Loss of calpain3b in Zebrafish, a Model of Limb-Girdle Muscular Dystrophy, Increases Susceptibility to Muscle Defects Due to Elevated Muscle Activity
Abstract
1. Introduction
2. Materials and Methods
2.1. Zebrafish Husbandry
2.2. In Vitro Transcription for mRNA Expression
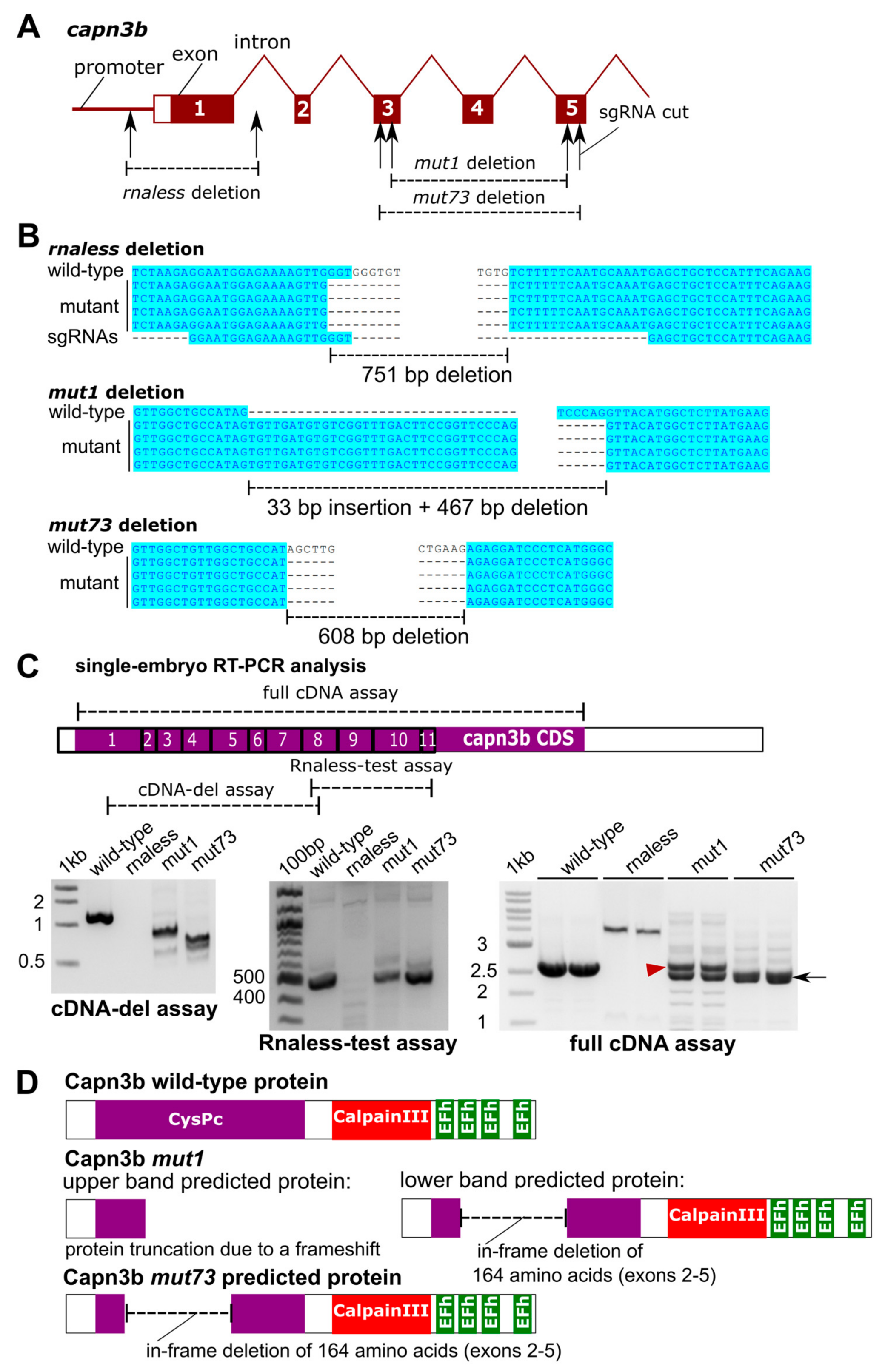
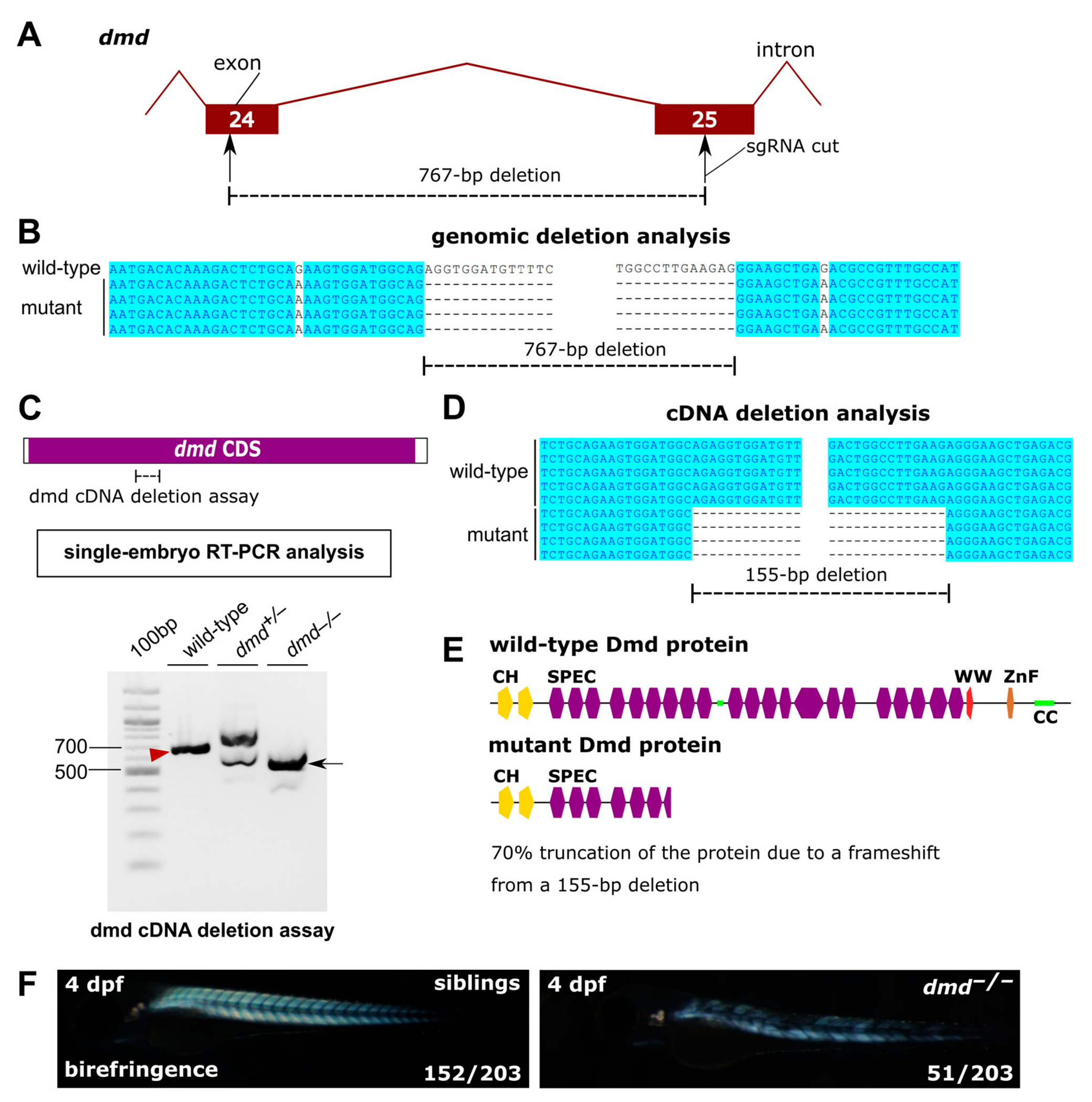
2.3. Generation of capn3b and dmd Mutants
2.4. RNA Extraction and cDNA Synthesis
2.5. Reverse Transcription-Polymerase Chain Reaction (RT-PCR) Analysis
2.6. Sanger Sequencing and Analysis
2.7. In Situ Hybridization
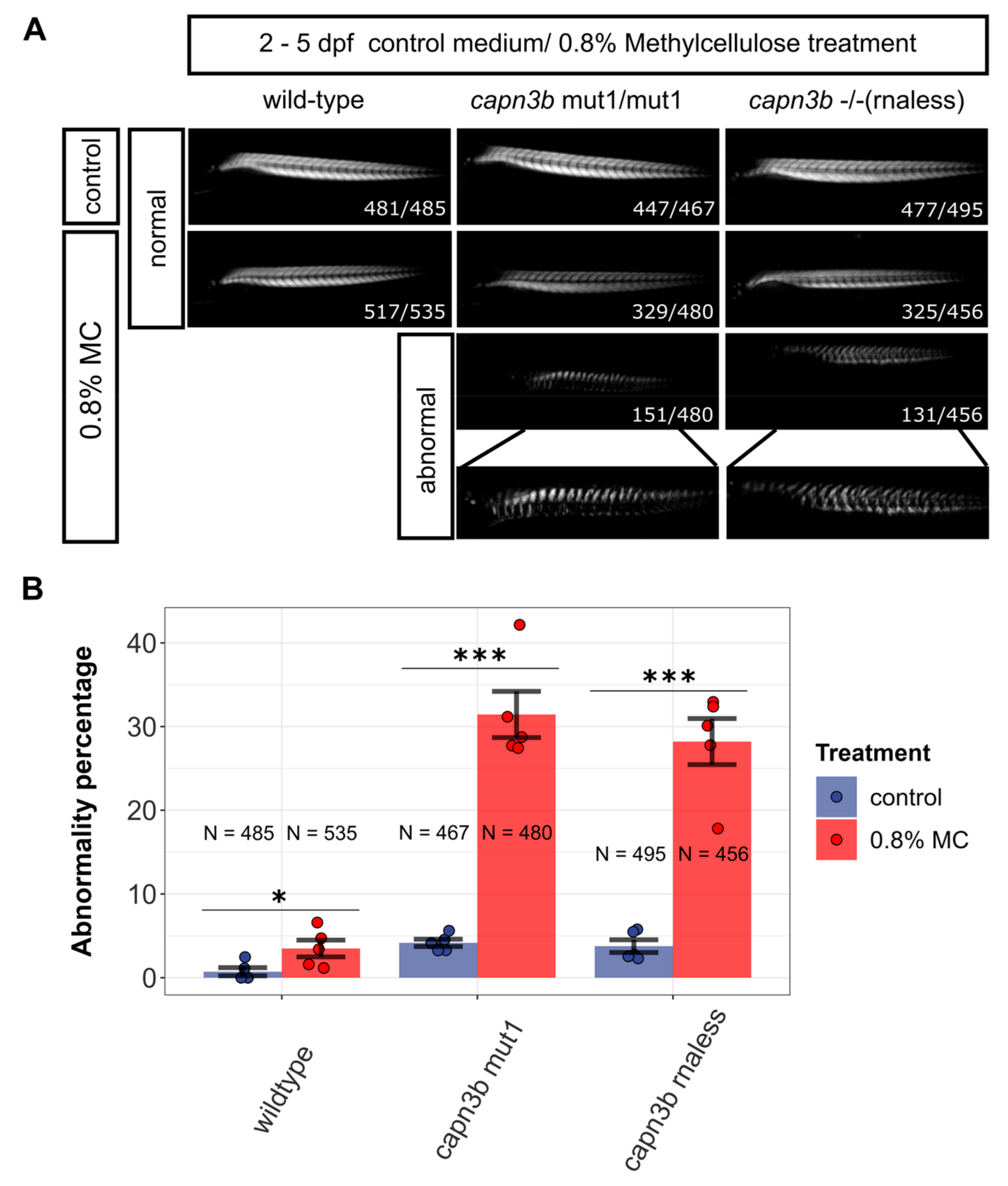
2.8. Methylcellulose Preparation and Treatment
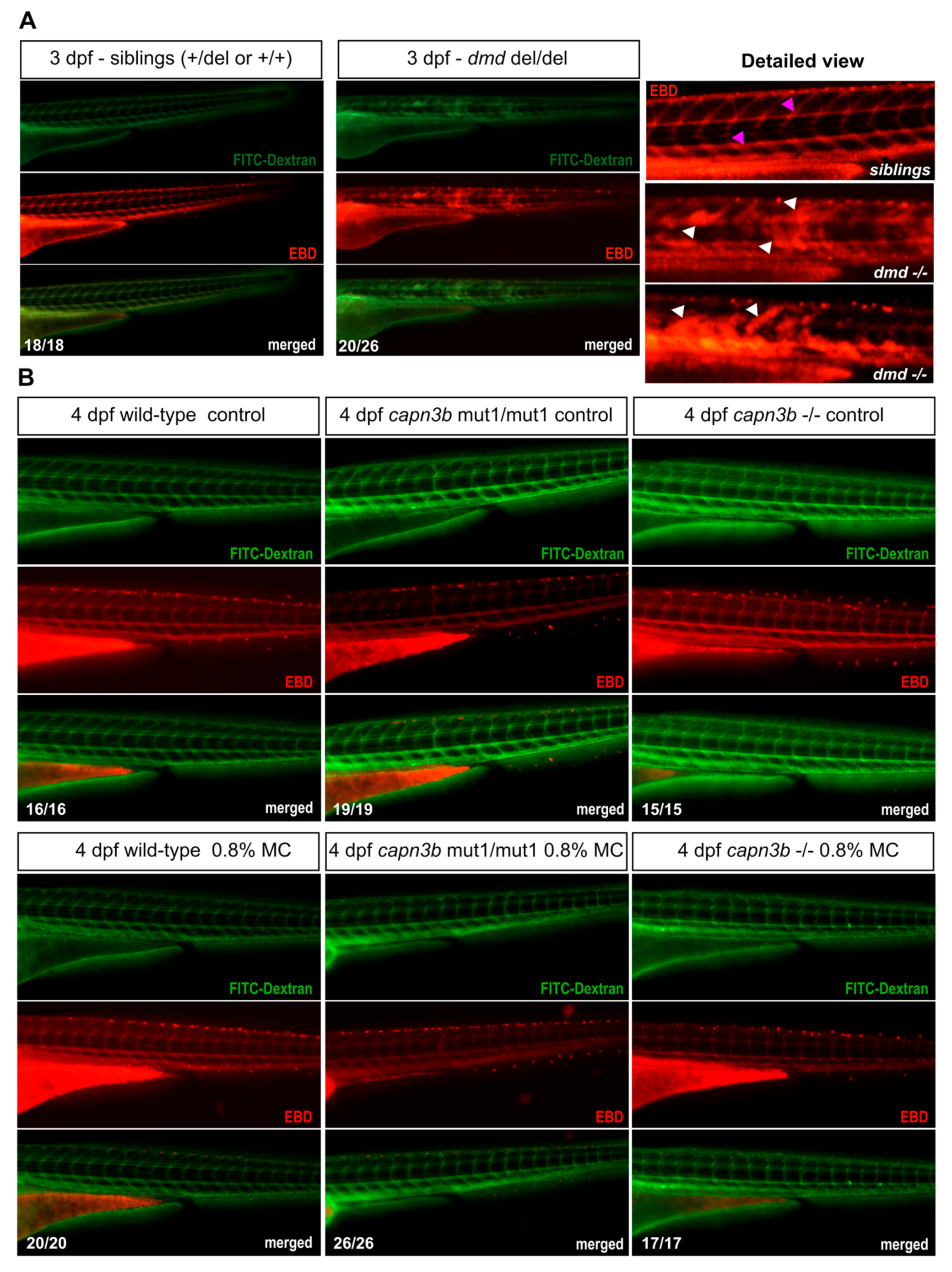
2.9. Evans Blue Dye Assay
2.10. Azinphos-Methyl Treatment
2.11. Birefringence Analysis
2.12. Statistical Analysis and Visualization of the Data
3. Results
3.1. Both RNA-Less and Partial Deletion capn3b Mutants Disrupt capn3b Expression, but the Mutants Have Normal Muscle Morpholog and Are Viable and Fertile
3.2. A Mutant in dmd Shows a Strong Muscular Dystrophy, Thus Serving as a Positive Control
3.3. Methylcellulose Incubation Reveals Increased Susceptibility of capn3b Mutants to Muscle Damage
3.4. No Evidence of Sarcolemmal Damage in capn3b Mutants but Disrupted Permeability in dmd Mutant Embryos
3.5. capn3b−/− Embryos Are More Susceptible to Muscle Damage Than Wild-Type after Treatment with Low Concentrations of Azinphos-Methyl
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ono, Y.; Ojima, K.; Shinkai-Ouchi, F.; Hata, S.; Sorimachi, H. An Eccentric Calpain, CAPN3/P94/Calpain-3. Biochimie 2016, 122, 169–187. [Google Scholar] [CrossRef] [PubMed]
- Richard, I.; Beckmann, J.S. Molecular Cloning of Mouse Canp3, the Gene Associated with Limb-Girdle Muscular Dystrophy 2A in Human. Mamm. Genome 1996, 7, 377–379. [Google Scholar] [CrossRef] [PubMed]
- Şahin, İ.O.; Özkul, Y.; Dündar, M. Current and Future Therapeutic Strategies for Limb Girdle Muscular Dystrophy Type R1: Clinical and Experimental Approaches. Pathophysiology 2021, 28, 238–249. [Google Scholar] [CrossRef] [PubMed]
- Ono, Y.; Shimada, H.; Sorimach, H.; Richard, I.; Saido, T.C.; Beckmann, J.S.; Ishiura, S.; Suzuki, K. Functional Defects of a Muscle-Specific Calpain, P94, Caused by Mutations Associated with Limb-Girdle Muscular Dystrophy Type 2A. J. Biol. Chem. 1998, 273, 17073–17078. [Google Scholar] [CrossRef]
- Taveau, M.; Bourg, N.; Sillon, G.; Roudaut, C.; Bartoli, M.; Richard, I. Calpain 3 Is Activated through Autolysis within the Active Site and Lyses Sarcomeric and Sarcolemmal Components. Mol. Cell. Biol. 2003, 23, 9127–9135. [Google Scholar] [CrossRef]
- Beckmann, J.S.; Spencer, M. Calpain 3, the “Gatekeeper” of Proper Sarcomere Assembly, Turnover and Maintenance. Neuromuscul. Disord. 2008, 18, 913–921. [Google Scholar] [CrossRef]
- Lasa-Elgarresta, J.; Mosqueira-Martín, L.; Naldaiz-Gastesi, N.; Sáenz, A.; de Munain, A.L.; Vallejo-Illarramendi, A. Calcium Mechanisms in Limb-Girdle Muscular Dystrophy with CAPN3 Mutations. Int. J. Mol. Sci. 2019, 20, 4548. [Google Scholar] [CrossRef]
- Toral-Ojeda, I.; Aldanondo, G.; Lasa-Elgarresta, J.; Lasa-Fernández, H.; Fernández-Torrón, R.; De Munain, A.L.; Vallejo-Illarramendi, A. Calpain 3 Deficiency Affects SERCA Expression and Function in the Skeletal Muscle. Expert Rev. Mol. Med. 2016, 18, e7. [Google Scholar] [CrossRef]
- Richard, I.; Roudaut, C.; Marchand, S.; Baghdiguian, S.; Herasse, M.; Stockholm, D.; Ono, Y.; Suel, L.; Bourg, N.; Sorimachi, H.; et al. Loss of Calpain 3 Proteolytic Activity Leads to Muscular Dystrophy and to Apoptosis-Associated IκBα/Nuclear Factor ΚB Pathway Perturbation in Mice. J. Cell Biol. 2000, 151, 1583–1590. [Google Scholar] [CrossRef]
- Kramerova, I.; Kudryashova, E.; Tidball, J.G.; Spencer, M.J. Null Mutation of Calpain 3 (P94) in Mice Causes Abnormal Sarcomere Formation in Vivo and in Vitro. Hum. Mol. Genet. 2004, 13, 1373–1388. [Google Scholar] [CrossRef]
- Kramerova, I.; Ermolova, N.; Eskin, A.; Hevener, A.; Quehenberger, O.; Armando, A.M.; Haller, R.; Romain, N.; Nelson, S.F.; Spencer, M.J. Failure to Up-Regulate Transcription of Genes Necessary for Muscle Adaptation Underlies Limb Girdle Muscular Dystrophy 2A (Calpainopathy). Hum. Mol. Genet. 2016, 25, 2194–2207. [Google Scholar] [CrossRef] [PubMed]
- Tao, T.; Shi, H.; Guan, Y.; Huang, D.; Chen, Y.; Lane, D.P.; Chen, J.; Peng, J. Def Defines a Conserved Nucleolar Pathway That Leads P53 to Proteasome-Independent Degradation. Cell Res. 2013, 23, 620–634. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Huang, D.; Shi, H.; Gao, C.; Wang, Y.; Peng, J. Capn3 Depletion Causes Chk1 and Wee1 Accumulation and Disrupts Synchronization of Cell Cycle Reentry during Liver Regeneration after Partial Hepatectomy. Cell Regen. 2020, 9, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ruan, H.; Sok, M.N.; Gao, C.; Hui, M.S.; Wu, W.; Zhang, Z.; Wen, Z.; Lane, D.P.; Peng, J. Loss of Function of Def Selectively Up-Regulates Δ113p53 Expression to Arrest Expansion Growth of Digestive Organs in Zebrafish. Genes Dev. 2005, 19, 2900–2911. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Huang, D.; Chen, F.; Gao, C.; Tao, T.; Shi, H.; Zhao, S.; Liao, Z.; Lo, L.J.; Wang, Y.; et al. Phosphorylation of Def Regulates Nucleolar P53 Turnover and Cell Cycle Progression through Def Recruitment of Calpain3. PLoS Biol. 2016, 14, e1002555. [Google Scholar] [CrossRef]
- El-Brolosy, M.A.; Kontarakis, Z.; Rossi, A.; Kuenne, C.; Günther, S.; Fukuda, N.; Kikhi, K.; Boezio, G.L.M.; Takacs, C.M.; Lai, S.-L.; et al. Genetic Compensation Triggered by Mutant MRNA Degradation. Nature 2019, 568, 193–197. [Google Scholar] [CrossRef]
- Westerfield, M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio Rerio), 4th ed.; University of Oregon Press: Eugene, OR, USA, 2000. [Google Scholar]
- White, R.M.; Sessa, A.; Burke, C.; Bowman, T.; LeBlanc, J.; Ceol, C.; Bourque, C.; Dovey, M.; Goessling, W.; Burns, C.E.; et al. Transparent Adult Zebrafish as a Tool for In Vivo Transplantation Analysis. Cell Stem Cell 2008, 2, 183–189. [Google Scholar] [CrossRef]
- Prykhozhij, S.V.; Steele, S.L.; Razaghi, B.; Berman, J.N. A Rapid and Effective Method for Screening, Sequencing and Reporter Verification of Engineered Frameshift Mutations in Zebrafish. Dis. Model. Mech. 2017, 10, 811–822. [Google Scholar] [CrossRef]
- Jao, L.; Wente, S.R.; Chen, W. Efficient Multiplex Biallelic Zebra Fi Sh Genome Editing Using a CRISPR Nuclease System. Proc. Natl. Acad. Sci. USA 2013, 110, 13904–13909. [Google Scholar] [CrossRef]
- Xu, H.; Xiao, T.; Chen, C.H.; Li, W.; Meyer, C.A.; Wu, Q.; Wu, D.; Cong, L.; Zhang, F.; Liu, J.S.; et al. Sequence Determinants of Improved CRISPR SgRNA Design. Genome Res. 2015, 25, 1147–1157. [Google Scholar] [CrossRef]
- Stemmer, M.; Thumberger, T.; Del Sol Keyer, M.; Wittbrodt, J.; Mateo, J.L. CCTop: An Intuitive, Flexible and Reliable CRISPR/Cas9 Target Prediction Tool. PLoS ONE 2015, 10, e0124633. [Google Scholar] [CrossRef] [PubMed]
- Meeker, N.D.; Hutchinson, S.A.; Ho, L.; Trede, N.S. Method for Isolation of PCR-Ready Genomic DNA from Zebrafish Tissues. Biotechniques 2007, 43, 610–614. [Google Scholar] [CrossRef]
- Lauter, G.; Söll, I.; Hauptmann, G. Two-Color Fluorescent in Situ Hybridization in the Embryonic Zebrafish Brain Using Differential Detection Systems. BMC Dev. Biol. 2011, 11, 43. [Google Scholar] [CrossRef]
- Smith, S.J.; Horstick, E.J.; Davidson, A.E.; Dowling, J. Analysis of Zebrafish Larvae Skeletal Muscle Integrity with Evans Blue Dye. J. Vis. Exp. 2015, 2015, 2–7. [Google Scholar] [CrossRef]
- Whitesell, T.R.; Chrystal, P.W.; Ryu, J.R.; Munsie, N.; Grosse, A.; French, C.R.; Workentine, M.L.; Li, R.; Zhu, L.J.; Waskiewicz, A.; et al. Foxc1 is Required for Embryonic Head Vascular Smooth Muscle Differentiation in Zebrafish. Dev. Biol. 2019, 453, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Etard, C.; Behra, M.; Fischer, N.; Hutcheson, D.; Geisler, R.; Strähle, U. The UCS Factor Steif/Unc-45b Interacts with the Heat Shock Protein Hsp90a during Myofibrillogenesis. Dev. Biol. 2007, 308, 133–143. [Google Scholar] [CrossRef]
- Wooddell, C.I.; Radley-Crabb, H.G.; Griffin, J.B.; Zhang, G. Myofiber Damage Evaluation by Evans Blue Dye Injection. Curr. Protoc. Mouse Biol. 2011, 1, 463–488. [Google Scholar] [CrossRef]
- Klüver, N.; Yang, L.; Busch, W.; Scheffler, K.; Renner, P.; Strähle, U.; Scholz, S. Transcriptional Response of Zebrafish Embryos Exposed to Neurotoxic Compounds Reveals a Muscle Activity Dependent Hspb11 Expression. PLoS ONE 2011, 6, e29063. [Google Scholar] [CrossRef]
- Shahid, M.; Takamiya, M.; Stegmaier, J.; Middel, V.; Gradl, M.; Kluver, N.; Mikut, R.; Dickmeis, T.; Scholz, S.; Rastegar, S.; et al. Zebrafish Biosensor for Toxicant Induced Muscle Hyperactivity. Sci. Rep. 2016, 6, 23768. [Google Scholar] [CrossRef]
- Anderson, J.L.; Mulligan, T.S.; Shen, M.-C.; Wang, H.; Scahill, C.M.; Tan, F.J.; Du, S.J.; Busch-Nentwich, E.M.; Farber, S.A. MRNA Processing in Mutant Zebrafish Lines Generated by Chemical and CRISPR-Mediated Mutagenesis Produces Unexpected Transcripts That Escape Nonsense-Mediated Decay. PLoS Genet. 2017, 13, e1007105. [Google Scholar] [CrossRef]
- Kok, F.O.; Shin, M.; Ni, C.-W.; Gupta, A.; Grosse, A.S.; van Impel, A.; Kirchmaier, B.C.; Peterson-Maduro, J.; Kourkoulis, G.; Male, I.; et al. Reverse Genetic Screening Reveals Poor Correlation between Morpholino-Induced and Mutant Phenotypes in Zebrafish. Dev. Cell 2015, 32, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Lawson, N.D. Reverse Genetics in Zebrafish: Mutants, Morphants, and Moving Forward. Trends Cell Biol. 2016, 26, 77–79. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Hromowyk, K.J.; Amacher, S.L.; Currie, P.D. Muscular Dystrophy Modeling in Zebrafish. Methods Cell Biol. 2017, 138, 347–380. [Google Scholar] [CrossRef] [PubMed]
- Lo, H.P.; Nixon, S.J.; Hall, T.E.; Cowling, B.S.; Ferguson, C.; Morgan, G.P.; Schieber, N.L.; Fernandez-Rojo, M.A.; Bastiani, M.; Floetenmeyer, M.; et al. The Caveolin-Cavin System Plays a Conserved and Critical Role in Mechanoprotection of Skeletal Muscle. J. Cell Biol. 2015, 210, 833–849. [Google Scholar] [CrossRef]
- Roostalu, U.; Strähle, U. In Vivo Imaging of Molecular Interactions at Damaged Sarcolemma. Dev. Cell 2012, 22, 515–529. [Google Scholar] [CrossRef]
- Ruparelia, A.A.; Oorschot, V.; Vaz, R.; Ramm, G.; Bryson-Richardson, R.J. Zebrafish Models of BAG3 Myofibrillar Myopathy Suggest a Toxic Gain of Function Leading to BAG3 Insufficiency. Acta Neuropathol. 2014, 128, 821–833. [Google Scholar] [CrossRef]
- Yano, T.; Tamura, K. The Making of Differences between Fins and Limbs. J. Anat. 2013, 222, 100–113. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prykhozhij, S.V.; Caceres, L.; Ban, K.; Cordeiro-Santanach, A.; Nagaraju, K.; Hoffman, E.P.; Berman, J.N. Loss of calpain3b in Zebrafish, a Model of Limb-Girdle Muscular Dystrophy, Increases Susceptibility to Muscle Defects Due to Elevated Muscle Activity. Genes 2023, 14, 492. https://doi.org/10.3390/genes14020492
Prykhozhij SV, Caceres L, Ban K, Cordeiro-Santanach A, Nagaraju K, Hoffman EP, Berman JN. Loss of calpain3b in Zebrafish, a Model of Limb-Girdle Muscular Dystrophy, Increases Susceptibility to Muscle Defects Due to Elevated Muscle Activity. Genes. 2023; 14(2):492. https://doi.org/10.3390/genes14020492
Chicago/Turabian StylePrykhozhij, Sergey V., Lucia Caceres, Kevin Ban, Anna Cordeiro-Santanach, Kanneboyina Nagaraju, Eric P. Hoffman, and Jason N. Berman. 2023. "Loss of calpain3b in Zebrafish, a Model of Limb-Girdle Muscular Dystrophy, Increases Susceptibility to Muscle Defects Due to Elevated Muscle Activity" Genes 14, no. 2: 492. https://doi.org/10.3390/genes14020492
APA StylePrykhozhij, S. V., Caceres, L., Ban, K., Cordeiro-Santanach, A., Nagaraju, K., Hoffman, E. P., & Berman, J. N. (2023). Loss of calpain3b in Zebrafish, a Model of Limb-Girdle Muscular Dystrophy, Increases Susceptibility to Muscle Defects Due to Elevated Muscle Activity. Genes, 14(2), 492. https://doi.org/10.3390/genes14020492






