Correlation between Parental Transcriptome and Field Data for the Characterization of Heterosis in Chinese Cabbage
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Heterosis Statistical Analysis
2.3. RNA Extraction, Library Construction and RNA-Seq
2.4. Differentially Expressed Genes Analysis
2.5. Transcriptome-Based Distance Analysis
2.6. Identifying Genes Correlated to PGW and MPH
2.7. GO and KEGG Enrichment Analysis
3. Results
3.1. Statistical Analysis of 10 Traits in Hybrids and Parents of Chinese Cabbage
3.2. Heterosis of 10 Traits in Hybrids of Chinese Cabbage
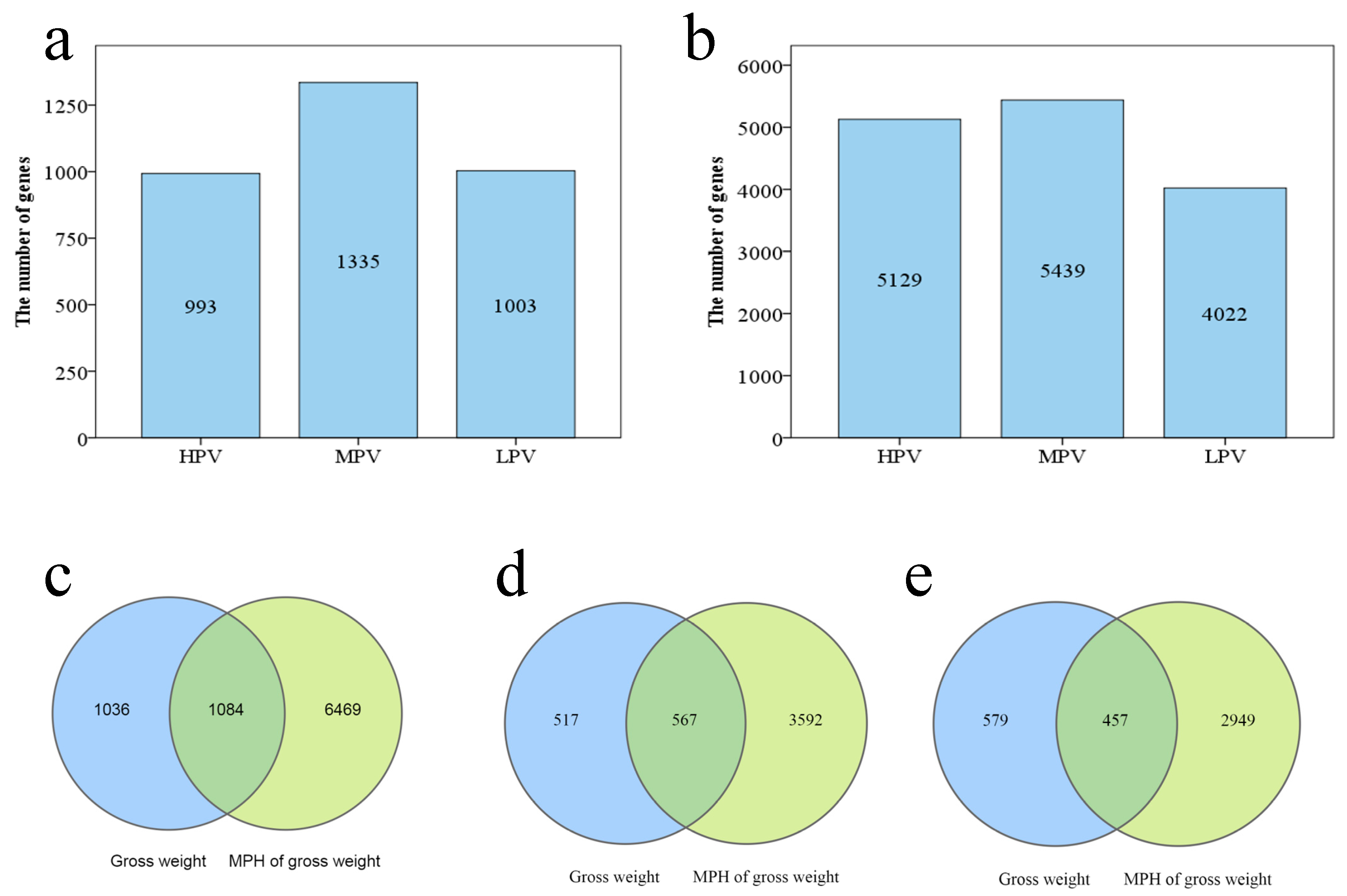
3.3. Correlation between the Parental DEGs Number and Hybrid Heterosis
3.4. Correlation between the Transcriptome-Based Distances and Heterosis with Traits
3.5. Genes Correlated to PGW and MPH
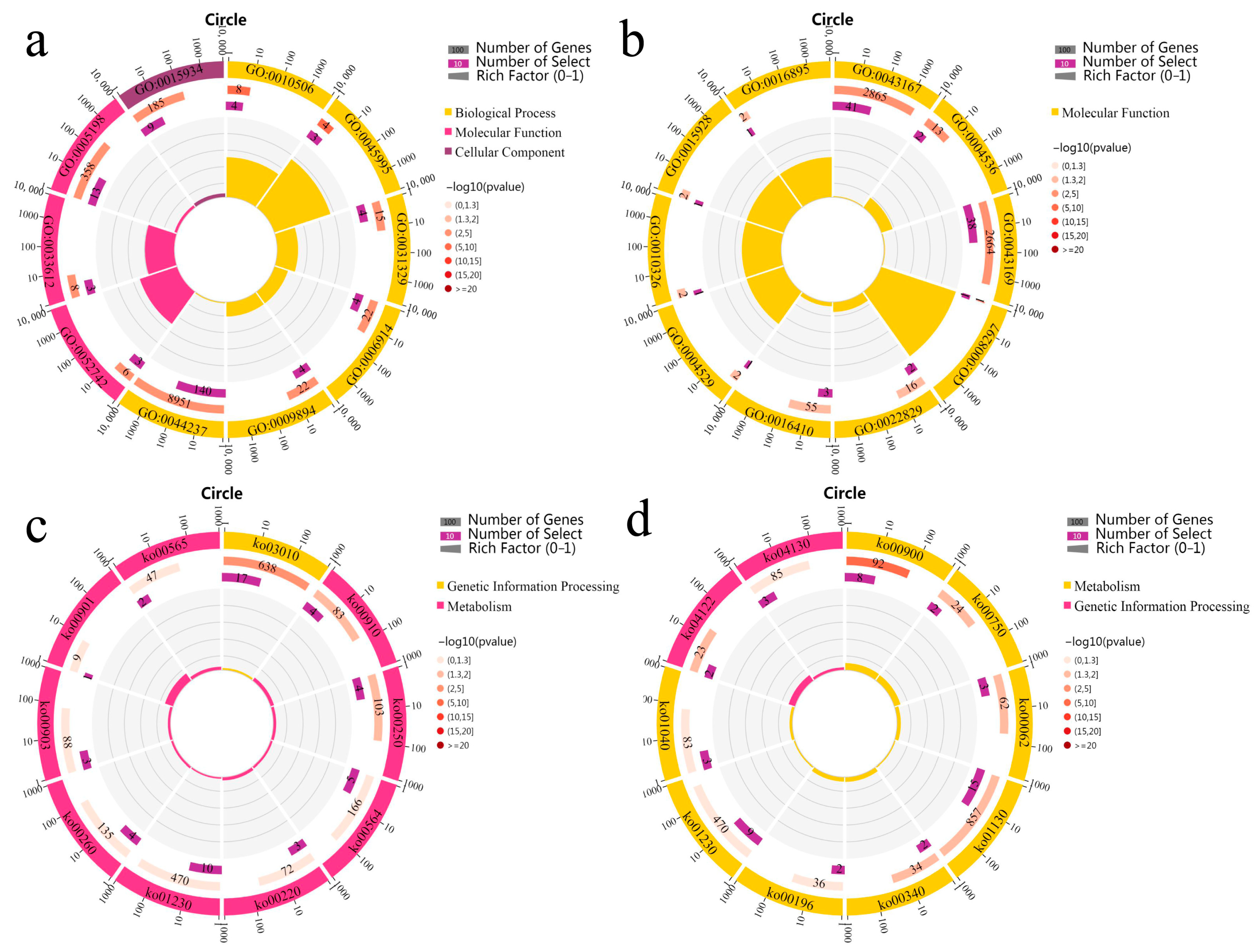
3.6. Enrichment Analysis of Genes Related to Heterosis
3.7. Metabolic Pathway Related to Heterosis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, X.; Liu, Y.; Zhang, Y.; Gu, R. Advances in Research on the Mechanism of Heterosis in Plants. Front. Plant Sci. 2021, 12, 745726. [Google Scholar] [CrossRef] [PubMed]
- Birchler, J.A.; Yao, H.; Chudalayandi, S.; Vaiman, D.; Veitia, R.A. Heterosis. Plant Cell 2010, 22, 2105–2112. [Google Scholar] [CrossRef]
- Rehman, A.U.; Dang, T.; Qamar, S.; Ilyas, A.; Fatema, R.; Kafle, M.; Hussain, Z.; Masood, S.; Iqbal, S.; Shahzad, K. Revisiting Plant Heterosis-From Field Scale to Molecules. Genes 2021, 12, 1688. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Gu, X.; Zhang, S.; Dong, S.; Miao, H.; Gebretsadik, K.; Bo, K. Molecular basis of heterosis and related breeding strategies reveal its importance in vegetable breeding. Hortic. Res. 2021, 8, 120. [Google Scholar] [CrossRef] [PubMed]
- Lippman, Z.B.; Zamir, D. Heterosis: Revisiting the magic. Trends Genet. 2007, 23, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Bruce, A.B. The mendelian theory of heredity and the augmentation of vigor. Science 1910, 32, 627–628. [Google Scholar] [CrossRef]
- Crow, J.F. 90 years ago: The beginning of hybrid maize. Genetics 1998, 148, 923–928. [Google Scholar] [CrossRef]
- East, E.M. Heterosis. Genetics 1936, 21, 375–397. [Google Scholar] [CrossRef]
- Shull, G.H. What Is “Heterosis”? Genetics 1948, 33, 439–446. [Google Scholar] [CrossRef]
- Richey, F.D. Mock-dominance and hybrid vigor. Science 1942, 96, 280–281. [Google Scholar] [CrossRef]
- Williams, W. Heterosis and the genetics of complex characters. Nature 1959, 184, 527–530. [Google Scholar] [CrossRef] [PubMed]
- Longin, C.F.; Mühleisen, J.; Maurer, H.P.; Zhang, H.; Gowda, M.; Reif, J.C. Hybrid breeding in autogamous cereals. Theor. Appl. Genet. 2012, 125, 1087–1096. [Google Scholar] [CrossRef]
- Gupta, P.K.; Balyan, H.S.; Gahlaut, V.; Saripalli, G.; Pal, B.; Basnet, B.R.; Joshi, A.K. Hybrid wheat: Past, present and future. Theor. Appl. Genet. 2019, 132, 2463–2483. [Google Scholar] [CrossRef] [PubMed]
- Dimitrijevic, A.; Horn, R. Sunflower Hybrid Breeding: From Markers to Genomic Selection. Front. Plant Sci. 2018, 8, 2238. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, Z.; Zhou, R.; Wang, Q.; Wang, L. Ratooning Annual Cotton (Gossypium spp.) for Perennial Utilization of Heterosis. Front. Plant Sci. 2020, 11, 554970. [Google Scholar] [CrossRef]
- Dunwell, J.M. Haploids in flowering plants: Origins and exploitation. Plant Biotechnol. J. 2010, 8, 377–424. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, R.; Uezono, K.; Ishikura, S.; Osabe, K.; Peacock, W.J.; Dennis, E.S. Recent research on the mechanism of heterosis is important for crop and vegetable breeding systems. Breed Sci. 2018, 68, 145–158. [Google Scholar] [CrossRef]
- Singh, S.; Dey, S.S.; Bhatia, R.; Kumar, R.; Behera, T.K. Current understanding of male sterility systems in vegetable Brassicas and their exploitation in hybrid breeding. Plant Reprod. 2019, 32, 231–256. [Google Scholar] [CrossRef]
- Schnable, P.S.; Springer, N.M. Progress toward understanding heterosis in crop plants. Annu. Rev. Plant Biol. 2013, 64, 71–88. [Google Scholar] [CrossRef]
- Cheng, S.H.; Zhuang, J.Y.; Fan, Y.Y.; Du, J.H.; Cao, L.Y. Progress in research and development on hybrid rice: A super-domesticate in China. Ann. Bot. 2007, 100, 959–966. [Google Scholar] [CrossRef]
- Knoch, D.; Werner, C.R.; Meyer, R.C.; Riewe, D.; Abbadi, A.; Lücke, S.; Snowdon, R.J.; Altmann, T. Multi-omics-based prediction of hybrid performance in canola. Theor. Appl. Genet. 2021, 134, 1147–1165. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Falke, K.C.; Thiemann, A.; Schrag, T.A.; Melchinger, A.E.; Scholten, S.; Frisch, M. Partial least squares regression, support vector machine regression, and transcriptome-based distances for prediction of maize hybrid performance with gene expression data. Theor. Appl. Genet. 2012, 124, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.; Thiemann, A.; Fu, J.; Schrag, T.A.; Scholten, S.; Melchinger, A.E. Transcriptome-based distance measures for grouping of germplasm and prediction of hybrid performance in maize. Theor. Appl. Genet. 2010, 120, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Thiemann, A.; Fu, J.; Schrag, T.A.; Melchinger, A.E.; Frisch, M.; Scholten, S. Correlation between parental transcriptome and field data for the characterization of heterosis in Zea mays L. Theor. Appl. Genet. 2010, 120, 401–413. [Google Scholar] [CrossRef]
- Pandey, S.K.; Dasgupta, T.; Rathore, A.; Vemula, A. Relationship of Parental Genetic Distance with Heterosis and Specific Combining Ability in Sesame (Sesamum indicum L.) Based on Phenotypic and Molecular Marker Analysis. Biochem. Genet. 2018, 56, 188–209. [Google Scholar] [CrossRef] [PubMed]
- Moll, R.H.; Lonnquist, J.H.; Fortuno, J.V.; Johnson, E.C. The Relationship of Heterosis and Genetic Divergence in Maize. Genetics 1965, 52, 139–144. [Google Scholar] [CrossRef]
- Hung, H.Y.; Browne, C.; Guill, K.; Coles, N.; Eller, M.; Garcia, A.; Lepak, N.; Melia-Hancock, S.; Oropeza-Rosas, M.; Salvo, S.; et al. The relationship between parental genetic or phenotypic divergence and progeny variation in the maize nested association mapping population. Heredity 2012, 108, 490–499. [Google Scholar] [CrossRef]
- Wei, X.; Zhang, J. The optimal mating distance resulting from heterosis and genetic incompatibility. Sci. Adv. 2018, 4, eaau5518. [Google Scholar] [CrossRef]
- Wu, J.W.; Hu, C.Y.; Shahid, M.Q.; Guo, H.B.; Zeng, Y.X.; Liu, X.D.; Lu, Y.G. Analysis on genetic diversification and heterosis in autotetraploid rice. Springerplus 2013, 2, 439. [Google Scholar] [CrossRef]
- Geng, X.; Qu, Y.; Jia, Y.; He, S.; Pan, Z.; Wang, L.; Du, X. Assessment of heterosis based on parental genetic distance estimated with SSR and SNP markers in upland cotton (Gossypium hirsutum L.). BMC Genom. 2021, 22, 123. [Google Scholar] [CrossRef]
- Singh, S.; Gupta, S.K. Formation of heterotic pools and understanding relationship between molecular divergence and heterosis in pearl millet [Pennisetum glaucum (L.) R.Br.]. PLoS ONE 2019, 14, e0207463. [Google Scholar] [CrossRef] [PubMed]
- Oakley, C.G.; Lundemo, S.; Agren, J.; Schemske, D.W. Heterosis is common and inbreeding depression absent in natural populations of Arabidopsis thaliana. J. Evol. Biol. 2019, 32, 592–603. [Google Scholar] [CrossRef]
- Stelkens, R.; Seehausen, O. Genetic distance between species predicts novel trait expression in their hybrids. Evolution 2009, 63, 884–897. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.S.; Yu, X.L.; Ye, W.Z.; Lu, G.; Xiang, X. Functional analysis of a novel male fertility CYP86MF gene in Chinese cabbage (Brassica campestris L. ssp. chinensis makino). Plant Cell. Rep. 2006, 24, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Ngo, V.D.; Jang, B.E.; Park, S.U.; Kim, S.J.; Kim, Y.J.; Chung, S.O. Estimation of functional components of Chinese cabbage leaves grown in a plant factory using diffuse reflectance spectroscopy. J. Sci. Food Agric. 2019, 99, 711–718. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Zhang, Z.; Zhang, L. Anthocyanin Accumulation, Antioxidant Ability and Stability, and a Transcriptional Analysis of Anthocyanin Biosynthesis in Purple Heading Chinese Cabbage (Brassica rapa L. ssp. pekinensis). J. Agric. Food Chem. 2016, 64, 132–145. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Tian, M.; Nie, S.; Zhang, L. Changes in Alternative Splicing Revealed Special Metabolic Pathways Related to Heterosis of Heading Chinese Cabbage. Horticulturae 2023, 9, 17. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef]
- Guo, M.; Rupe, M.A.; Yang, X.; Crasta, O.; Zinselmeier, C.; Smith, O.S.; Bowen, B. Genome-wide transcript analysis of maize hybrids: Allelic additive gene expression and yield heterosis. Theor. Appl. Genet. 2006, 113, 831–845. [Google Scholar] [CrossRef]
- Stokes, D.; Fraser, F.; Morgan, C.; O’Neill, C.M.; Dreos, R.; Magusin, A.; Szalma, S.; Bancroft, I. An association transcriptomics approach to the prediction of hybridperformance. Mol. Breed. 2010, 26, 91–106. [Google Scholar]
- Lecompte, O.; Ripp, R.; Thierry, J.C.; Moras, D.; Poch, O. Comparative analysis of ribosomal proteins in complete genomes: An example of reductive evolution at the domain scale. Nucleic Acids Res. 2002, 30, 5382–5390. [Google Scholar] [PubMed]
- Degenhardt, R.F.; Bonham-Smith, P.C. Arabidopsis ribosomal proteins RPL23aA and RPL23aB are differentially targeted to the nucleolus and are disparately required for normal development. Plant Physiol. 2008, 147, 128–142. [Google Scholar] [PubMed]

| Male Parent | A | B | C | D | E | F | G | H | |
|---|---|---|---|---|---|---|---|---|---|
| Female Parent | |||||||||
| A | AB | / | AD | AE | AF | AG | AH | ||
| B | BA | BC | BD | BE | BF | BG | BH | ||
| C | CA | CB | CD | CE | CF | CG | CH | ||
| D | DA | DB | DC | DE | DF | DG | DH | ||
| E | EA | EB | EC | ED | EF | EG | EH | ||
| F | FA | FB | FC | FD | FE | / | FH | ||
| G | / | GB | GC | GD | GE | GF | GH | ||
| H | HA | HB | HC | HD | HE | HF | HG | ||
| Trait | Parents | Hybrids | ||||
|---|---|---|---|---|---|---|
| Mean Value | Range of Variation | Variance Coefficient | Mean Value | Range of Variation | Variance Coefficient | |
| PGW | 2.13 ± 0.96 | 0.53–3.53 | 44.95 | 3.75 ± 0.99 | 1.40–6.50 | 26.33 |
| NOL | 8.63 ± 2.50 | 5.00–11.67 | 29.03 | 9.72 ± 2.05 | 6.33–17.00 | 21.13 |
| LOL | 38.36 ± 8.95 | 23.50–51.33 | 23.33 | 44.47 ± 5.59 | 32.67–56.00 | 12.57 |
| WOL | 24.40 ± 5.91 | 15.63–34.00 | 24.21 | 30.37 ± 3.76 | 22.00–37.50 | 12.39 |
| LHH | 27.25 ± 6.58 | 19.33–37.50 | 24.16 | 29.62 ± 5.13 | 19.00–42.14 | 17.32 |
| LHW | 17.76 ± 2.31 | 13.40–21.50 | 13.02 | 21.19 ± 2.29 | 13.00–26.50 | 10.83 |
| HW | 1.39 ± 0.65 | 0.46–2.49 | 46.59 | 2.57 ± 2.29 | 0.52–4.22 | 26.34 |
| LNH | 34.92 ± 4.82 | 27.67–41.33 | 13.81 | 39.14 ± 5.89 | 23.00–51.33 | 15.06 |
| PW | 54.63 ± 12.06 | 34.00–66.33 | 22.08 | 65.91 ± 8.32 | 45.50–93.67 | 12.62 |
| PH | 37.08 ± 6.91 | 29.33–49.00 | 18.64 | 39.25 ± 5.46 | 28.00–52.33 | 13.91 |
| Traits | The Number of Genes | ||
|---|---|---|---|
| UP | DOWN | ALL | |
| PGW | 0.091 | 0.054 | 0.096 |
| NOL | −0.152 | −0.134 | −0.189 |
| LOL | 0.018 | −0.076 | −0.037 |
| WOL | 0.051 | −0.109 | −0.035 |
| LHH | 0.047 | 0.001 | 0.032 |
| LHW | −0.121 | −0.117 | −0.157 |
| HW | 0.133 | 0.079 | 0.141 |
| LNH | 0.508 ** | −0.074 | 0.298 * |
| PW | 0 | −0.014 | −0.009 |
| PH | 0.155 | −0.048 | 0.075 |
| Traits | The Number of Genes | ||
|---|---|---|---|
| UP | DOWN | ALL | |
| PGW | 0.340 * | 0.104 | 0.298 * |
| NOL | 0.028 | 0.013 | 0.027 |
| LOL | 0.437 ** | 0.252 | 0.459 ** |
| WOL | 0.298 * | −0.017 | 0.192 |
| LHH | 0.350 * | 0.254 | 0.401 ** |
| LHW | 0.327 * | 0.234 | 0.372 ** |
| HW | 0.354 ** | 0.109 | 0.311 * |
| LNH | 0.556 ** | 0.002 | 0.379 ** |
| PW | 0.255 | 0.132 | 0.259 |
| PH | 0.437 ** | 0.153 | 0.395 ** |
| Binary | Euclidean | |
|---|---|---|
| PGW | 0.109 | 0.347 * |
| NOL | −0.191 | 0.082 |
| LOL | −0.029 | 0.245 |
| WOL | −0.025 | 0.207 |
| LHH | 0.041 | 0.449 ** |
| LHW | −0.147 | 0.15 |
| HW | 0.154 | 0.398 ** |
| LNH | 0.298 * | 0.337 * |
| PW | 0.001 | 0.189 |
| PH | 0.081 | 0.301 * |
| Binary | Euclidean | |
|---|---|---|
| PGW | 0.301 * | 0.384 ** |
| NOL | 0.02 | 0.095 |
| LOL | 0.461 ** | 0.559 ** |
| WOL | 0.196 | 0.162 |
| LHH | 0.404 ** | 0.566 ** |
| LHW | 0.378 ** | 0.393 ** |
| HW | 0.316 * | 0.394 ** |
| LNH | 0.379 ** | 0.263 |
| PW | 0.258 | 0.352 ** |
| PH | 0.399 ** | 0.401 ** |
| GeneID | Symbol | PGW | MPH of PGW | ||
|---|---|---|---|---|---|
| cor | Q Value | cor | Q Value | ||
| BraA01g015640.3C | RPL7AB | 0.3184 | 0.0202 | 0.351 | 0.01 |
| BraA02g038190.3C | RPP2C | 0.3917 | 0.0037 | 0.378 | 0.0053 |
| BraA03g010340.3C | RPL10AC | 0.4304 | 0.0013 | 0.4291 | 0.0013 |
| BraA03g020910.3C | RPL23A | 0.318 | 0.0203 | 0.7465 | 1.39 × 10−10 |
| BraA03g047490.3C | RPL15A | 0.3514 | 0.0099 | 0.3096 | 0.0241 |
| BraA04g006490.3C | RPL24B | 0.3209 | 0.0191 | 0.4076 | 0.0025 |
| BraA04g014040.3C | RPS10B | 0.3328 | 0.0149 | 0.4193 | 0.0018 |
| BraA05g028570.3C | RPL30B | 0.366 | 0.007 | 0.4511 | 0.0007 |
| BraA06g029850.3C | RPL6 | 0.4174 | 0.0019 | 0.4623 | 0.0005 |
| BraA06g032890.3C | 0.3173 | 0.0206 | 0.4261 | 0.0015 | |
| BraA07g012190.3C | RPL17B | 0.3825 | 0.0047 | 0.3783 | 0.0052 |
| BraA07g018220.3C | RPL10AB | 0.4172 | 0.0019 | 0.5375 | 3.32 × 10−5 |
| BraA07g022360.3C | RPS26B | 0.3703 | 0.0063 | 0.4319 | 0.0012 |
| BraA07g031310.3C | RPL17B | 0.3695 | 0.0065 | 0.4108 | 0.0022 |
| BraA08g004460.3C | RPL18 | 0.3652 | 0.0072 | 0.5671 | 9.55 × 10−6 |
| BraA09g019250.3C | ARP1 | 0.2793 | 0.0428 | 0.357 | 0.0087 |
| BraA09g022110.3C | RPL32A | 0.3953 | 0.0034 | 0.4913 | 0.0002 |
| GeneID | Symbol | PGW | MPH of PGW | ||
|---|---|---|---|---|---|
| cor | Q Value | cor | Q Value | ||
| BraA01g044250.3C | IPP2 | −0.4225 | 0.0016 | −0.3671 | 0.0069 |
| BraA02g023510.3C | HMG1 | −0.3538 | 0.0094 | −0.3694 | 0.0065 |
| BraA02g028120.3C | HMGS | −0.3386 | 0.0131 | −0.5465 | 2.30× 10−5 |
| BraA03g011760.3C | −0.353 | 0.0095 | −0.3878 | 0.0041 | |
| BraA06g027360.3C | FLCY | −0.3564 | 0.0088 | −0.5517 | 1.85 × 10−5 |
| BraA07g002120.3C | −0.3555 | 0.009 | −0.4595 | 0.0005 | |
| BraA08g025620.3C | ICMEL1 | −0.391 | 0.0038 | −0.5731 | 7.30 × 10−6 |
| BraA09g014810.3C | ISPF | −0.3647 | 0.0073 | −0.3582 | 0.0084 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, R.; Tian, M.; He, Q.; Zhang, L. Correlation between Parental Transcriptome and Field Data for the Characterization of Heterosis in Chinese Cabbage. Genes 2023, 14, 776. https://doi.org/10.3390/genes14040776
Li R, Tian M, He Q, Zhang L. Correlation between Parental Transcriptome and Field Data for the Characterization of Heterosis in Chinese Cabbage. Genes. 2023; 14(4):776. https://doi.org/10.3390/genes14040776
Chicago/Turabian StyleLi, Ru, Min Tian, Qiong He, and Lugang Zhang. 2023. "Correlation between Parental Transcriptome and Field Data for the Characterization of Heterosis in Chinese Cabbage" Genes 14, no. 4: 776. https://doi.org/10.3390/genes14040776
APA StyleLi, R., Tian, M., He, Q., & Zhang, L. (2023). Correlation between Parental Transcriptome and Field Data for the Characterization of Heterosis in Chinese Cabbage. Genes, 14(4), 776. https://doi.org/10.3390/genes14040776






