DNA Polymerase Delta Exhibits Altered Catalytic Properties on Lysine Acetylation
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Substrate Design
2.3. Purification of Recombinant Proteins
2.4. Construction, Expression, and Purification of the Catalytic Domain of h-p300
2.5. Acetylation of Recombinant h-Pol δ
2.6. Pol δ Extension, Strand Displacement, and Structure Resolution Assays
2.7. Exonuclease Assays
2.8. Quantification of Polymerization and Exonuclease Products
2.9. Mammalian Cell Culture
2.10. IP and Western Blot Analysis of h-pol δ
2.11. Analysis of the Acetylation Status of h-pol δ by Mass Spectroscopy
3. Results
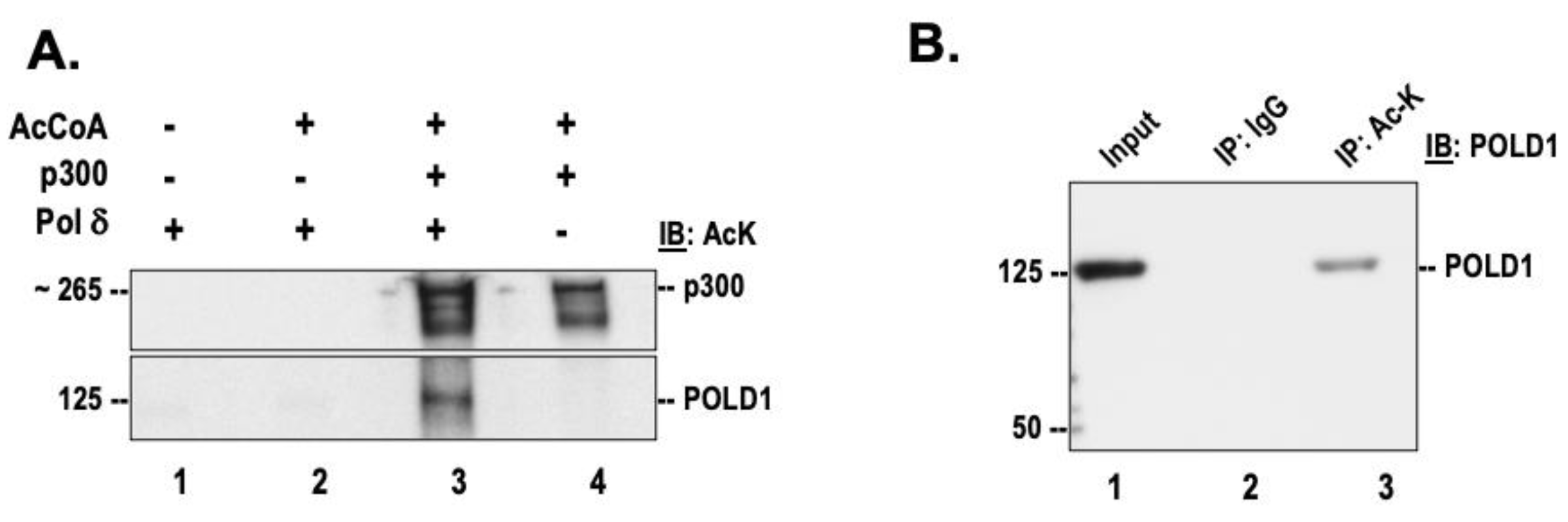
3.1. DNA Polymerase δ Is Acetylated In Vitro and In Vivo
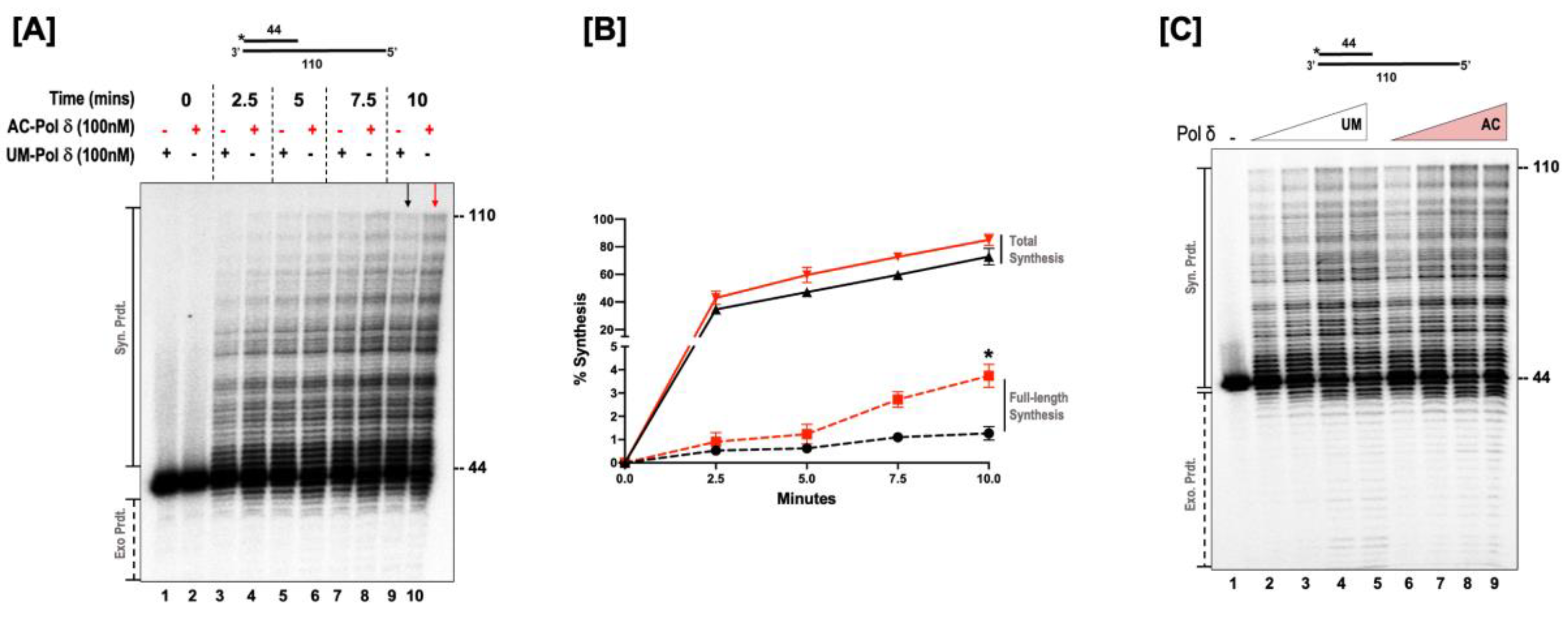
3.2. Acetylation Stimulates the Synthesis Activity of DNA Polymerase δ
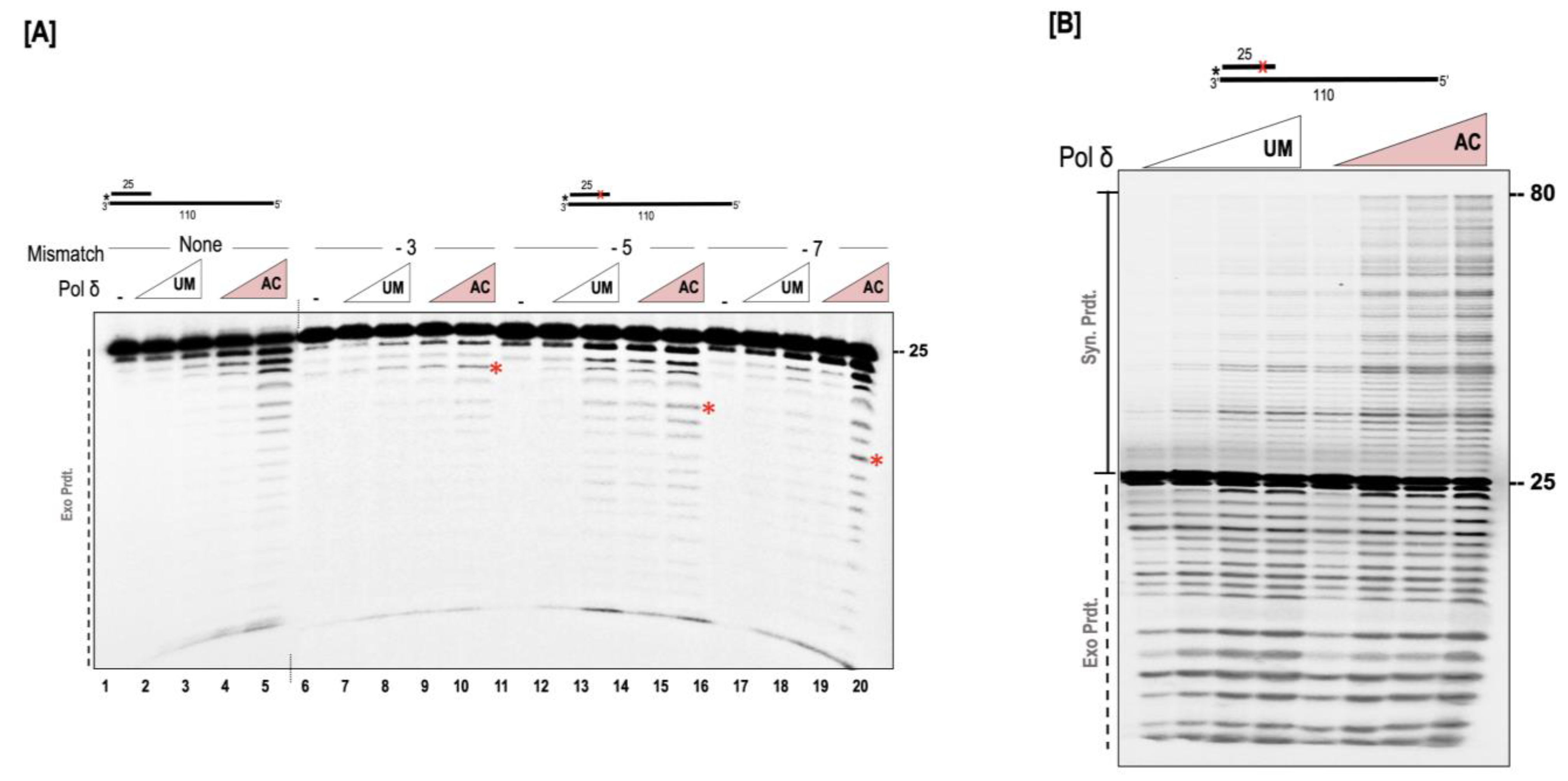
3.3. Acetylation Stimulates the Strand Displacement Activity of Pol δ
3.4. Acetylation Enhances the Ability of Pol δ to Resolve Complex Secondary Structures
3.5. Replication Accessory Proteins Can Stimulate Acetylated Polymerase
3.6. Acetylated Polymerase Displays Higher Exonuclease Activity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barnes, R.; Eckert, K. Maintenance of Genome Integrity: How Mammalian Cells Orchestrate Genome Duplication by Coordinating Replicative and Specialized DNA Polymerases. Genes 2017, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Pursell, Z.F.; Isoz, I.; Lundstrom, E.B.; Johansson, E.; Kunkel, T.A. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science 2007, 317, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Nick McElhinny, S.A.; Gordenin, D.A.; Stith, C.M.; Burgers, P.M.; Kunkel, T.A. Division of labor at the eukaryotic replication fork. Mol. Cell 2008, 30, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Burgers, P.M. Polymerase dynamics at the eukaryotic DNA replication fork. J. Biol. Chem. 2009, 284, 4041–4045. [Google Scholar] [CrossRef]
- Kunkel, T.A.; Burgers, P.M. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 2008, 18, 521–527. [Google Scholar] [CrossRef]
- Thompson, J.A.; Marzahn, M.R.; O’Donnell, M.; Bloom, L.B. Replication factor C is a more effective proliferating cell nuclear antigen (PCNA) opener than the checkpoint clamp loader, Rad24-RFC. J. Biol. Chem. 2012, 287, 2203–2209. [Google Scholar] [CrossRef]
- Zheng, L.; Shen, B. Okazaki fragment maturation: Nucleases take centre stage. J. Mol. Cell Biol. 2011, 3, 23–30. [Google Scholar] [CrossRef]
- Balakrishnan, L.; Gloor, J.W.; Bambara, R.A. Reconstitution of eukaryotic lagging strand DNA replication. Methods 2010, 51, 347–357. [Google Scholar] [CrossRef]
- Pike, J.E.; Henry, R.A.; Burgers, P.M.; Campbell, J.L.; Bambara, R.A. An alternative pathway for Okazaki fragment processing: Resolution of fold-back flaps by Pif1 helicase. J. Biol. Chem. 2010, 285, 41712–41723. [Google Scholar] [CrossRef]
- Reijns, M.A.M.; Kemp, H.; Ding, J.; de Proce, S.M.; Jackson, A.P.; Taylor, M.S. Lagging-strand replication shapes the mutational landscape of the genome. Nature 2015, 518, 502–506. [Google Scholar] [CrossRef]
- Williams, J.S.; Kunkel, T.A. Ribonucleotides in DNA: Origins, repair and consequences. DNA Repair 2014, 19, 27–37. [Google Scholar] [CrossRef]
- Fazlieva, R.; Spittle, C.S.; Morrissey, D.; Hayashi, H.; Yan, H.; Matsumoto, Y. Proofreading exonuclease activity of human DNA polymerase delta and its effects on lesion-bypass DNA synthesis. Nucleic Acids Res. 2009, 37, 2854–2866. [Google Scholar] [CrossRef]
- Zhou, Y.; Meng, X.; Zhang, S.; Lee, E.Y.C.; Lee, M.Y.W.T. Characterization of Human DNA Polymerase Delta and Its Subassemblies Reconstituted by Expression in the Multibac System. PLoS ONE 2012, 7, e39156. [Google Scholar] [CrossRef]
- Lee, M.; Wang, X.; Zhang, S.; Zhang, Z.; Lee, E.Y.C. Regulation and Modulation of Human DNA Polymerase delta Activity and Function. Genes 2017, 8, 190. [Google Scholar] [CrossRef]
- Rahmeh, A.A.; Zhou, Y.; Xie, B.; Li, H.; Lee, E.Y.; Lee, M.Y. Phosphorylation of the p68 subunit of Pol delta acts as a molecular switch to regulate its interaction with PCNA. Biochemistry 2012, 51, 416–424. [Google Scholar] [CrossRef]
- Moritz, A.; Li, Y.; Guo, A.; Villen, J.; Wang, Y.; MacNeill, J.; Kornhauser, J.; Sprott, K.; Zhou, J.; Possemato, A.; et al. Akt-RSK-S6 kinase signaling networks activated by oncogenic receptor tyrosine kinases. Sci. Signal. 2010, 3, ra64. [Google Scholar] [CrossRef]
- Choudhary, C.; Kumar, C.; Gnad, F.; Nielsen, M.L.; Rehman, M.; Walther, T.C.; Olsen, J.V.; Mann, M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 2009, 325, 834–840. [Google Scholar] [CrossRef]
- Balakrishnan, L.; Stewart, J.; Polaczek, P.; Campbell, J.L.; Bambara, R.A. Acetylation of Dna2 endonuclease/helicase and flap endonuclease 1 by p300 promotes DNA stability by creating long flap intermediates. J. Biol. Chem. 2010, 285, 4398–4404. [Google Scholar] [CrossRef]
- Naryzhny, S.N.; Lee, H. The post-translational modifications of proliferating cell nuclear antigen: Acetylation, not phosphorylation, plays an important role in the regulation of its function. J. Biol. Chem. 2004, 279, 20194–20199. [Google Scholar] [CrossRef]
- Hasan, S.; Stucki, M.; Hassa, P.O.; Imhof, R.; Gehrig, P.; Hunziker, P.; Hubscher, U.; Hottiger, M.O. Regulation of human flap endonuclease-1 activity by acetylation through the transcriptional coactivator p300. Mol. Cell 2001, 7, 1221–1231. [Google Scholar] [CrossRef]
- Pike, J.E.; Burgers, P.M.; Campbell, J.L.; Bambara, R.A. Pif1 helicase lengthens some Okazaki fragment flaps necessitating Dna2 nuclease/helicase action in the two-nuclease processing pathway. J. Biol. Chem. 2009, 284, 25170–25180. [Google Scholar] [CrossRef] [PubMed]
- Edwards, D.N.; Machwe, A.; Wang, Z.; Orren, D.K. Intramolecular telomeric G-quadruplexes dramatically inhibit DNA synthesis by replicative and translesion polymerases, revealing their potential to lead to genetic change. PLoS ONE 2014, 9, e80664. [Google Scholar] [CrossRef] [PubMed]
- Henricksen, L.A.; Umbricht, C.B.; Wold, M.S. Recombinant replication protein A: Expression, complex formation, and functional characterization. J. Biol. Chem. 1994, 269, 11121–11132. [Google Scholar] [CrossRef] [PubMed]
- Podust, L.M.; Podust, V.N.; Sogo, J.M.; Hubscher, U. Mammalian DNA polymerase auxiliary proteins: Analysis of replication factor C-catalyzed proliferating cell nuclear antigen loading onto circular double-stranded DNA. Mol. Cell. Biol. 1995, 15, 3072–3081. [Google Scholar] [CrossRef] [PubMed]
- Podust, V.N.; Tiwari, N.; Stephan, S.; Fanning, E. Replication factor C disengages from proliferating cell nuclear antigen (PCNA) upon sliding clamp formation, and PCNA itself tethers DNA polymerase delta to DNA. J. Biol. Chem. 1998, 273, 31992–31999. [Google Scholar] [CrossRef]
- Bordoli, L.; Husser, S.; Luthi, U.; Netsch, M.; Osmani, H.; Eckner, R. Functional analysis of the p300 acetyltransferase domain: The PHD finger of p300 but not of CBP is dispensable for enzymatic activity. Nucleic Acids Res. 2001, 29, 4462–4471. [Google Scholar] [CrossRef]
- Maksimoska, J.; Segura-Pena, D.; Cole, P.A.; Marmorstein, R. Structure of the p300 histone acetyltransferase bound to acetyl-coenzyme A and its analogues. Biochemistry 2014, 53, 3415–3422. [Google Scholar] [CrossRef]
- Akimov, V.; Barrio-Hernandez, I.; Hansen, S.V.F.; Hallenborg, P.; Pedersen, A.K.; Bekker-Jensen, D.B.; Puglia, M.; Christensen, S.D.K.; Vanselow, J.T.; Nielsen, M.M.; et al. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat. Struct. Mol. Biol. 2018, 25, 631–640. [Google Scholar] [CrossRef]
- Mertins, P.; Qiao, J.W.; Patel, J.; Udeshi, N.D.; Clauser, K.R.; Mani, D.R.; Burgess, M.W.; Gillette, M.A.; Jaffe, J.D.; Carr, S.A. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat. Methods 2013, 10, 634–637. [Google Scholar] [CrossRef]
- Kim, W.; Bennett, E.J.; Huttlin, E.L.; Guo, A.; Li, J.; Possemato, A.; Sowa, M.E.; Rad, R.; Rush, J.; Comb, M.J.; et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 2011, 44, 325–340. [Google Scholar] [CrossRef]
- Danielsen, J.M.; Sylvestersen, K.B.; Bekker-Jensen, S.; Szklarczyk, D.; Poulsen, J.W.; Horn, H.; Jensen, L.J.; Mailand, N.; Nielsen, M.L. Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol. Cell. Proteom. MCP 2011, 10, M110.003590. [Google Scholar] [CrossRef]
- Wagner, S.A.; Beli, P.; Weinert, B.T.; Nielsen, M.L.; Cox, J.; Mann, M.; Choudhary, C. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteom. MCP 2011, 10, M111.013284. [Google Scholar] [CrossRef]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015, 43, D512–D520. [Google Scholar] [CrossRef]
- Billon, P.; Li, J.; Lambert, J.P.; Chen, Y.; Tremblay, V.; Brunzelle, J.S.; Gingras, A.C.; Verreault, A.; Sugiyama, T.; Couture, J.F.; et al. Acetylation of PCNA Sliding Surface by Eco1 Promotes Genome Stability through Homologous Recombination. Mol. Cell 2017, 65, 78–90. [Google Scholar] [CrossRef]
- Rossi, M.L.; Bambara, R.A. Reconstituted Okazaki fragment processing indicates two pathways of primer removal. J. Biol. Chem. 2006, 281, 26051–26061. [Google Scholar] [CrossRef]
- Langston, L.D.; O’Donnell, M. DNA polymerase delta is highly processive with proliferating cell nuclear antigen and undergoes collision release upon completing DNA. J. Biol. Chem. 2008, 283, 29522–29531. [Google Scholar] [CrossRef]
- Drosopoulos, W.C.; Vierra, D.A.; Kenworthy, C.A.; Coleman, R.A.; Schildkraut, C.L. Dynamic Assembly and Disassembly of the Human DNA Polymerase delta Holoenzyme on the Genome In Vivo. Cell Rep. 2020, 30, 1329–1341. [Google Scholar] [CrossRef]
- Sparks, M.A.; Singh, S.P.; Burgers, P.M.; Galletto, R. Complementary roles of Pif1 helicase and single stranded DNA binding proteins in stimulating DNA replication through G-quadruplexes. Nucleic Acids Res. 2019, 47, 8595–8605. [Google Scholar] [CrossRef]
- Sparks, M.A.; Burgers, P.M.; Galletto, R. Pif1, RPA, and FEN1 modulate the ability of DNA polymerase delta to overcome protein barriers during DNA synthesis. J. Biol. Chem. 2020, 295, 15883–15891. [Google Scholar] [CrossRef]
- Lee, S.H. The 3′-5′ exonuclease of human DNA polymerase delta (pol delta) is regulated by pol delta accessory factors and deoxyribonucleoside triphosphates. Nucleic Acids Res. 1993, 21, 1935–1939. [Google Scholar] [CrossRef]
- Lancey, C.; Tehseen, M.; Raducanu, V.S.; Rashid, F.; Merino, N.; Ragan, T.J.; Savva, C.G.; Zaher, M.S.; Shirbini, A.; Blanco, F.J.; et al. Structure of the processive human Pol delta holoenzyme. Nat. Commun. 2020, 11, 1109. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.H.; Garg, P.; Stith, C.M.; Al-Refai, H.; Sterling, J.F.; Murray, L.J.; Kunkel, T.A.; Resnick, M.A.; Burgers, P.M.; Gordenin, D.A. The multiple biological roles of the 3′-->5′ exonuclease of Saccharomyces cerevisiae DNA polymerase delta require switching between the polymerase and exonuclease domains. Mol. Cell. Biol. 2005, 25, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Hasan, S.; El-Andaloussi, N.; Hardeland, U.; Hassa, P.O.; Burki, C.; Imhof, R.; Schar, P.; Hottiger, M.O. Acetylation regulates the DNA end-trimming activity of DNA polymerase beta. Mol. Cell 2002, 10, 1213–1222. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.Y.; Tan, C.K.; So, A.G.; Downey, K.M. Purification of deoxyribonucleic acid polymerase delta from calf thymus: Partial characterization of physical properties. Biochemistry 1980, 19, 2096–2101. [Google Scholar] [CrossRef]
- Swan, M.K.; Johnson, R.E.; Prakash, L.; Prakash, S.; Aggarwal, A.K. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta. Nat. Struct. Mol. Biol. 2009, 16, 979–986. [Google Scholar] [CrossRef]
- Balakrishnan, L.; Bambara, R.A. Eukaryotic lagging strand DNA replication employs a multi-pathway mechanism that protects genome integrity. J. Biol. Chem. 2011, 286, 6865–6870. [Google Scholar] [CrossRef]
- Balakrishnan, L.; Bambara, R.A. Okazaki fragment metabolism. Cold Spring Harb. Perspect. Biol. 2013, 5, a010173. [Google Scholar] [CrossRef]
- Waga, S.; Stillman, B. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 1998, 67, 721–751. [Google Scholar] [CrossRef]
- Pavlov, Y.I.; Frahm, C.; Nick McElhinny, S.A.; Niimi, A.; Suzuki, M.; Kunkel, T.A. Evidence that errors made by DNA polymerase alpha are corrected by DNA polymerase delta. Curr. Biol. CB 2006, 16, 202–207. [Google Scholar] [CrossRef]
- Chen, L.F.; Mu, Y.; Greene, W.C. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J. 2002, 21, 6539–6548. [Google Scholar] [CrossRef]
- Fan, W.; Luo, J. SIRT1 regulates UV-induced DNA repair through deacetylating XPA. Mol. Cell 2010, 39, 247–258. [Google Scholar] [CrossRef]
- Gu, W.; Roeder, R.G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 1997, 90, 595–606. [Google Scholar] [CrossRef]
- Martinez-Balbas, M.A.; Bauer, U.M.; Nielsen, S.J.; Brehm, A.; Kouzarides, T. Regulation of E2F1 activity by acetylation. EMBO J. 2000, 19, 662–671. [Google Scholar] [CrossRef]
- Voineagu, I.; Narayanan, V.; Lobachev, K.S.; Mirkin, S.M. Replication stalling at unstable inverted repeats: Interplay between DNA hairpins and fork stabilizing proteins. Proc. Natl. Acad. Sci. USA 2008, 105, 9936–9941. [Google Scholar] [CrossRef]
- Leach, D.R. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. BioEssays News Rev. Mol. Cell. Dev. Biol. 1994, 16, 893–900. [Google Scholar] [CrossRef]
- Simone, R.; Fratta, P.; Neidle, S.; Parkinson, G.N.; Isaacs, A.M. G-quadruplexes: Emerging roles in neurodegenerative diseases and the non-coding transcriptome. FEBS Lett. 2015, 589, 1653–1668. [Google Scholar] [CrossRef]
- Huppert, J.L.; Balasubramanian, S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007, 35, 406–413. [Google Scholar] [CrossRef]
- Huppert, J.L.; Balasubramanian, S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005, 33, 2908–2916. [Google Scholar] [CrossRef]
- Zhao, J.; Bacolla, A.; Wang, G.; Vasquez, K.M. Non-B DNA structure-induced genetic instability and evolution. Cell. Mol. Life Sci. CMLS 2010, 67, 43–62. [Google Scholar] [CrossRef]
- Kamath-Loeb, A.S.; Loeb, L.A.; Johansson, E.; Burgers, P.M.; Fry, M. Interactions between the Werner syndrome helicase and DNA polymerase delta specifically facilitate copying of tetraplex and hairpin structures of the d(CGG)n trinucleotide repeat sequence. J. Biol. Chem. 2001, 276, 16439–16446. [Google Scholar] [CrossRef]
- Sfeir, A.; Kosiyatrakul, S.T.; Hockemeyer, D.; MacRae, S.L.; Karlseder, J.; Schildkraut, C.L.; de Lange, T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 2009, 138, 90–103. [Google Scholar] [CrossRef] [PubMed]



| Primer | Length | Sequence |
|---|---|---|
| Upstream (5′-3′) | ||
| U1 | 44 | GTCCACCCGACGCCACCTCCTGCCTTCAATGTGCTGGGATCCTA |
| U2 | 25 | GTCCACCCGACGCCACCTCCTGCCT |
| U3 | 25 | GTCCACCCGACGCCACCTCCTGACT |
| U4 | 25 | GTCCACCCGACGCCACCTCCCGCCT |
| U5 | 25 | GTCCACCCGACGCCACCTACTGCCT |
| Downstream (5′-3′) | ||
| D1 | 60 | AGACGAATTCCGGATACGACGGCCAGTGCCGACCGTGCCAGCCTAAATTTCAATCCACCC |
| D2 | 60 | GGGCGACTCCCGGGGGCGCCGGCCCGTGCCGGCCGTGCCGGCCTCCCTGTCAACCCACCC |
| D3 | 60 | AAACTAATTCTGAATAAGAAAAAAAGTGTTTAACTTAAAAGCCTAAATTTAAATCCAAAA |
| Template (3′-5′) | ||
| T1 | 110 | CAGGTGGGCTGCGGTGGAGGACGGAAGTTACACGACCCTAGGATGTTGGTTCTGCTTAAGGCCTATGCTGCCGGTCACGGCTGGCACGGTCGGATTTAAAGTTAGGTGGG |
| T2 | 110 | CAGGTGGGCTGCGGTGGAGGACGGAAGTTACACGACCCTAGGATGTTGGTCCCGCTGAGGGCCCCCGCGGCCGGGCACGGCCGGCACGGCCGGAGGGACAGTTGGGTGGG |
| T3 | 110 | CAGGTGGGCTGCGGTGGAGGACGGAAGTTACACGACCCTAGGATGTTGGTTTTGATTAAGACTTATTCTTTTTTTCACAAATTGAATTTTCGGATTTAAATTTAGGTTTT |
| T4 | 80 | CAGGTGGGCTGCGGTGGAGGACGGAAGTTACACGACCCTAGGATGTGGTTCTGCCGGACGAGTATTATCTCGTCCGGTAAAGT |
| T5 | 110 | CAGGTGGGCTGCGGTGGAGGACGGAAGTTACACGACCCTAGGATGTTGGTTAGGGTTAGGGTTAGGGTTAGGGGTCACGGCTGGCACGGTCGGATTTAAAGTTAGGTGGG |
| T6 | 110 | CAGGTGGGCTGCGGTGGAGGACGGAAGTTACACGACCCTAGGATGTTGGTTCTGTCCAGAGCTCCGGACGAGTATTATCTCGTCCGGAGCTCTGGATAAAGTTAGGTGGG |
| T7 | 80 | CAGGTGGGCTGCGGTGGAGGACGGAAGTTACACGACCCTAGGATGTTGGTTCTGCTTAAGGCCTATGCTGCCGGTCACGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Njeri, C.; Pepenella, S.; Battapadi, T.; Bambara, R.A.; Balakrishnan, L. DNA Polymerase Delta Exhibits Altered Catalytic Properties on Lysine Acetylation. Genes 2023, 14, 774. https://doi.org/10.3390/genes14040774
Njeri C, Pepenella S, Battapadi T, Bambara RA, Balakrishnan L. DNA Polymerase Delta Exhibits Altered Catalytic Properties on Lysine Acetylation. Genes. 2023; 14(4):774. https://doi.org/10.3390/genes14040774
Chicago/Turabian StyleNjeri, Catherine, Sharon Pepenella, Tripthi Battapadi, Robert A. Bambara, and Lata Balakrishnan. 2023. "DNA Polymerase Delta Exhibits Altered Catalytic Properties on Lysine Acetylation" Genes 14, no. 4: 774. https://doi.org/10.3390/genes14040774
APA StyleNjeri, C., Pepenella, S., Battapadi, T., Bambara, R. A., & Balakrishnan, L. (2023). DNA Polymerase Delta Exhibits Altered Catalytic Properties on Lysine Acetylation. Genes, 14(4), 774. https://doi.org/10.3390/genes14040774




