Abstract
Translational Bioinformatics (TBI) is defined as the union of translational medicine and bioinformatics. It emerges as a major advance in science and technology by covering everything, from the most basic database discoveries, to the development of algorithms for molecular and cellular analysis, as well as their clinical applications. This technology makes it possible to access the knowledge of scientific evidence and apply it to clinical practice. This manuscript aims to highlight the role of TBI in the study of complex diseases, as well as its application to the understanding and treatment of cancer. An integrative literature review was carried out, obtaining articles through several websites, among them: PUBMED, Science Direct, NCBI-PMC, Scientific Electronic Library Online (SciELO), and Google Academic, published in English, Spanish, and Portuguese, indexed in the referred databases and answering the following guiding question: “How does TBI provide a scientific understanding of complex diseases?” An additional effort is aimed at the dissemination, inclusion, and perpetuation of TBI knowledge from the academic environment to society, helping the study, understanding, and elucidating of complex disease mechanics and their treatment.
1. Introduction
Computational biology, in recent decades, has gradually become more relevant in biological sciences [1], due to the significant decrease in cost of next-generation technologies and the progressive insertion of bioinformatics in medicine and translational research, playing a significant role in enhancing various biological studies [1,2].
These technologies have been adopted worldwide by numerous research groups and are being used to identify new Mendelian disease genes, while next-generation sequencing (NGS) is reaching routine clinical diagnostics [2]. However, translating the genome to the clinic depends on cross-referencing large amounts of data and various omics, as well as considering the environmental influence in these processes. Translational Bioinformatics’s (TBI) main objective is to translate results obtained from data processing into clinical practice. This requires the effort of a multidisciplinary team and the creation of computational models capable of enabling such a task [3].
Although its precise origin is uncertain, bioinformatics emerged from two main developments: (i) the increased understanding of the form and function of biological molecules, starting from results obtained from 1930s electrophoresis technique, and the consecutive discoveries involving DNA, RNA and protein structure; (ii) parallelly, the increase in computational power, such as mainframe applications (in the 1950s) and, later, modern workstations, as well as a continuous growth in the number of bioinformatics professionals, allowing for the understanding of numerous biological problems [4].
The identification of genes associated with human diseases is a determining factor for disease diagnosis in medicine [5]. In recent years several high-throughput techniques have been used to search for genes that cause common diseases, such as gene expression profiling, single nucleotide polymorphism (SNP) typing, whole genome sequencing, exome, total RNA sequencing (RNAseq), and protein sequencing [6]. However, there is a deadlock related to the use of such techniques, as they usually produce hundreds of candidate genes related to the disease being investigated [6]. To solve this problem, computational approaches, particularly network-based approaches, have been developed to efficiently determine disease-associated genes from existing biomedical networks [5], enabling data clustering and integration in order to build an accurate understand how cells work [4].
The creation of databases with biological information was the most important development to support the emergence of Bioinformatics. In the 1970s, structural biologists, using X-ray crystallography techniques, built the Protein Data Bank (PDB) specifying the Cartesian coordinates of the structures they elucidated, and made the PDB available to the public. As the ability to sequence DNA molecules became more affordable, DNA databases increased in number and quality. In the mid-1980s, the GenBank database was developed as a gene sequence information repository from the National Institutes of Health, an annotated collection of all publicly available DNA sequences [4].
Advances in research methods have resulted in experiments that generate vast amounts of data, which pose new challenges for researchers in storing, analyzing, and sharing data efficiently and appropriately [4]. In this context, the concept of Big Data arises, referring to a complex and large amount of data defined by: (i) large volume; (ii) diversity: stored data format can be unstructured and structured text, images, metadata, among others; (iii) speed of analytical processing; and (iv) data veracity or uncertainty: very large data collections can often combine several sources of varying reliability and trustworthiness [7]. In biological sciences, Big Data has been employed in enhancing curation, efficient analysis, and accurate interpretation, as well as accurate modeling and prediction of complex data [1].
In the age of Big Data, TBI can produce new methods and research knowledge about complex diseases, most notably cancer. To enable accurate and interpretable predictions, computational models based on protein structure analysis, methylation, expression, and activity of cancer hallmark signaling pathways are increasingly becoming the focus of current studies [8].
This review aims to guide, inform, and disseminate the understanding of TBI as a means to advance modern medicine. It highlights the benefits of understanding complex diseases, enabling training, specialization and abilities needed by various health professionals. We hope to answer the question: “How does TBI apply to and provide scientific understanding of complex diseases?”, and serve as a guide for those interested in this area.
2. TBI: From Definition to Application
TBI is a discipline that has come to the forefront of science in recent years because of the many beneficial results in solving and treating complex diseases. According to Tenenbaum [9], TBI has become an important discipline in the era of personalized medicine, broadening the insights and hypotheses about previously untestable foci of study.
TBI is considered a hybrid field of study incorporating basic and clinical research, also being called biomedical informatics. Other fields also bridge basic and clinical research, such as imaging informatics, clinical informatics and public health informatics. As stated by Sarkar [10], these fields supported the transfer and integration of knowledge in the main domains of translational medicine, ranging from molecules to populations.
According to the American Medical Informatics Association (AMIA), TBI has succeeded in developing storage, analytical, and interpretive methods capable of optimizing the transformation of increasingly voluminous biomedical and genomic data into predictive data for the development of preventive medicine [9].
TBI is crucial for moving basic biological discoveries from the research bench to the patient care setting (clinical research), using clinical information to understand basic biology. Likewise, TBI focuses on patient care, including the creation of new diagnoses, prognoses, prevention strategies, and therapies based on biological discoveries (Table 1) [11].
Systems biology is a modern and advanced field of study, focusing on understanding the behavior of entire biological systems, not just individual components. It uses methods ranging from qualitative network diagrams that link molecular and phenotypic entities, to formal quantitative models based on differential equations [11].
TBI research has started to develop Translational Biomedical knowledge in order to form a common language among researchers from different fields, connecting genomic information with phenotypic correlation analysis, enabling this increasingly transdisciplinary field [10].
In the last two decades, along with TBI and systems biology, the analysis and modeling of biomedical data with a network structure has emerged, supported by numerous network-based learning methods that have been developed in order to select one that learns and adapts to clinical application, resulting in highly meaningful, accessible, low-cost, and less time-consuming data analysis (Table 1) [12].


Table 1.
TBI application in complex diseases.
Table 1.
TBI application in complex diseases.
| Authors | Study Description |
|---|---|
| Ahmed [13]; Kang; Ko; Mersha [14]; Savoska; Ristevski; Trajkovik [15]. | Integration of collective and individualized clinical data with patient-specific multi-omic data, AI algorithms and cloud electronic health record databases. |
| Baruah; Deka; Mahanta [16]. | Multidisciplinary cooperation between laboratory and clinical researchers, aiming to meet community needs. |
| Bellazzi et al. [17]. | Clinical bioinformatics (CBI) seeks to integrate molecular and clinical data, using bioinformatics to understand molecular mechanisms and potential therapies. |
| Bruggemann et al. [18]. | Pharmacogenomics provides personalized patient care by selecting specific drugs for diseases, such as non-small cell lung cancer. |
| Han; Liu [19]. | AI unravelling latent data behavior and generating new insights and optimal strategies in decision-making. |
| Liu et al. [20]. | TBI used in reproductive medicine. |
| Liu; Wang; Lai [21]. | Single-cell total RNA sequencing (scRNA-seq) and bulk total RNA sequencing (RNA-seq) associated with machine learning for detection of tumor origin. |
| Lussier; Butte; Hunter [22]. | TBI effects on: (i) availability and cost reduction of molecular measurements; (ii) accessibility to measurements of health and disease status; (iii) sharing data and molecular tools; (iv) interpretation of new clinical molecular discoveries; (v) research funding. |
| Mitra et al. [23]. | In silico analysis of acquired, archived, and retrieved biological data, as well as dynamic molecular docking experiments affecting pharmacology, biotechnology, bioengineering and education. |
| Sheikh; Ramlal; Khan [24]. | Predictive modeling of antineoplastic drugs through computational analysis of microarrays. |
| Tang et al. [25]. | TBI encompassing issues of equity and inclusion, by means of phenotyping applications, characterization of disease subtypes, predictive modeling, biomarker discovery and selection of better treatments. |
| Torkamannia; Omidi; Ferdousi [26]. | Combined pharmacotherapy with synergistic/additive effect as a powerful treatment strategy for complex diseases. |
| Yang et al. [27]. | High-throughput technologies/TBI in the biological interpretation of Big Data, uncovering mechanistic landscapes of complex diseases. |
3. Translational Biomedical Knowledge
Translational Biomedical Knowledge and TBI have been shown to be highly efficient in developing new insights and proposing new hypotheses that were previously untestable. The 2019 Yearbook of the International Medical Informatics Association (IMIA) demonstrates TBI trends, on various aspects of bioinformatics methods and techniques to advance clinical care [28,29].
This translational knowledge has enabled work to be done at a low cost and in less time, because much of the recent discussion from bench to bedside research has been focused on how to allocate limited resources in support of science, in order to generate transformative clinical impact [30]. According to Shameer et al. [31], by characterizing health status through individual translational knowledge, earlier identification of true and personalized pathological changes can occur, avoiding unnecessary testing after accidental findings.
This knowledge can turn genomic data into clinically actionable knowledge, allowing for the understanding of complex drug interactions. Shameer et al. [31] explain that by integrating data from deep molecular profiling technologies (genomic, transcriptomic, proteomic or metabolomic), collectively defined as multi-omics data with clinical information, authors were able to explain some of the clinical variations between individuals and improve the use of health monitoring data for prediction, diagnosis, and development of intelligent clinical decision support systems, helping a more comprehensive understanding of clinical pharmacodynamics, pharmacokinetics, and the molecular mechanisms underlying drug effects, achieving relevant clinical utility and lower treatment side effects [32].
4. Precision Medicine and TBI
The objective of precision medicine is to use genetic data to improve health care decision-making [33,34]. Considering the growing volume of data and information generated, TBI can contribute considerably to the evolution of precision medicine. Since the primary goal of precision medicine is to find a dynamic treatment regimen that works well in future patients, Kosorok and Laber [34] highlight the usefulness of machine learning methods.
Recent studies have pointed to omics data importance and the potential in precision medicine. Xiao and colleagues [35] demonstrated how combined omics data from triple negative breast cancer (TNBC) has linked its metabolome and genomics. Authors pointed out the importance of TNBC metabolomic data for its precision treatment [35]. Milluzzo and collaborators [36], in a review paper addressing the clinical management of patients with diabetes and cancer, highlight the importance of a personalized, patient-centered, multidisciplinary and shared approach in the treatment of complex diseases, leading to the optimization of human and financial resources, and obtaining better clinical results [36].
Complex and voluminous datasets can be used through TBI to provide information that will enable better clinical outcomes. Qazi and Raza [37] point out that TBI has the potential to evolve traditional domains of biomedical sciences to improve health care infrastructure management and regulation [37].
5. Omics Revolution in Complex Diseases
The revolution in molecular biology caused the need to analyze previously unprecedented large amounts of data, requiring biology to make a transition from a more qualitative science to a true data science [38]. Organizing, integrating, and understanding have become the watchwords for multi-omics studies associated with understanding the biology of complex diseases. While the number of multi-omics studies has increased rapidly in recent years, the diversity of methods for integrating these data remains limited, being purely data driven. In order to advance in this field, there is a need to capitalize on the extensive prior knowledge about component interactions, and create a mechanistic understanding tool [39].
With the integration of omics approaches, the unbiased analysis of the whole genome (genomics), transcriptome (transcriptomics), proteome (proteomics), and metabolome (metabolomics) from different types of samples has been achieved [40]. The integration of the genome and transcriptome has offered an unprecedented opportunity to determine unambiguous genotype-phenotype correlation, such as the integration of genomic variation (e.g., single nucleotide polymorphism–SNP) and transcriptional alteration in single cells [41].
The integration of transcriptome with proteome has allowed phenotypes to be fully defined based on gene expression, cellular metabolism, structural dynamics, and signal transduction [41]. In the case of metabolomics integration, it was possible to perform targeted or unbiased identification of endogenous metabolites from body fluids, correlating metabolism, epigenomic changes, and proteomics [40]. There has been a transition from focusing on a small number of genes at a time, to simultaneously measuring as many of these cellular components as possible, significantly elevating the biological datasets collected simultaneously [38].
According to Subramanian et al. [42], the addressable biological questions on the vision of multi-omics data integration are broadly categorized into three different case studies: (i) disease subtyping and classification; (ii) predicting biomarkers for various applications, including diagnostics; (iii) gaining insights into disease biology.
The subtyping and classification of samples based on their omics profiles, especially cancer, are heterogeneous due to the remarkable differences in disease progression in affected individuals. Therefore, identifying the underlying subtypes of a disease or classifying samples into known subgroups makes it possible to understand disease etiology and identify appropriate interventions for patients belonging to different subgroups. Biomolecules are tightly interconnected, providing the flow of information of biological processes. Understanding the mechanistic details of disease biology is critical to the diagnosis and development of new therapeutic interventions [42].
Among the complex diseases addressed in this manuscript, cancer stands out as the main purpose of our review. To achieve this goal, we have searched https://clinicaltrials.gov (accessed on 9 January 2023) and 109 studies were found for the terms “Omics” and “Cancer”. Figure 1 shows the distribution of clinical trials around the globe (Asia, Europe and the United States). The most relevant trials are shown in Table 2.
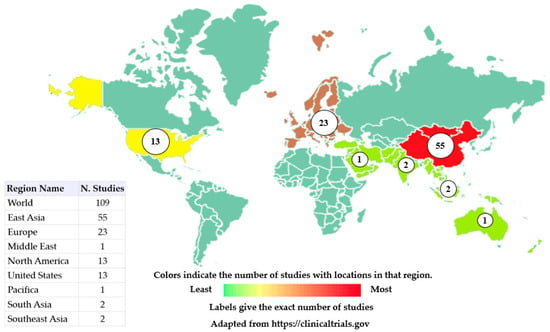
Figure 1.
Global distribution of clinical trials using terms Omics and Cancer. Clinical studies that have omics and cancer as keywords are shown here. Studies in East Asia correspond to 50.46% of clinical trials, European studies correspond to 21.10%, and United States correspond to 11.92% of all clinical trials. Colors indicate the number of studies located in the region, ranging from green (least studies) to red (most studies). Studies with no location are not included in the counts or on the map and studies with multiple locations are included in all corresponding regions.

Table 2.
Most relevant clinical trials using the terms Omics” and “Cancer”.
6. Integrating Complex Diseases through TBI
Ontologies are used to document new knowledge from biological and biomedical research, from classical biochemical experiments to omics experiments. These ontologies are created, maintained, and extended by experts, with the goal of providing a unified annotation schema that is human- and machine-readable [43]. According to The Gene Ontology Consortium [44], the ontology encompasses three divisions, these being: (i) molecular function (the activity of a gene product at the molecular level), (ii) cellular component (the location of a gene product’s activity in relation to biological structures), and (iii) biological process (a larger biological program in which the molecular function of a gene is used). According to these divisions, ontology enables complex answers to be obtained in a simplified way, being treatable by both man and machine, in order to offer reliable data according to the system addressed.
Manda [45] addressed the use of association rule mining focused on predicting annotations, becoming crucial to discover new relationships between ontologies and other applications. Wang [46] and Quan [47] demonstrated the use of ontology data in pathway analysis (organizing and eliminating pathway data redundancy), which are crucial for understanding physiology and pathogenesis of diseases. Schriml et al. [48] applied a human disease ontology (DO) to provide disease classification by formal semantic rules to specifically express meaningful disease models, aiming to include mechanistically inferred multiple-disease classifications, enabling new insights into related diseases, such as the heterogeneity of genetic diseases and the multicellular origin of cancer.
7. Application of TBI in Complex Diseases
Applications of TBI are evidenced by Roy, Singh and Gupta [49], who highlighted an innovative study against pancreatic ductal adenocarcinoma, through an integrated analysis of DNA methylation and gene expression datasets aiming at better mechanistic and molecular insights that can be correlated with clinical data. These authors provide valuable results for prognostic improvements, personalized treatment and delineation of the heterogeneous landscape of pancreatic ductal adenocarcinoma, and may enable personalized therapies and risk prediction.
According to Liu et al. [50], a new connection of TBI with validation through in vitro, in vivo, and patient-derived samples has proven possible in the face of breast cancer biomarker discovery. They used circulating non-coding RNA as a source of new biomarkers for non-invasive screening. As a consequence, hsa-miR-423-5p expression in plasma and blood exosomes of breast cancer patients was observed to be abnormally high compared to healthy controls. Encoding genes regulated by hsa-miR-423-5p were widely distributed in signaling pathways associated with tumors in silico.
Cai et al. [51] incorporated improvements in analytical methods for the detection of differentially expressed genes (DEGs) between two different phenotypes with limited sample sizes, enabling improvements in reaching vital clues for cancer treatment. Chen et al. have developed a highly effective tool to accurately detect and visualize gene fusions, which play an important role in cancer.
Yu, Zhao and Gao [52] used miRNA data on target genes and disease tissue specificity, as well as information from the Food and Drug Administration, to construct drug-miRNA-disease networks, and potential disease treatment prediction, which was employed in breast cancer cases, detecting new potential drugs for treatment. Zeng et al. [53] built a co-expression network using a gastric cancer model, and performed enrichment analyses to identify key unique genes, suggesting them as likely biomarkers of cell subtype. Likewise, Zhou et al. [54] demonstrated green tea data on suppression of proliferation pathways in cancer, as well as positive regulation for certain miRNAs, in a study combing miRNA, mRNA, pathway and network analysis.
TBI has highlighted several new and improved studies for other complex diseases, such as rare diseases, metabolic syndrome, and pandemic diseases, and has served as a pillar for new research on biological networks, such as co-expression networks and multi-omics analyses. In this context, Akgün et al. [55] addressed the importance of TBI for rare disease analyses and preservation of genomic data for these patients, due to the difficulty of obtaining biological data.
Immel et al. [56] portrayed the use of genomic DNA analysis from buried victims who had been afflicted by the medieval plague. Given its devastating effect, the second plague pandemic caused by Yersinia. pestis was a strong candidate to exert selective pressure on the human immune response. Thus, the authors developed a study that indicated that the differences in allele frequencies of HLA genes involved in innate and adaptive immunity (responsible for extracellular and intracellular responses) to pathogenic bacteria (such as Yersinia. pestis) may have been affected by the historical epidemics that occurred in Europe in the past.
Recently, humanity experienced a major pandemic caused by the SARS-CoV-2 virus, which, due to the lack of available drugs or vaccines, made the rapid virus spread and progress throughout the world, causing many deaths. TBI proved to be useful and innovative by performing a crosstalk between molecular modeling techniques, molecular docking, and in vitro testing, as described by Pooja et al. [57], who made the ability of in silico studies to provide candidate molecules for antiviral drug development explicit.
Jaballah et al. [58] coupled in silico studies via biological networks with molecular analyses for understanding menopausal hormonal changes associated with the onset of metabolic syndrome (MS) and its consequences for type 2 diabetes (T2DM) and cardiovascular disease (CVD). To achieve this goal, the authors exploited a TBI approach to detect common genetic signatures for MS, DM2 and CVD, and menopausal status, and, through enrichment analysis, provided core genes that may play a key role in menopausal status and influence the risks of MS, DM2, and CVD.
Based on Djeddi et al. [59], TBI has also been used in multi-omics approaches finding different molecular signatures common to diverse diseases and therapeutic strategies. Huang et al. [60] highlighted the high demand of using gene co-expression network (GCN) mining, in such way that TBI identified gene modules with correlated expression profiles. These interactions have made it possible to discover new latent genetic interactions, and new gene functions, and to extract molecular features of certain disease groups, finding new disease biomarkers.
Thalor et al. [61] highlighted the impact of a translational approach on the elucidation of potential gene signatures, such as genes associated with MAPK, PI3-AkT, Wnt, TGF-β and other signal transduction pathways, to demonstrate new molecular hypotheses about the metastasis process related to triple negative breast cancer (TNBC). Ullah et al. [62] highlighted the ability of bioinformatics to provide better diagnostic and therapeutic markers for colorectal cancer, being able to identify the effectiveness of SRY-Box Transcription Factor 9 (SOX9) in improving colorectal cancer prognosis.
Kaur [63] applied a TBI approach to the study of gliomas to obtain better molecular classification of different grades of glioma and demonstrated the need for in-depth assessments on critical genes for cancer development. Yi et al. [64] applied TBI on mantle cell lymphoma by including sequencing of patient longitudinal samples and RNA-seq data, obtaining genetic subsets that could guide a clinical understanding of cancer clonal evolution.
Yu et al. [65] have highlighted how connecting Bioinformatics with the clinic allows for the construction of a novel framework to evaluate and select assays to monitor cancer. They used NGS with large gene panels of somatic cancer mutations of circulating tumor DNA (ctDNA). In parallel, Xu et al. [66] pointed to the application of this area on the current COVID-19 pandemic research, in which TBI have helped understand potential disease mechanisms, and effective and less toxic treatments.
Last, but not least, Battineni et al. [67] emphasized an improvement on sample storage for future research through the use of TBI, because this area provides a crucial means of applying artificial intelligence on data analysis, disease diagnosis, prediction and classification of pathological findings. These numerous applications underscore the unequivocal innovative role of TBI in various areas of medicine in an interdisciplinary and multidisciplinary way, as symbolized in Figure 2.
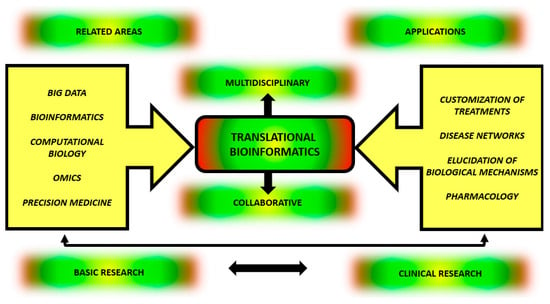
Figure 2.
Integration between origin and outcomes of Translational bioinformatics. TBI applied to the study of complex diseases involves basic research and clinical research in a reciprocal, collaborative and multidisciplinary way. For correct understanding and treating complex multifactorial diseases, there is a need to integrate information from multiple areas, such as Big Data, bioinformatics, computational biology, molecular biology associated with omics data and precision medicine. By doing so, this knowledge can be applied to treatment customization, understanding disease networks, elucidating biological processes and developing translational pharmacology.
7.1. Relationship between Next-Generation Sequencing (NGS) and TBI in the Study of Complex Diseases
The constant evolution of sequencing technologies has motivated modern bioinformatics, shaping the area of basic and clinical research with advanced techniques based on sophisticated computation, artificial intelligence, machine, and deep learning [68].
The development and application of next-generation sequencing platforms enabled bioinformatics improvements primarily through sequencing automation methods, mainly due to the needs of personal genomes and metagenomics projects [68].
Sequencing is subdivided into generations: (i) first generation (Sanger sequencing), provides high precision and helps in validating NGS discoveries, but has a low yield; (ii) second generation, with high throughput, short reading length, low cost, difficult sample preparation, clinical applications and PCR amplification; (iii) third generation, highlights the absence of PCR amplification, requires less initial material, longer reading lengths, very low cost and low error rate during library preparation, but permeates a relatively high sequencing error rate and a small number of algorithms/tools for final analysis; and (iv) fourth generation, ultra-fast scanning of the entire genome, enabling sample spatial distribution reads (in situ sequencing) [69].
Among numerous applications of this crosstalk between NGS and bioinformatics, clinical genetics has achieved numerous improvements regarding the analysis of hundreds of genes at an unprecedented speed and low cost, applying bioinformatics algorithms to deal with complex and heterogeneous disorders by combining information from multiple omics sources (such as genome, transcriptome, proteome and epigenome), to develop new machine learning algorithms, aiming at improving NGS utility and performance, achieving superior clinical diagnostics and opening new therapeutic paths [70].
In addition, third-generation sequencing has demonstrated innovative translational solutions for the diagnosis of infectious diseases (pathogen detection and characterization of mixed microbial communities) [71], cancers and other diseases, identifying a large number of disease variants in the human genome [72], using a relatively low-cost platform, fast response time and easy-to-use bioinformatics pipelines [71], thus, generating an increase in disease molecular diagnostic accuracy, using unique and real-time molecular sequencing technologies [73].
Exome sequencing has been providing an increase in disease diagnostic yield, identifying new pathogenic genetic variants [74].
Bioinformatics coupled with high computational power through cloud platforms, offers scalability, safety and performance [75], integrating diagnostic and therapeutic tools for genomic and pharmacogenomic discoveries, in order to provide routine medical care, design specific drugs and personalized genome tests [76].
7.2. Multi-Omics, Single Cells, and TBI in the Study of Complex Diseases
The advances provided by sequencing associated with precision medicine have driven new paradigm shifts in clinical practice and basic research. The union of collective and individualized clinical data with patient-specific multi-omics data has resulted in new therapeutic strategies [13]. Multi-omics approaches allow for the integration of data from various platforms, in a multifaceted view of disease processes [77].
Single-cell omics provides a basis for data-driven reconstruction of cell lineage hierarchies, deepening the understanding of the underlying mechanisms that govern health and disease [77,78]. Bioinformatics plays a crucial role in the interpretation and analysis of single-cell data results. To achieve this, five complementary strategies are applied: (i) combine; (ii) separate; (iii) split; (iv) convert; and (v) predict [78].
While new research methodologies have been developed, new computational resources, along with new algorithms, mathematical models and new tools, have been used in multi-omics single-cell studies to uncover new information about complex diseases, such as cancer [77,78,79]. Nam, Chaligne and Landau [80] highlighted the multidimensional incorporation of omics with single cells in cancer, promoting better understanding of tumor evolution, unveiling the cell-to-cell genetic diversity, epigenetic profiles, spatial distributions, and microenvironment interactions.
8. Conclusions
TBI has the mission to investigate tumor biology from different perspectives, using novel approaches, combining cellular and molecular biology techniques, bioinformatics, and clinical data. By these means, it is possible to better understand and characterize tumor evolution, as well as develop new strategies for disease detection, control, and treatment. Research in this area aims to evaluate a large amount of data and understand molecular alterations in the cell genome that lead to the tumor phenotype, and evaluate the effects of these alterations on cell signaling and metabolic pathways, as well as the interaction of tumor biology with the immune system and its microenvironment. In the face of new discoveries in TBI, biomarkers for clinical and therapeutic use can be identified.
TBI research is characterized by being multidisciplinary, and it brings together expertise from different areas such as cell biology, molecular biology, genetics, immunology, virology, biochemistry, bioinformatics, and medicine, applying it in the pharmaceutical and biotechnology industry, in oncology translational research, and in the development of mathematical and computational models that can bring forward better treatment and quality of life for cancer patients.
Given the information presented, it appears that a joint effort is needed to increase the dissemination of knowledge about TBI from academia to society, highlighting the great benefit and impact of this area on the study, understanding, and elucidation of mechanisms for the treatment of complex diseases, especially cancer. In addition, it is important that higher education institutions offer students integrated and interdisciplinary training, bringing together the areas of computing, mathematics, statistics, biology, medicine, and other areas of health, aiming to train professionals to be able to act and develop research projects in TBI integrating basic and clinical research through diverse and dynamic content.
We hope to have provided an efficient guide for the dissemination of academic knowledge in this area for numerous audiences, especially students and health professionals.
Author Contributions
Conceptualization, M.C.C. and D.D.M.; methodology, M.C.C., D.D.M., C.V.C., D.d.A.D., C.C.A., I.P.P. and R.F.R.B.; data curation, M.C.C., D.D.M., B.G.d.O.B., C.R.V., C.C.A. and F.M.G.; writing—original draft preparation, M.C.C., D.D.M., D.G.B., R.F.R.B., G.M.S., L.S.L., T.S.L. and I.D.L.; review and editing, M.C.C., D.D.M., E.d.V.W.d.S., F.d.P., L.N.R.A. and R.S.d.R.T.; translation, G.M.S., L.S.L., T.S.L. and I.D.L.; supervision, M.C.C., E.F.d.C., D.D.M. and I.D.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We would like to thank Ufes Writing Center (Caesa) for their support and support for their free translation services for this article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lin, H.; Peng, S.; Huang, J. Special issue on computational resources and methods in biological sciences. Int. J. Biol. Sci. 2018, 14, 807–810. [Google Scholar] [CrossRef] [PubMed]
- Robinson, P.N. Genomic data sharing for translational research and diagnostics. Genome Med. 2014, 6, 78. [Google Scholar] [CrossRef]
- Londin, E.R.; Barash, C.I. What is Translational Bioinformatics? Appl. Transl. Genom. 2015, 6, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Mooney, S.D.; Tenenbaum, J.D.; Altman, R.B. Translational Bioinformatics. In Biomedical Informatics: Computer Applications in Health Care and Biomedicine; Springer: Berlin/Heidelberg, Germany, 2014; pp. 867–911. [Google Scholar]
- Peng, J.; Bai, K.; Shang, X.; Wang, G.; Xue, H.; Jin, S.; Cheng, L.; Wang, Y.; Chen, J. Predicting disease-related genes using integrated biomedical networks. BMC Genom. 2017, 18, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kimmel, C.; Visweswaran, S. An algorithm for network-based gene prioritization that encodes knowledge both in nodes and in links. PLoS ONE 2013, 8, e79564. [Google Scholar] [CrossRef] [PubMed]
- Bellazzi, R. Big data and biomedical informatics: A challenging opportunity. Yearb. Med. Inform. 2014, 23, 8–13. [Google Scholar] [CrossRef]
- Azuaje, F. Computational models for predicting drug responses in cancer research. Brief. Bioinform. 2017, 18, 820–829. [Google Scholar] [CrossRef]
- Tenenbaum, J.D. Translational Bioinformatics: Past, present, and future. Genom. Proteom. Bioinform. 2016, 14, 31–41. [Google Scholar] [CrossRef]
- Sarkar, I.N. Biomedical informatics and translational medicine. J. Transl. Med. 2010, 8, 22. [Google Scholar] [CrossRef]
- Altman, R.B. TBI: Linking the molecular world to the clinical world. Clin. Pharmacol. Ther. 2012, 91, 994–1000. [Google Scholar] [CrossRef]
- Su, C.; Tong, J.; Zhu, Y.; Cui, P.; Wang, F. Network embedding in biomedical data science. Brief. Bioinform. 2020, 21, 182–197. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, Z. Multi-omics strategies for personalized and predictive medicine: Past, current, and future translational opportunities. Emerg. Top. Life Sci. 2022, 6, 215–225. [Google Scholar] [CrossRef]
- Kang, M.; Ko, E.; Mersha, T.B. A roadmap for multi-omics data integration using deep learning. Brief. Bioinform. 2022, 23, bbab454. [Google Scholar] [CrossRef] [PubMed]
- Savoska, S.; Ristevski, B.; Trajkovik, V. Personal Health Record Data-Driven Integration of Heterogeneous Data. In Data-Driven Approach for Bio-Medical and Healthcare; Springer: Singapore, 2023; 21p. [Google Scholar]
- Baruah, C.; Deka, B.; Mahanta, S. A Review of Recent Advances in TBI and Systems Biomedicine. Inf. Retr. Bioinform. 2022, 1, 37–62. [Google Scholar]
- Bellazzi, R.; Masseroli, M.; Murphy, S.; Shabo, A.; Romano, P. Clinical Bioinformatics: Challenges and opportunities. BMC Bioinform. 2012, 13, S1. [Google Scholar] [CrossRef] [PubMed]
- Bruggemann, L.; Falls, Z.; Mangione, W.; Schwartz, S.A.; Battaglia, S.; Aalinkeel, R.; Mahajan, S.D.; Samudrala, R. Multiscale Analysis and Validation of Effective Drug Combinations Targeting Driver KRAS Mutations in Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2023, 24, 997. [Google Scholar] [CrossRef]
- Han, H.; Liu, X. The challenges of explainable AI in biomedical data science. BMC Bioinform. 2022, 22, 1–3. [Google Scholar] [CrossRef]
- Liu, K.; Zhang, Y.; Martin, C.; Ma, X.; Shen, B. Translational Bioinformatics for Human Reproductive Biology Research: Examples, Opportunities and Challenges for a Future Reproductive Medicine. Int. J. Mol. Sci. 2023, 24, 4. [Google Scholar] [CrossRef]
- Liu, H.-P.; Wang, D.; Lai, H.-M. Can we infer tumor presence of single cell transcriptomes and their tumor of origin from bulk transcriptomes by machine learning? Comput. Struct. Biotechnol. J. 2022, 20, 2672–2679. [Google Scholar] [CrossRef]
- Lussier, Y.A.; Butte, A.J.; Hunter, L. Current methodologies for TBI. J. Biomed. Inform. 2010, 43, 355. [Google Scholar] [CrossRef]
- Mitra, D.; Mitra, D.; Bensaad, M.S.; Sinha, S.; Pant, K.; Pant, M.; Priyadarshini, A.; Singh, P.; Dassamiour, S.; Hambaba, L.; et al. Evolution of Bioinformatics and its impact on modern bio-science in the twenty-first century: Special attention to pharmacology, plant science and drug discovery. Comput. Toxicol. 2022, 24, 100248. [Google Scholar] [CrossRef]
- Sheikh, K.; Ramlal, A.; Khan, F.N. Computational Resources for Oncology Research: A Comprehensive Analysis. In Computational Intelligence in Oncology; Springer: Singapore, 2022; Volume 1016, pp. 65–92. [Google Scholar]
- Tang, A.; Woldemariam, S.; Roger, J.; Sirota, M. Translational Bioinformatics to Enable Precision Medicine for All: Elevating Equity across Molecular, Clinical, and Digital Realms. Yearb. Med. Inform. 2022, 31, 106–115. [Google Scholar] [CrossRef]
- Torkamannia, A.; Omidi, Y.; Ferdousi, R. A review of machine learning approaches for drug synergy prediction in cancer. Brief. Bioinform. 2022, 23, bbac075. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Dunker, A.K.; Liu, J.S.; Qin, X.; Arabnia, H.R.; Yang, W.; Niemierko, A.; Chen, Z.; Luo, Z.; Wang, L.; et al. Advances in Translational Bioinformatics facilitate revealing the landscape of complex disease mechanisms. BMC Bioinform. 2014, 15, I1. [Google Scholar] [CrossRef]
- Smaïl-Tabbone, M.; Rance, B. Contributions from the 2018 literature on bioinformatics and translational informatics. Yearb. Med. Inform. 2019, 28, 190–193. [Google Scholar] [CrossRef]
- Smaïl-Tabbone, M.; Rance, B. Contributions from the 2019 Literature on Bioinformatics and Translational Informatics. Yearb. Med. Inform. 2020, 29, 188–192. [Google Scholar] [CrossRef]
- Hutchins, B.I.; Davis, M.T.; Meseroll, R.A.; Santangelo, G.M. Predicting translational progress in biomedical research. PLoS Biol. 2019, 17, e3000416. [Google Scholar] [CrossRef]
- Shameer, K.; Badgeley, M.A.; Miotto, R.; Glicksberg, B.S.; Morgan, J.W.; Dudley, J.T. Translational Bioinformatics in the era of real-time biomedical, health care and wellness data streams. Brief. Bioinform. 2017, 18, 105–124. [Google Scholar] [CrossRef]
- Wu, H.Y.; Shendre, A.; Zhang, S.; Zhang, P.; Wang, L.; Zeruesenay, D.; Rocha, L.M.; Shatkay, H.; Quinney, S.K.; Ning, X.; et al. Translational knowledge discovery between drug interactions and pharmacogenetics. Clin. Pharmacol. Ther. 2020, 107, 886–902. [Google Scholar] [CrossRef]
- König, I.R.; Fuchs, O.; Hansen, G.; von Mutius, E.; Kopp, M.V. What is precision medicine? Eur. Respir. J. 2017, 50, 4. [Google Scholar] [CrossRef]
- Kosorok, M.R.; Laber, E.B. Precision medicine. Annu. Rev. Stat. Its Appl. 2019, 6, 263–286. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Ma, D.; Yang, Y.S.; Yang, F.; Ding, J.H.; Gong, Y.; Jiang, L.; Ge, L.P.; Wu, S.Y.; Yu, Q.; et al. Comprehensive metabolomics expands precision medicine for triple-negative breast cancer. Cell Res. 2022, 32, 477–490. [Google Scholar] [CrossRef] [PubMed]
- Milluzzo, A.; Vigneri, P.; Martorana, F.; Vigneri, R.; Sciacca, L. Type 2 diabetes and cancer: Problems and suggestions for best patient management. Explor. Med. 2020, 1, 184–204. [Google Scholar] [CrossRef]
- Qazi, S.; Raza, K. Translational Bioinformatics in healthcare: Past, present, and future. In Translational Bioinformatics in Healthcare and Medicine; Academic Press: Cambridge, MA, USA, 2021; Volume 13, p. 12. [Google Scholar]
- Djordjevic, M.; Rodic, A.; Graovac, S. From biophysics to ‘omics and systems biology. Eur. Biophys. J. 2019, 48, 413–424. [Google Scholar] [CrossRef]
- Noor, E.; Cherkaoui, S.; Sauer, U. Biological insights through omics data integration. Curr. Opin. Syst. Biol. 2019, 15, 39–47. [Google Scholar] [CrossRef]
- Parsons, J.; Francavilla, C. ′Omics approaches to explore the breast cancer landscape. Front. Cell Dev. Biol. 2020, 7, 395. [Google Scholar] [CrossRef]
- Peng, A.; Mao, X.; Zhong, J.; Fan, S.; Hu, Y. Single-Cell Multi-Omics and Its Prospective Application in Cancer Biology. Proteomics 2020, 20, 1900271. [Google Scholar] [CrossRef]
- Subramanian, I.; Verma, S.; Kumar, S.; Jere, A.; Anamika, K. Multi-omics data integration, interpretation, and its application. Bioinform. Biol. Insights 2020, 14, 1177932219899051. [Google Scholar] [CrossRef]
- Hinderer III, E.W.; Flight, R.M.; Dubey, R.; MacLeod, J.N.; Moseley, H.N. Advances in gene ontology utilization improve statistical power of annotation enrichment. PLoS ONE 2019, 14, e0220728. [Google Scholar] [CrossRef]
- Gene Ontology Consortium. The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef]
- Manda, P. Data mining powered by the gene ontology. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2020, 10, e1359. [Google Scholar] [CrossRef]
- Wang, L.L. Ontology-driven pathway data integration. Ph.D. Dissertation, University of Washington, Seattle, WA, USA, 2019. [Google Scholar]
- Wang, L.; Wang, B.; Quan, Z. Identification of aberrantly methylateddifferentially expressed genes and gene ontology in prostate cancer. Mol. Med. Rep. 2020, 21, 744–758. [Google Scholar] [PubMed]
- Schriml, L.M.; Mitraka, E.; Munro, J.; Tauber, B.; Schor, M.; Nickle, L.; Felix, V.; Jeng, L.; Bearer, C.; Lichenstein, R.; et al. Human Disease Ontology 2018 update: Classification, content and workflow expansion. Nucleic Acids Res. 2019, 47, D955–D962. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Singh, A.P.; Gupta, D. Unsupervised subtyping and methylation landscape of pancreatic ductal adenocarcinoma. Heliyon 2021, 7, e06000. [Google Scholar] [CrossRef]
- Liu, D.; Li, B.; Shi, X.; Zhang, J.; Chen, A.M.; Xu, J.; Wang, W.; Huang, K.; Gao, J.; Zheng, Z.; et al. Cross-platform genomic identification and clinical validation of breast cancer diagnostic biomarkers. Aging 2021, 13, 4258. [Google Scholar] [CrossRef]
- Cai, Z.; Guldbrandtsen, B.; Lund, M.S.; Sahana, G. Prioritizing candidate genes post-GWAS using multiple sources of data for mastitis resistance in dairy cattle. BMC Genom. 2018, 19, 656. [Google Scholar] [CrossRef]
- Yu, L.; Zhao, J.; Gao, L. Predicting potential drugs for breast cancer based on miRNA and tissue specificity. Int. J. Biol. Sci. 2018, 14, 971–982. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, X.; Zheng, L.; Zhao, H.; Li, W.; Wang, B.; Xue, L.; Tian, Y.; Xie, Y. Construction and analysis of the tumor-specific mRNA–miRNA–lncRNA network in gastric cancer. Front. Pharmacol. 2020, 11, 1112. [Google Scholar] [CrossRef]
- Zhou, H.; Chen, J.X.; Yang, C.S.; Yang, M.Q.; Deng, Y.; Wang, H. Gene regulation mediated by microRNAs in response to green tea polyphenol EGCG in mouse lung cancer. BMC Genom. 2014, 15, S3. [Google Scholar] [CrossRef]
- Akgün, M.; Ünal, A.B.; Ergüner, B.; Pfeifer, N.; Kohlbacher, O. Identifying disease-causing mutations with privacy protection. Bioinformatics 2020, 36, 5205–5213. [Google Scholar] [CrossRef]
- Immel, A.; Key, F.M.; Szolek, A.; Barquera, R.; Robinson, M.K.; Harrison, G.F.; Palmer, W.H.; Spyrou, M.A.; Susat, J.; Krause-Kyora, B.; et al. Analysis of genomic DNA from medieval plague victims suggests long-term effect of Yersinia pestis on human immunity genes. Mol. Biol. Evol. 2021, 38, 4059–4076. [Google Scholar] [CrossRef]
- Pooja, M.; Reddy, G.J.; Hema, K.; Dodoala, S.; Koganti, B. Unravelling high-affinity binding compounds towards transmembrane protease serine 2 enzyme in treating SARS-CoV-2 infection using molecular modelling and docking studies. Eur. J. Pharmacol. 2021, 890, 173688. [Google Scholar]
- Jaballah, A.; Soltani, I.; Bahia, W.; Dandana, A.; Hasni, Y.; Miled, A.; Ferchichi, S. The Relationship Between Menopause and Metabolic Syndrome: Experimental and Bioinformatics Analysis. Biochem. Genet. 2021, 59, 1558–1581. [Google Scholar] [CrossRef] [PubMed]
- Djeddi, S.; Reiss, D.; Menuet, A.; Freismuth, S.; de Carvalho Neves, J.; Djerroud, S.; Massana-Muñoz, X.; Sosson, A.S.; Kretz, C.; Raffelsberger, W.; et al. Multi-omics comparisons of different forms of centronuclear myopathies and the effects of several therapeutic strategies. Mol. Ther. 2021, 29, 2514–2534. [Google Scholar] [CrossRef]
- Huang, Z.; Han, Z.; Wang, T.; Shao, W.; Xiang, S.; Salama, P.; Rizkalla, M.; Huang, K.; Zhang, J. TSUNAMI: Translational Bioinformatics tool suite for network analysis and mining. Genom. Proteom. Bioinform. 2021, 19, 1023–1031. [Google Scholar] [CrossRef] [PubMed]
- Thalor, A.; Joon, H.K.; Singh, G.; Roy, S.; Gupta, D. Machine learning assisted analysis of breast cancer gene expression profiles reveals novel potential prognostic biomarkers for triple-negative breast cancer. Comput. Struct. Biotechnol. J. 2022, 20, 1618–1631. [Google Scholar] [CrossRef] [PubMed]
- Ullah, M.A.; Islam, N.N.; Sarkar, B.; Araf, Y.; Zohora, U.S.; Rahman, M.S. Bioinformatics analysis on the prognostic significance of SOX9 gene and its transcriptional product in colorectal cancer. Hum. Gene 2022, 33, 201043. [Google Scholar] [CrossRef]
- Kaur, P. Cancer informatics survey of different grades and subtypes of glioma (P14-9.003). Neurology 2022, 98, 825. [Google Scholar]
- Yi, S.; Yan, Y.; Jin, M.; Bhattacharya, S.; Wang, Y.; Wu, Y.; Yang, L.; Gine, E.; Clot, G.; Chen, L.; et al. Genomic and transcriptomic profiling reveals distinct molecular subsets associated with outcomes in mantle cell lymphoma. J. Clin. Investig. 2022, 132, 3. [Google Scholar] [CrossRef]
- Yu, L.; Lopez, G.; Rassa, J.; Wang, Y.; Basavanhally, T.; Browne, A.; Huang, C.P.; Dorsey, L.; Jen, J.; Hersey, S. Direct comparison of circulating tumor DNA sequencing assays with targeted large gene panels. PLoS ONE 2022, 17, e0266889. [Google Scholar] [CrossRef]
- Xu, H.; Buckeridge, D.L.; Wang, F.; Tarczy-Hornoch, P. Novel Informatics Approaches to COVID-Research: From methods to applications. J. Biomed. Inform. 2022, 129, 104028. [Google Scholar] [CrossRef] [PubMed]
- Battineni, G.; Hossain, M.A.; Chintalapudi, N.; Amenta, F. A Survey on the Role of Artificial Intelligence in Biobanking Studies. Diagnostics 2022, 12, 1179. [Google Scholar] [CrossRef] [PubMed]
- Branco, I.; Choupina, A. Bioinformatics: New tools and applications in life science and personalized medicine. Appl. Microbiol. Biotechnol. 2021, 105, 937–951. [Google Scholar] [CrossRef] [PubMed]
- Pervez, M.T.; Abbas, S.H.; Moustafa, M.F.; Aslam, N.; Shah, S.S.M. A Comprehensive Review of Performance of Next-Generation Sequencing Platforms. BioMed Res. Int. 2022, 2022, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Pereira, R.; Oliveira, J.; Sousa, M. Bioinformatics and computational tools for next-generation sequencing analysis in clinical genetics. J. Clin. Med. 2020, 9, 132. [Google Scholar] [CrossRef]
- Petersen, L.M.; Martin, I.W.; Moschetti, W.E.; Kershaw, C.M.; Tsongalis, G.J. Third-generation sequencing in the clinical laboratory: Exploring the advantages and challenges of nanopore sequencing. J. Clin. Microbiol. 2019, 58, e01315-19. [Google Scholar] [CrossRef]
- Suzuki, Y. Advent of a new sequencing era: Long-read and on-site sequencing. J. Hum. Genet. 2020, 65, 1. [Google Scholar] [CrossRef]
- Xiao, T.; Zhou, W. The third generation sequencing: The advanced approach to genetic diseases. Transl. Pediatr. 2020, 9, 163. [Google Scholar] [CrossRef]
- Klee, E.W.; Cousin, M.A.; Pinto e Vairo, F.; Morales-Rosado, J.A.; Macke, E.L.; Jenkinson, W.G.; Ferrer, A.; Schultz-Rogers, L.E.; Olson, R.J.; Oliver, G.R.; et al. Impact of integrated translational research on clinical exome sequencing. Genet. Med. 2021, 23, 498–507. [Google Scholar] [CrossRef]
- Lightbody, G.; Haberland, V.; Browne, F.; Taggart, L.; Zheng, H.; Parkes, E.; Blayney, J.K. Review of applications of high-throughput sequencing in personalized medicine: Barriers and facilitators of future progress in research and clinical application. Brief. Bioinform. 2019, 20, 1795–1811. [Google Scholar] [CrossRef]
- Sarma, M.K.; Ningthoujam, R.; Panda, M.K.; Babu, P.J.; Srivastava, A.; Das, M.; Singh, Y.D. Translational healthcare system through bioinformatics. In Translational Bioinformatics Applications in Healthcare; CRC Press: Boca Raton, FL, USA, 2021; Volume 1, pp. 3–21. [Google Scholar]
- Hu, Y.; An, Q.; Sheu, K.; Trejo, B.; Fan, S.; Guo, Y. Single cell multi-omics technology: Methodology and application. Front. Cell Dev. Biol. 2018, 6, 28. [Google Scholar] [CrossRef] [PubMed]
- Bock, C.; Farlik, M.; Sheffield, N.C. Multi-omics of single cells: Strategies and applications. Trends Biotechnol. 2016, 34, 605–608. [Google Scholar] [CrossRef] [PubMed]
- Kashima, Y.; Sakamoto, Y.; Kaneko, K.; Seki, M.; Suzuki, Y.; Suzuki, A. Single-cell sequencing techniques from individual to multiomics analyses. Exp. Mol. Med. 2020, 52, 1419–1427. [Google Scholar] [CrossRef]
- Nam, A.S.; Chaligne, R.; Landau, D.A. Integrating genetic and non-genetic determinants of cancer evolution by single-cell multi-omics. Nat. Rev. Genet. 2021, 22, 3–18. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).