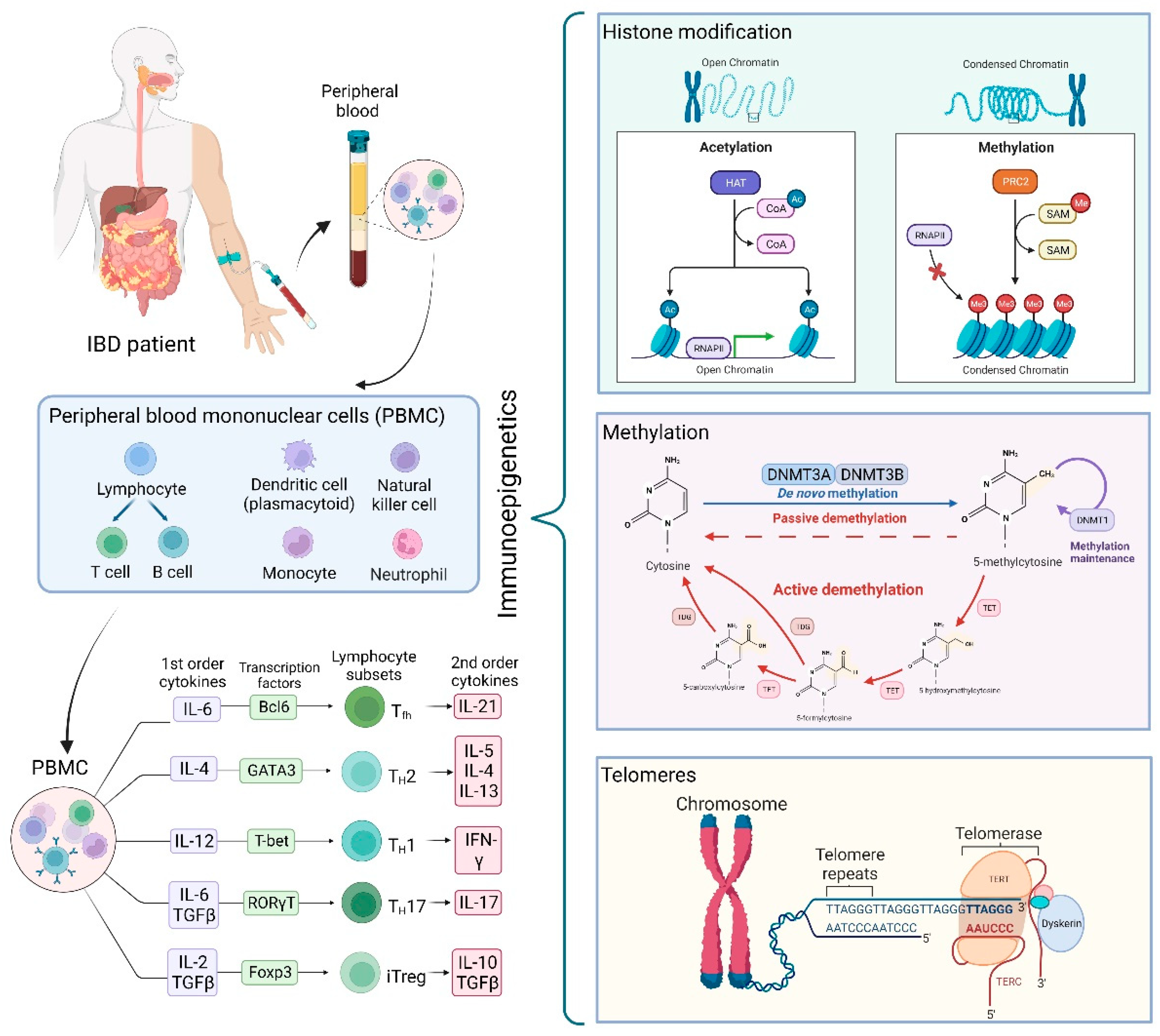
Immunoepigenetic Regulation of Inflammatory Bowel Disease: Current Insights into Novel Epigenetic Modulations of the Systemic Immune Response
Abstract
1. Introduction
1.1. Genetic Analysis
1.2. Epigenetics
2. Histone Modifications and Chromatin Organizers as Influencers of the Impaired Immune System in IBD
2.1. Histone Modification and Immune Mediators of Inflammation in IBD
2.2. Histone Modification and Immune Cells in IBD
3. Methylation as a Mechanism for Imprinting Altered Key Elements in IBD
Impaired Methylation Results in the Alteration of Immune Factors
4. Telomeres in the Context of the Immune System and IBD
5. Other Aspects to Consider
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gonzalez-Jaramillo, V.; Portilla-Fernandez, E.; Glisic, M.; Voortman, T.; Ghanbari, M.; Bramer, W.; Chowdhury, R.; Nijsten, T.; Dehghan, A.; Franco, O.H.; et al. Epigenetics and inflammatory markers: A systematic review of the current evidence. Int. J. Inflam. 2019, 8, 6273680. [Google Scholar] [CrossRef] [PubMed]
- Scarpa, M.; Stylianou, E. Epigenetics: Concepts and relevance to IBD pathogenesis. Inflamm. Bowel Dis. 2012, 18, 1982–1996. [Google Scholar] [CrossRef] [PubMed]
- Kuenzig, M.E.; Fung, S.G.; Marderfeld, L.; Mak, J.W.Y.; Kaplan, G.G.; Ng, S.C.; Wilson, D.C.; Cameron, F.; Henderson, P.; Kotze, P.G.; et al. InsightScope pediatric IBD epidemiology group. Twenty-first century trends in the global epidemiology of pediatric-onset inflammatory bowel disease: Systematic review. Gastroenterology 2022, 162, 1147–1159.e4. [Google Scholar] [CrossRef]
- Zhao, M.; Feng, R.; Ben-Horin, S.; Zhuang, X.; Tian, Z.; Li, X.; Ma, R.; Mao, R.; Qiu, Y.; Chen, M. Systematic review with meta-analysis: Environmental and dietary differences of inflammatory bowel disease in Eastern and Western populations. Aliment. Pharmacol. Ther. 2022, 55, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Meddens, C.A.; van der List, A.C.J.; Nieuwenhuis, E.E.S.; Mokry, M. Non-coding DNA in IBD: From sequence variation in DNA regulatory elements to novel therapeutic potential. Gut 2019, 68, 928–941. [Google Scholar] [CrossRef]
- Moret-Tatay, I.; Cerrillo, E.; Hervas, D.; Iborra, M.; Saez-González, E.; Forment, J.; Tortosa, L.; Nos, P.; Gadea, J.; Beltran, B. Specific plasma microRNA signatures in predicting and confirming Crohn’s disease recurrence: Role and pathogenic implications. Clin. Transl. Gastroenterol. 2021, 12, e00416. [Google Scholar] [CrossRef]
- Stylianou, E. Recent advances in the etiopathogenesis of inflammatory bowel disease: The role of omics. Mol. Diagn. Ther. 2018, 22, 11–23. [Google Scholar] [CrossRef]
- Blumberg, R.S. Environment and genes: What is the interaction? Dig. Dis. 2016, 34, 20–26. [Google Scholar] [CrossRef]
- Fiocchi, C.; Iliopoulos, D. What’s new in IBD therapy: An “omics network” approach. Pharmacol. Res. 2020, 159, 104886. [Google Scholar] [CrossRef]
- Ray, G.; Longworth, M.S. Epigenetics, DNA organization, and inflammatory bowel disease. Inflamm. Bowel Dis. 2019, 25, 235–247. [Google Scholar] [CrossRef]
- Seyed Tabib, N.S.; Madgwick, M.; Sudhakar, P.; Verstockt, B.; Korcsmaros, T.; Vermeire, S. Big data in IBD: Big progress for clinical practice. Gut 2020, 69, 1520–1532. [Google Scholar] [CrossRef] [PubMed]
- Cleynen, I.; Boucher, G.; Jostins, L.; Schumm, L.P.; Zeissig, S.; Ahmad, T.; Andersen, V.; Andrews, J.M.; Annese, V.; Brand, S.; et al. Inherited determinants of Crohn’s disease and ulcerative colitis phenotypes: A genetic association study. Lancet 2016, 387, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Cardinale, C.J.; March, M.E.; Lin, X.; Liu, Y.; Spruce, L.A.; Bradfield, J.P.; Wei, Z.; Seeholzer, S.H.; Grant, S.F.A.; Hakonarson, H. Regulation of Janus kinase 2 by an inflammatory bowel disease causal non-coding single nucleotide polymorphism. J. Crohns Colitis 2020, 14, 646–653. [Google Scholar] [CrossRef] [PubMed]
- Venkateswaran, S.; Denson, L.A.; Jurickova, I.; Dodd, A.; Zwick, M.E.; Cutler, D.J.; Kugathasan, S.; Okou, D.T. Neutrophil GM-CSF signaling in inflammatory bowel disease patients is influenced by non-coding genetic variants. Sci. Rep. 2019, 9, 9168. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Li, X.; Liu, S.; Zhang, Y.; Zhang, D. Toll-like receptors and inflammatory bowel disease. Front. Immunol. 2018, 9, 72. [Google Scholar] [CrossRef] [PubMed]
- Weissman, S.M. Personalized medicine: A new horizon for medical therapy. Precis. Clin. Med. 2018, 1, 1–2. [Google Scholar] [CrossRef]
- Natasha, G.; Zilbauer, M. Epigenetics in IBD: A conceptual framework for disease pathogenesis. Frontline Gastroenterol. 2022, 13, e22–e27. [Google Scholar]
- Zeng, Z.; Mukherjee, A.; Zhang, H. From genetics to epigenetics, roles of epigenetics in inflammatory bowel disease. Front. Genet. 2019, 10, 1017. [Google Scholar] [CrossRef]
- Moret-Tatay, I.; Iborra, M.; Cerrillo, E.; Tortosa, L.; Nos, P.; Beltran, B. Possible biomarkers in blood for Crohn’s disease: Oxidative stress and microRNAs-current evidences and further aspects to unravel. Oxid. Med. Cell. Longev. 2016, 2016, 2325162. [Google Scholar] [CrossRef]
- Alemany-Cosme, E.; Saez-Gonzalez, E.; Moret, I.; Mateos, B.; Iborra, M.; Nos, P.; Sandoval, J.; Beltran, B. Oxidative stress in the pathogenesis of Crohn’s disease and the interconnection with immunological response, microbiota, external environmental factors, and epigenetics. Antioxidants 2021, 10, 64. [Google Scholar] [CrossRef]
- Rath, S.; Hawsawi, Y.M.; Alzahrani, F.; Khan, M.I. Epigenetic regulation of inflammation: The metabolomics connection. Semin. Cell. Dev. Biol. 2022, 17, 00270–00271. [Google Scholar] [CrossRef] [PubMed]
- Castelo-Branco, P.; Jeronimo, C. Present and future perspectives for targeting histone modifications in therapy. In Histone Modifications in Therapy; Academic Press: Cambridge, MA, USA, 2020; pp. 415–457. [Google Scholar]
- Renaude, E.; Kroemer, M.; Loyon, R.; Binda, D.; Borg, C.; Guittaut, M.; Hervouet, E.; Peixoto, P. The fate of Th17 cells is shaped by epigenetic modifications and remodeled by the tumor microenvironment. Int. J. Mol. Sci. 2020, 21, 1673. [Google Scholar] [CrossRef]
- Xu, J.; Xu, H.M.; Yang, M.F.; Liang, Y.J.; Peng, Q.Z.; Zhang, Y.; Tian, C.M.; Wang, L.S.; Yao, J.; Nie, Y.Q.; et al. New insights into the epigenetic regulation of inflammatory bowel disease. Front. Pharmacol. 2022, 13, 813659. [Google Scholar] [CrossRef] [PubMed]
- Geremia, A.; Biancheri, P.; Allan, P.; Corazza, G.R.; Di Sabatino, A. Innate and adaptive immunity in inflammatory bowel disease. Autoimmun. Rev. 2014, 13, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Cao, X. Epigenetic regulation of the innate immune response to infection. Nat. Rev. Immunol. 2019, 19, 417–432. [Google Scholar] [CrossRef] [PubMed]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef] [PubMed]
- Hugot, J.P.; Chamaillard, M.; Zouali, H.; Lesage, S.; Cézard, J.P.; Belaiche, J.; Almer, S.; Tysk, C.; O’Morain, C.A.; Gassull, M.; et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 2001, 411, 599–603. [Google Scholar] [CrossRef]
- Ueno, A.; Jeffery, L.; Kobayashi, T.; Hibi, T.; Ghosh, S.; Jijon, H. Th17 plasticity and its relevance to inflammatory bowel disease. J. Autoimmun. 2018, 87, 38–49. [Google Scholar] [CrossRef]
- Kole, A.; Maloy, K.J. Control of intestinal inflammation by interleukin-10. Curr. Top Microbiol. Immunol. 2014, 380, 19–38. [Google Scholar]
- Bai, A.H.; Wu, W.K.; Xu, L.; Wong, S.H.; Go, M.Y.; Chan, A.W.; Harbord, M.; Zhang, S.; Chen, M.; Wu, J.C.; et al. Dysregulated lysine acetyltransferase 2B promotes inflammatory bowel disease pathogenesis through transcriptional repression of interleukin-10. J. Crohns Colitis 2016, 6, 726–734. [Google Scholar] [CrossRef]
- Zheng, Z.; Huang, G.; Gao, T.; Huang, T.; Zou, M.; Zou, Y.; Duan, S. Epigenetic changes associated with interleukin-10. Front. Immunol. 2020, 11, 1105. [Google Scholar] [CrossRef]
- Igalouzene, R.; Hernandez-Vargas, H.; Benech, N.; Guyennon, A.; Bauché, D.; Barrachina, C.; Dubois, E.; Marie, J.C.; Soudja, S.M. SMAD4 TGF-β-independent function preconditions naive CD8+ T cells to prevent severe chronic intestinal inflammation. J. Clin. Investig. 2022, 132, e151020. [Google Scholar] [CrossRef]
- Yang, Y.; Guan, J.; Shaikh, A.S.; Liang, Y.; Sun, L.; Wang, M.; Li, D.; Qiu, C.; Li, X. Histone acetyltransferase MOF affects the progression of DSS-induced colitis. Cell Physiol. Biochem. 2018, 47, 2159–2169. [Google Scholar] [CrossRef] [PubMed]
- Baine, I.; Basu, S.; Ames, R.; Sellers, R.S.; Macian, F. Helios induces epigenetic silencing of IL2 gene expression in regulatory T cells. J. Immunol. 2013, 190, 1008–1016. [Google Scholar] [CrossRef] [PubMed]
- Edwards, A.J.; Pender, S.L. Histone deacetylase inhibitors and their potential role in inflammatory bowel diseases. Biochem. Soc. Trans. 2011, 39, 1092–1095. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.-I.; Cho, H.; Jeon, R.; Sung, M.-K. Therapeutic efficacy of novel HDAC inhibitors SPA3052 and SPA3074 against intestinal inflammation in a murine model of colitis. Pharmaceuticals 2022, 15, 1515. [Google Scholar] [CrossRef]
- Ali, M.N.; Choijookhuu, N.; Takagi, H.; Srisowanna, N.; Nguyen Nhat Huynh, M.; Yamaguchi, Y.; Synn Oo, P.; Tin Htwe Kyaw, M.; Sato, K.; Yamaguchi, R.; et al. The HDAC inhibitor, SAHA, prevents colonic inflammation by suppressing pro-inflammatory cytokines and chemokines in DSS-induced colitis. Acta Histochem. Cytochem. 2018, 51, 33–40. [Google Scholar] [CrossRef]
- Joosse, M.E.; Charbit-Henrion, F.; Boisgard, R.; Raatgeep, R.H.C.; Lindenbergh-Kortleve, D.J.; Costes, L.M.M.; Nugteren, S.; Guegan, N.; Parlato, M.; Veenbergen, S.; et al. Duplication of the IL2RA locus causes excessive IL-2 signaling and may predispose to very early onset colitis. Mucosal Immunol. 2021, 14, 1172–1182. [Google Scholar] [CrossRef]
- Yuan, C.W.; Sun, X.L.; Qiao, L.C.; Xu, H.X.; Zhu, P.; Chen, H.J.; Yang, B.L. Non-SMC condensin I complex subunit D2 and non-SMC condensin II complex subunit D3 induces inflammation via the IKK/NF-κB pathway in ulcerative colitis. World J. Gastroenterol. 2019, 25, 6813–6822. [Google Scholar] [CrossRef]
- Vieujean, S.; Caron, B.; Haghnejad, V.; Jouzeau, J.Y.; Netter, P.; Heba, A.C.; Ndiaye, N.C.; Moulin, D.; Barreto, G.; Danese, S.; et al. Impact of the exposome on the epigenome in inflammatory bowel disease patients and animal models. Int. J. Mol. Sci. 2022, 23, 7611. [Google Scholar] [CrossRef]
- López-Muñoz, P.; Beltran, B.; Saez-González, E.; Alba, A.; Nos, P.; Iborra, M. Influence of vitamin D deficiency on inflammatory markers and clinical disease activity in IBD patients. Nutrients 2019, 11, 1059. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Yang, M.F.; Liang, Y.J.; Xu, J.; Xu, H.M.; Nie, Y.Q.; Wang, L.S.; Yao, J.; Li, D.F. Immunology of inflammatory bowel disease: Molecular mechanisms and therapeutics. J. Inflamm. Res. 2022, 15, 1825–1844. [Google Scholar] [CrossRef] [PubMed]
- Däbritz, J.; Menheniott, T.R. Linking immunity, epigenetics, and cancer in inflammatory bowel disease. Inflamm. Bowel Dis. 2014, 20, 1638–1654. [Google Scholar] [CrossRef] [PubMed]
- Nakase, H.; Sato, N.; Mizuno, N.; Ikawa, Y. The influence of cytokines on the complex pathology of ulcerative colitis. Autoimmun. Rev. 2022, 21, 103017. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.W.; Fu, S.H.; Chien, M.W.; Hsu, C.Y.; Lin, M.H.; Dong, J.L.; Lu, R.J.; Lee, Y.J.; Chen, P.Y.; Wang, C.H.; et al. Blimp-1 molds the epigenetic architecture of IL-21-mediated autoimmune diseases through an autoregulatory circuit. JCI Insight 2022, 7, e151614. [Google Scholar] [CrossRef]
- Sarmento, O.F.; Svingen, P.A.; Xiong, Y.; Sun, Z.; Bamidele, A.O.; Mathison, A.J.; Smyrk, T.C.; Nair, A.A.; Gonzalez, M.M.; Sagstetter, M.R.; et al. The Role of the histone methyltransferase enhancer of zeste homolog 2 (EZH2) in the pathobiological mechanisms underlying inflammatory bowel disease (IBD). J. Biol. Chem. 2017, 292, 706–722. [Google Scholar] [CrossRef]
- Bamidele, A.O.; Svingen, P.A.; Sagstetter, M.R.; Sarmento, O.F.; Gonzalez, M.; Braga Neto, M.B.; Kugathasan, S.; Lomberk, G.; Urrutia, R.A.; Faubion, W.A., Jr. Disruption of FOXP3-EZH2 interaction represents a pathobiological mechanism in intestinal inflammation. Cell Mol. Gastroenterol. Hepatol. 2018, 7, 55–71. [Google Scholar] [CrossRef]
- Liu, Y.; Peng, J.; Sun, T.; Li, N.; Zhang, L.; Ren, J.; Yuan, H.; Kan, S.; Pan, Q.; Li, X.; et al. Epithelial EZH2 serves as an epigenetic determinant in experimental colitis by inhibiting TNFα-mediated inflammation and apoptosis. Proc. Natl. Acad. Sci. USA 2017, 114, E3796–E3805. [Google Scholar] [CrossRef]
- Huang, M.; Wang, Q.; Long, F.; Di, Y.; Wang, J.; Zhun Zhu, Y.; Liu, X. Jmjd3 regulates inflammasome activation and aggravates DSS-induced colitis in mice. FASEB J. 2020, 34, 4107–4119. [Google Scholar] [CrossRef]
- Doñas, C.; Neira, J.; Osorio-Barrios, F.; Carrasco, M.; Fernández, D.; Prado, C.; Loyola, A.; Pacheco, R.; Rosemblatt, M. The demethylase inhibitor GSK-J4 limits inflammatory colitis by promoting de novo synthesis of retinoic acid in dendritic cells. Sci. Rep. 2021, 11, 1342. [Google Scholar] [CrossRef]
- Ghiboub, M.; Koster, J.; Craggs, P.D.; Li Yim, A.Y.F.; Shillings, A.; Hutchinson, S.; Bingham, R.P.; Gatfield, K.; Hageman, I.L.; Yao, G.; et al. Modulation of macrophage inflammatory function through selective inhibition of the epigenetic reader protein SP140. BMC Biol. 2022, 20, 182. [Google Scholar] [CrossRef]
- Fraschilla, I.; Amatullah, H.; Rahman, R.U.; Jeffrey, K.L. Immune chromatin reader SP140 regulates microbiota and risk for inflammatory bowel disease. Cell Host Microbe 2022, 30, 1370–1381. [Google Scholar] [CrossRef] [PubMed]
- Camacho-Ordonez, N.; Ballestar, E.; Timmers, H.T.M.; Grimbacher, B. What can clinical immunology learn from inborn errors of epigenetic regulators? J. Allergy Clin. Immunol. 2021, 147, 1602–1618. [Google Scholar] [CrossRef] [PubMed]
- Bullwinkel, J.; Lüdemann, A.; Debarry, J.; Singh, P.B. Epigenotype switching at the CD14 and CD209 genes during differentiation of human monocytes to dendritic cells. Epigenetics 2011, 6, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Kamada, N.; Hisamatsu, T.; Okamoto, S.; Chinen, H.; Kobayashi, T.; Sato, T.; Sakuraba, A.; Kitazume, M.T.; Sugita, A.; Koganei, K.; et al. Unique CD14 intestinal macrophages contribute to the pathogenesis of Crohn disease via IL-23/IFN-γ axis. J. Clin. Investig. 2008, 118, 2269–2280. [Google Scholar]
- Moret-Tatay, I.; Cerrillo, E.; Saez-Gonzalez, E.; Hervás, D.; Iborra, M.; Sandoval, J.; Buso, E.; Tortosa, L.; Nos, P.; Beltran, B. Identification of epigenetic methylation signatures with clinical value in Crohn’s disease. Clin. Transl. Gastroenterol. 2019, 10, e00083. [Google Scholar] [CrossRef]
- Joustra, V.W.; Li Yim, A.Y.F.; de Bruyn, J.R.; Duijvestein, M.; Hageman, I.L.; de Jonge, W.J.; Henneman, P.; Wildenberg, M.; D’Haens, G. Peripheral blood dna methylation profiles do not predict endoscopic post-operative recurrence in Crohn’s disease patients. Int. J. Mol. Sci. 2022, 23, 10467. [Google Scholar] [CrossRef]
- Kalla, R.; Adams, A.T.; Nowak, J.K.; Bergemalm, D.; Vatn, S.; Ventham, N.T.; Kennedy, N.A.; Ricanek, P.; Lindstrom, J.; IBD-Character Consortium; et al. Analysis of systemic epigenetic alterations in inflammatory bowel disease: Defining geographical, genetic, and immune-inflammatory influences on the circulating methylome. J. Crohns Colitis 2022, jjac127. [Google Scholar] [CrossRef]
- Hornschuh, M.; Wirthgen, E.; Wolfien, M.; Singh, K.P.; Wolkenhauer, O.; Däbritz, J. The role of epigenetic modifications for the pathogenesis of Crohn’s disease. Clin. Epigenet. 2021, 13, 108. [Google Scholar] [CrossRef]
- Weng, N.P.; Araki, Y.; Subedi, K. The molecular basis of the memory T cell response: Differential gene expression and its epigenetic regulation. Nat. Rev. Immunol. 2012, 12, 306–315. [Google Scholar] [CrossRef]
- Magro, F.; Araujo, F.; Pereira, P.; Meireles, E.; Diniz-Ribeiro, M.; Velosom, F.T. Soluble selectins, sICAM, sVCAM, and angiogenic proteins in different activity groups of patients with inflammatory bowel disease. Dig. Dis. Sci. 2004, 49, 1265–1274. [Google Scholar] [CrossRef] [PubMed]
- Bisping, G.; Lügering, N.; Lütke-Brintrup, S.; Pauels, H.G.; Schürmann, G.; Domschke, W.; Kucharzik, T. Patients with inflammatory bowel disease (IBD) reveal increased induction capacity of intracellular interferon-gamma (IFN-γ) in peripheral CD8+ lymphocytes co-cultured with intestinal epithelial cells. Clin. Exp. Immunol. 2001, 123, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Henning, A.N.; Roychoudhuri, R.; Restifo, N.P. Epigenetic control of CD8+ T cell differentiation. Nat. Rev. Immunol. 2018, 18, 340–356. [Google Scholar] [CrossRef]
- Jones, G.R.; Brown, S.L.; Phythian-Adams, A.T.; Ivens, A.C.; Cook, P.C.; MacDonald, A.S. The methyl-CpG-binding protein Mbd2 regulates susceptibility to experimental colitis via control of CD11c+ cells and colonic epithelium. Front. Immunol. 2020, 11, 183. [Google Scholar] [CrossRef] [PubMed]
- Rosati, E.; Rios Martini, G.; Pogorelyy, M.V.; Minervina, A.A.; Degenhardt, F.; Wendorff, M.; Sari, S.; Mayr, G.; Fazio, A.; Dowds, C.M.; et al. A novel unconventional T cell population enriched in Crohn’s disease. Gut 2022, 71, 2194–2204. [Google Scholar] [CrossRef]
- McDermott, E.; Ryan, E.J.; Tosetto, M.; Gibson, D.; Burrage, J.; Keegan, D.; Byrne, K.; Crowe, E.; Sexton, G.; Malone, K.; et al. DNA methylation profiling in inflammatory bowel disease provides new insights into disease pathogenesis. J. Crohns Colitis 2016, 10, 77–86. [Google Scholar] [CrossRef]
- Vlantis, K.; Polykratis, A.; Welz, P.S.; van Loo, G.; Pasparakis, M.; Wullaert, A. TLR-independent anti-inflammatory function of intestinal epithelial TRAF6 signalling prevents DSS-induced colitis in mice. Gut 2016, 65, 935–943. [Google Scholar] [CrossRef]
- Da-Silva, N.; Arasaradnam, R.; Getliffe, K.; Sung, E.; Oo, Y.; Nwokolo, C. Altered mRNA expression of telomere binding proteins (TPP1, POT1, RAP1, TRF1 and TRF2) in ulcerative colitis and Crohn’s disease. Dig. Liver Dis. 2010, 42, 544–548. [Google Scholar] [CrossRef]
- Melicher, D.; Buzas, E.I.; Falus, A. Genetic and epigenetic trends in telomere research: A novel way in immunoepigenetics. Cell. Mol. Life Sci. 2015, 72, 4095–4109. [Google Scholar] [CrossRef]
- Chakravarti, D.; Lee, R.; Multani, A.S.; Santoni, A.; Keith, Z.; Hsu, W.H.; Chang, K.; Reyes, L.; Rashid, A.; Wu, C.J.; et al. Telomere dysfunction instigates inflammation in inflammatory bowel disease. Proc. Natl. Acad. Sci. USA 2021, 118, e2024853118. [Google Scholar] [CrossRef]
- Takagi, S.; Kinouchi, Y.; Chida, M.; Hiwatashi, N.; Noguchi, M.; Takahashi, S.; Shimosegawa, T. Strong telomerase activity of B lymphocyte from mesenteric lymph nodes of patients with inflammatory bowel disease. Dig. Dis. Sci. 2003, 48, 2091–2094. [Google Scholar] [CrossRef] [PubMed]
- Chakravarti, D.; DePinho, R.A. Telomere dysfunction as an initiator of inflammation: Clues to an age-old mystery. J. Inflamm. Bowel Dis. Disord. 2021, 6. [Google Scholar]
- Salk, J.J.; Bansal, A.; Lai, L.A.; Crispin, D.A.; Ussakli, C.H.; Horwitz, M.S.; Bronner, M.P.; Brentnall, T.A.; Loeb, L.A.; Rabinovitch, P.S.; et al. Clonal expansions and short telomeres are associated with neoplasia in early-onset, but not late-onset, ulcerative colitis. Inflamm. Bowel Dis. 2013, 19, 2593–2602. [Google Scholar] [CrossRef] [PubMed]
- Truta, B.; Wohler, E.; Sobreira, N.; Datta, L.W.; Brant, S.R. Role of telomere shortening in anticipation of inflammatory bowel disease. World J. Gastrointest. Pharmacol. Ther. 2020, 11, 69–78. [Google Scholar] [CrossRef]
- Alghoul, Z.; Yang, C.; Merlin, D. The current status of molecular biomarkers for inflammatory bowel disease. Biomedicines 2022, 10, 1492. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bastida, G.; Mínguez, A.; Nos, P.; Moret-Tatay, I. Immunoepigenetic Regulation of Inflammatory Bowel Disease: Current Insights into Novel Epigenetic Modulations of the Systemic Immune Response. Genes 2023, 14, 554. https://doi.org/10.3390/genes14030554
Bastida G, Mínguez A, Nos P, Moret-Tatay I. Immunoepigenetic Regulation of Inflammatory Bowel Disease: Current Insights into Novel Epigenetic Modulations of the Systemic Immune Response. Genes. 2023; 14(3):554. https://doi.org/10.3390/genes14030554
Chicago/Turabian StyleBastida, Guillermo, Alejandro Mínguez, Pilar Nos, and Inés Moret-Tatay. 2023. "Immunoepigenetic Regulation of Inflammatory Bowel Disease: Current Insights into Novel Epigenetic Modulations of the Systemic Immune Response" Genes 14, no. 3: 554. https://doi.org/10.3390/genes14030554
APA StyleBastida, G., Mínguez, A., Nos, P., & Moret-Tatay, I. (2023). Immunoepigenetic Regulation of Inflammatory Bowel Disease: Current Insights into Novel Epigenetic Modulations of the Systemic Immune Response. Genes, 14(3), 554. https://doi.org/10.3390/genes14030554




