GO:0015840 | urea transport | BP | 0.047565 | 2 |
| GO:0019755 | one-carbon compound transport | BP | 0.047565 | 2 |
| GO:0071918 | urea transmembrane transport | BP | 0.047565 | 2 |
| KEGG Pathway | Rich Factor | p-Value | Gene Number |
|---|---|---|---|
| Ribosome | 0.211073 | 4.03 × 10−12 | 61 |
| Biosynthesis of amino acids | 0.25 | 0.000259 | 19 |
| Glycolysis/gluconeogenesis | 0.241935 | 0.00275 | 15 |
| Comparison | KEGG Pathway | Rich Factor | p-Value | Gene Number |
|---|---|---|---|---|
| M0 vs. M3 | 2-oxocarboxylic acid metabolism | 0.176471 | 0.02762 | 3 |
| M0 vs. M6 | Ubiquitin-mediated proteolysis | 0.05036 | 0.025317 | 7 |
| lncRNA id | log2FoldChange | mRNA id | Gene Name | log2FoldChange |
|---|---|---|---|---|
| M0 vs. M3 | ||||
| TCONS_00074191 | 12.80220475 | ENSCHIG00000007982 | METTL11B | −3.86509 |
| TCONS_00136308 | 13.66725117 | XLOC_087192 | XLOC_087192 | 1.83846 |
| TCONS_00074190 | 11.40821788 | ENSCHIG00000007982 | METTL11B | −3.86509 |
| M0 vs. M6 | ||||
| TCONS_00135868 | −15.24605461 | ENSCHIG00000017944 | NRG4 | −1.63975 |
| TCONS_00045045 | 14.24586981 | ENSCHIG00000024367 | NBEA | 1.245331 |
| TCONS_00078361 | 13.30525325 | ENSCHIG00000007982 | METTL11B | −4.7664 |
| TCONS_00135347 | 12.15929256 | ENSCHIG00000017618 | PDE8A | −1.74242 |
| TCONS_00214300 | −4.18296294 | ENSCHIG00000013176 | OR2AP1 | −4.19294 |
| TCONS_00167695 | −11.75820204 | ENSCHIG00000020411 | A1CF | −12.2245 |
| TCONS_00045031 | 13.47115691 | ENSCHIG00000024367 | NBEA | 1.245331 |
| TCONS_00142330 | −10.67270908 | ENSCHIG00000017389 | ATP2B2 | −2.93845 |
| M6 vs. M3 | ||||
| TCONS_00190312 | −12.28261621 | ENSCHIG00000007731 | NGF | −4.65694 |
| TCONS_00182938 | 13.70600879 | ENSCHIG00000015813 | GRM5 | 9.54066 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Ye, J.; Lin, X.; Xue, H.; Zou, X.; Liu, G.; Deng, M.; Sun, B.; Guo, Y.; Liu, D.; et al. Identification of Key Functional Genes and LncRNAs Influencing Muscle Growth and Development in Leizhou Black Goats. Genes 2023, 14, 881. https://doi.org/10.3390/genes14040881
Zhao X, Ye J, Lin X, Xue H, Zou X, Liu G, Deng M, Sun B, Guo Y, Liu D, et al. Identification of Key Functional Genes and LncRNAs Influencing Muscle Growth and Development in Leizhou Black Goats. Genes. 2023; 14(4):881. https://doi.org/10.3390/genes14040881
Chicago/Turabian StyleZhao, Xiuhui, Junning Ye, Xunkai Lin, Huiwen Xue, Xian Zou, Guangbin Liu, Ming Deng, Baoli Sun, Yongqing Guo, Dewu Liu, and et al. 2023. "Identification of Key Functional Genes and LncRNAs Influencing Muscle Growth and Development in Leizhou Black Goats" Genes 14, no. 4: 881. https://doi.org/10.3390/genes14040881
APA StyleZhao, X., Ye, J., Lin, X., Xue, H., Zou, X., Liu, G., Deng, M., Sun, B., Guo, Y., Liu, D., & Li, Y. (2023). Identification of Key Functional Genes and LncRNAs Influencing Muscle Growth and Development in Leizhou Black Goats. Genes, 14(4), 881. https://doi.org/10.3390/genes14040881






