Making the Genome Huge: The Case of Triatoma delpontei, a Triatominae Species with More than 50% of Its Genome Full of Satellite DNA
Abstract
1. Introduction
2. Material and Methods
2.1. Samples and DNA Extraction
2.2. Genome Sequencing and Graph-Based Clustering of Sequencing Reads
2.3. Chromosome Preparation and Physical Mapping by Fluorescence In Situ Hybridization
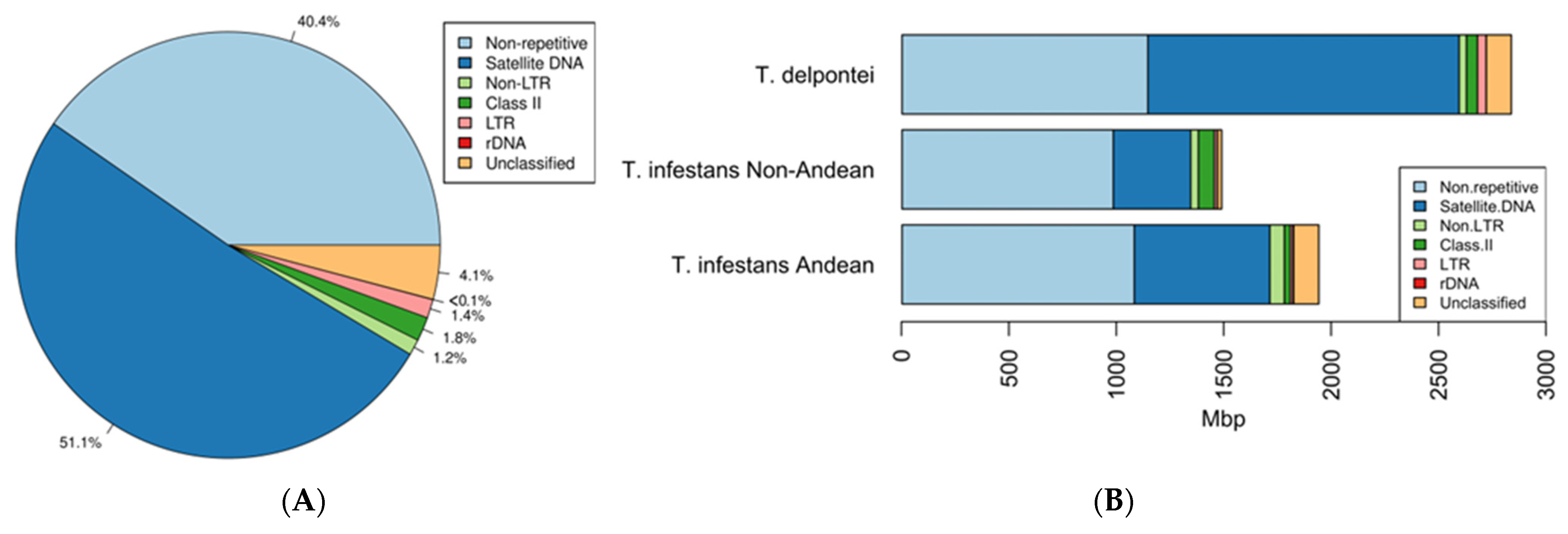
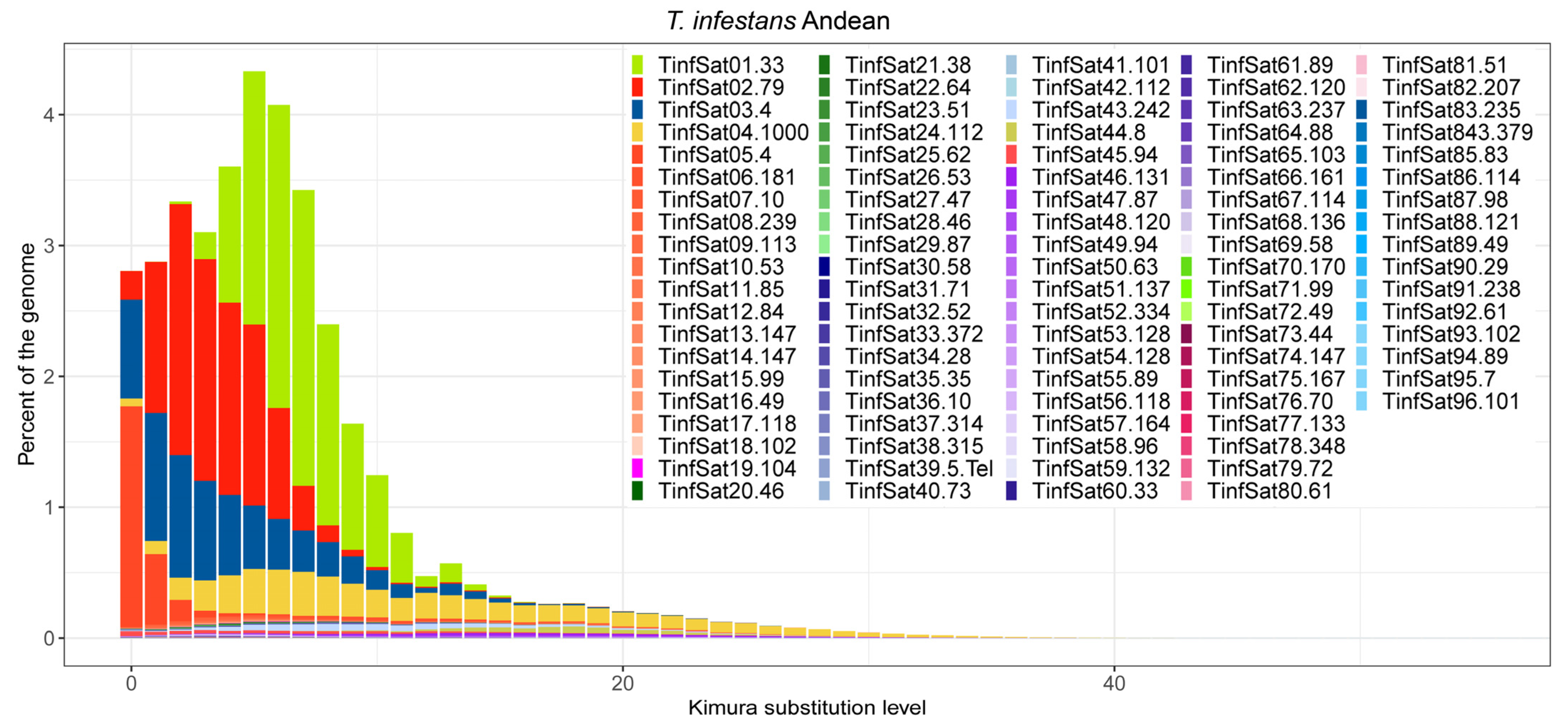
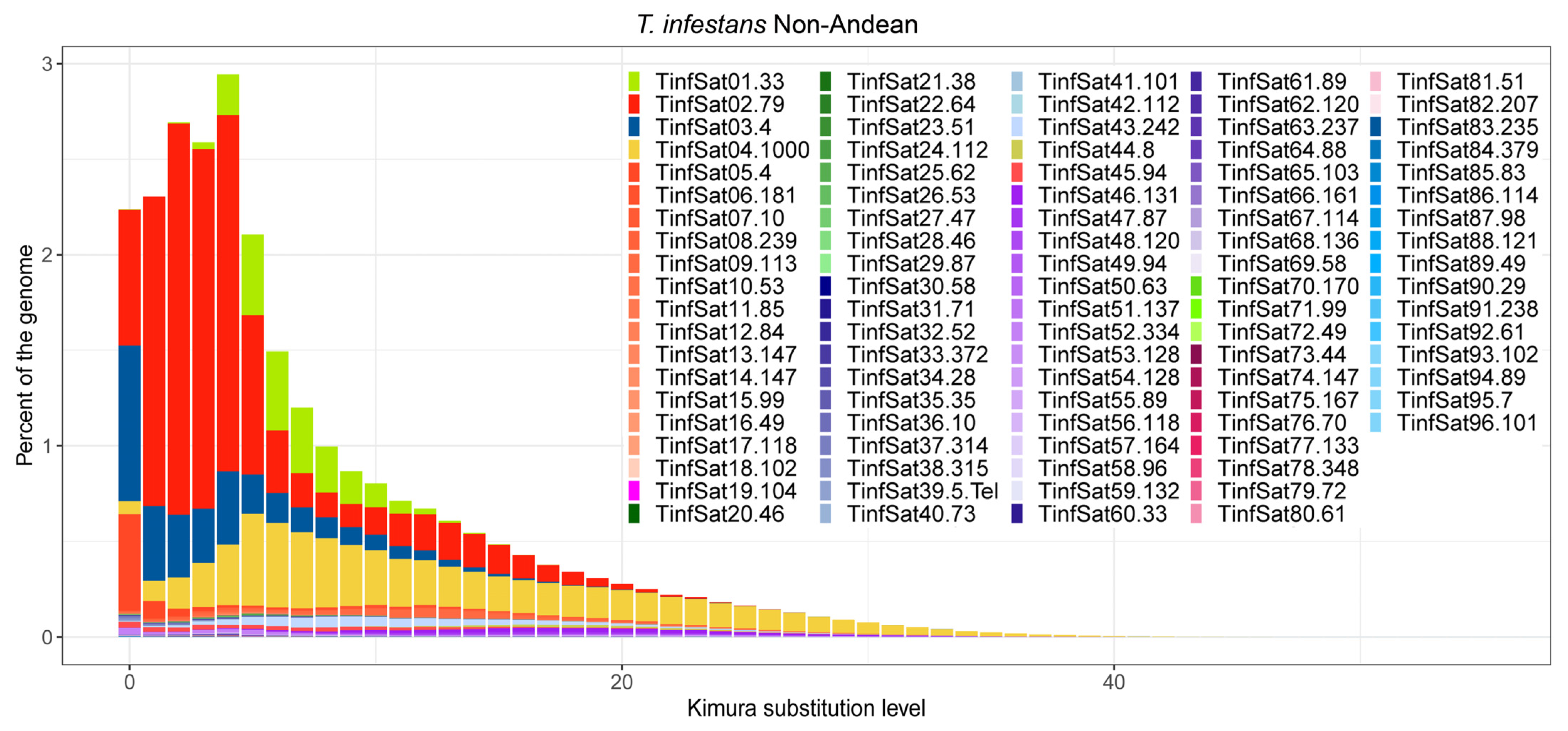
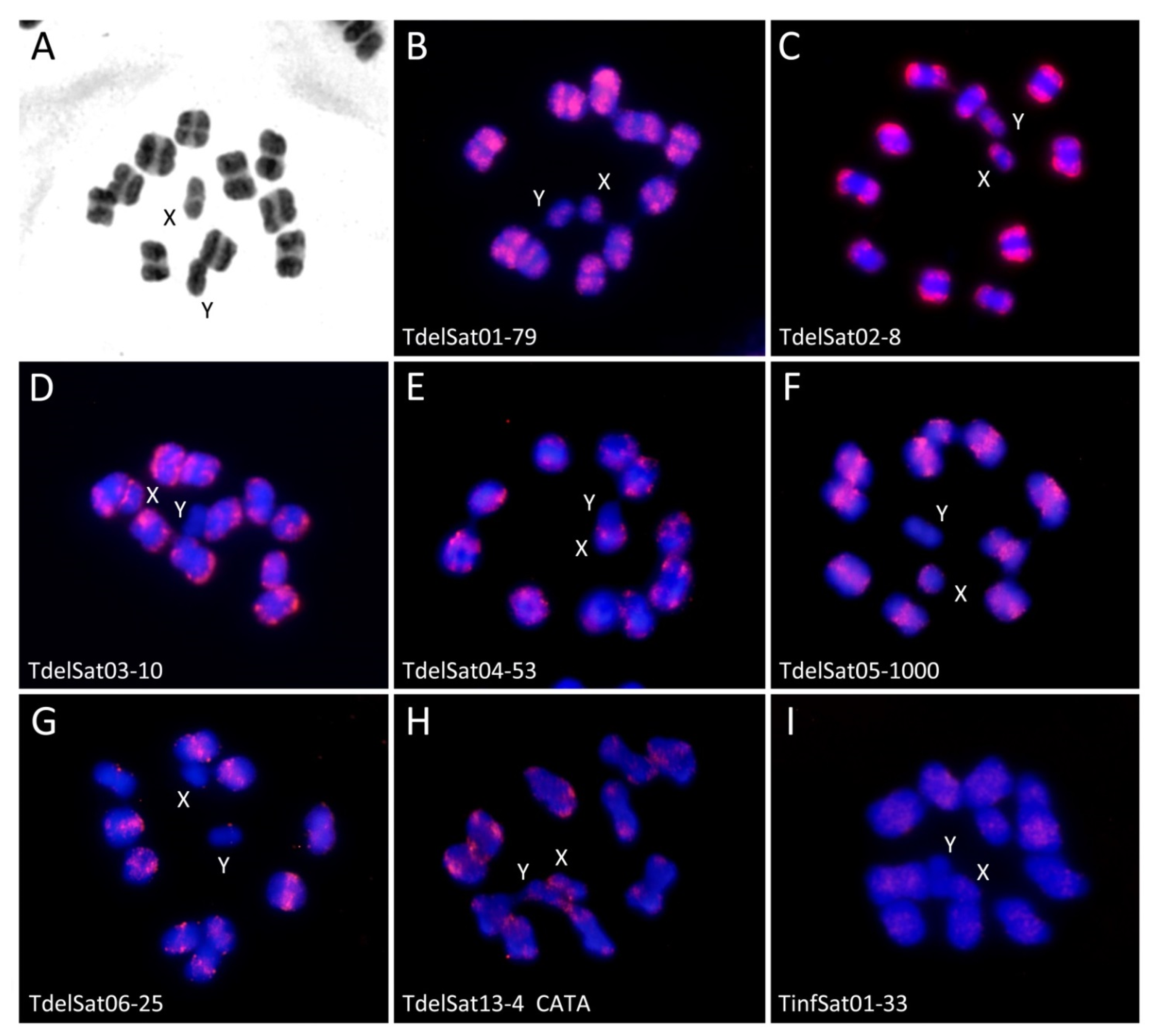
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gil-Santana, H.R.; Chavez, T.; Pita, S.; Panzera, F.; Galvão, C. Panstrongylus noireaui, a remarkable new species of Triatominae (Hemiptera, Reduviidae) from Bolivia. ZooKeys 2022, 1104, 203–225. [Google Scholar] [CrossRef]
- Oliveira Correia, J.P.S.; Gil-Santana, H.R.; Dale, C.; Galvão, C. Triatoma guazu Lent and Wygodzinsky is a junior synonym of Triatoma williami Galvão, Souza and Lima. Insects 2022, 13, 591. [Google Scholar] [CrossRef] [PubMed]
- WHO—World Health Organization. Chagas Disease (Also Known as American trypanosomiasis); WHO: Geneva, Switzerland, 2022; Available online: https://www.who.int/news-room/fact-sheets/detail/chagas-disease-(american-trypanosomiasis) (accessed on 1 December 2022).
- Abad-Franch, F.; Monteiro, F.A. Molecular research and the control of Chagas disease vectors. An. Acad. Bras. Ciênc. 2005, 77, 437–454. [Google Scholar] [CrossRef] [PubMed]
- Dias, J.C.P. Southern Cone initiative for the elimination of domestic populations of Triatoma infestans and the interruption of transfusional Chagas disease. Historical aspects, present situation, and perspectives. Mem. Inst. Oswaldo Cruz 2007, 102, 11–18. [Google Scholar] [CrossRef]
- Panzera, F.; Dujardin, J.P.; Nicolini, P.; Caraccio, M.N.; Rose, V.; Tellez, T.; Bermúdez, H.; Bargues, M.D.; Mas-Coma, S.; O’Connor, J.E.; et al. Genomic changes of Chagas disease vector, South America. Emerg. Infect. Dis. 2004, 10, 438–446. [Google Scholar] [CrossRef]
- Panzera, F.; Ferreiro, M.J.; Pita, S.; Calleros, L.; Pérez, R.; Basmadjián, Y.; Guevara, Y.; Brenière, S.F.; Panzera, Y. Evolutionary and dispersal history of Triatoma infestans, main vector of Chagas disease, by chromosomal markers. Infect. Genet. Evol. 2014, 27, 105–113. [Google Scholar] [CrossRef]
- Bargues, M.D.; Klisiowicz, D.R.; Panzera, F.; Noireau, F.; Marcilla, A.; Perez, R.; Rojas, M.G.; O’Connor, J.E.; Gonzalez-Candelas, F.; Galvão, C.; et al. Origin and phylogeography of the Chagas disease main vector Triatoma infestans based on nuclear rDNA sequences and genome size. Infect. Genet. Evol. 2006, 6, 46–62. [Google Scholar] [CrossRef]
- Waleckx, E.; Salas, R.; Huamán, N.; Buitrago, R.; Bosseno, M.-F.; Aliaga, C.; Barnabé, C.; Rodriguez, R.; Zoveda, F.; Monje, M.; et al. New insights on the Chagas disease main vector Triatoma infestans (Reduviidae, Triatominae) brought by the genetic analysis of Bolivian sylvatic populations. Infect. Genet. Evol. 2011, 11, 1045–1057. [Google Scholar] [CrossRef]
- Moriconi, D.E.; Macedo-Lopes, C.; Sartorio, A.; Juárez, M.P.; Girotti, J.R.; Calderón-Fernández, G.M. Chemotaxonomy of five South American species of the Triatoma genus (Hemiptera: Reduviidae: Triatominae) based on their cuticle hydrocarbon pattern. J. Med. Entomol. 2022, 59, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Lent, H.; Wygodzinsky, P. Revision of the Triatominae (Hemiptera, Reduviidae) and their significance as vector of Chagas disease. Bull. Am. Mus. Nat. Hist. 1979, 163, 123–520. [Google Scholar]
- Monteiro, F.A.; Weirauch, C.; Felix, M.; Lazoski, C.; Abad-Franch, F. Evolution, systematics, and biogeography of the Triatominae, vectors of Chagas disease. Adv. Parasitol. 2018, 99, 265–344. [Google Scholar] [CrossRef] [PubMed]
- Kieran, T.J.; Gordon, E.R.L.; Zaldívar-Riverón, A.; Ibarra-Cerdeña, C.N.; Glenn, T.C.; Weirauch, C. Ultraconserved elements reconstruct the evolution of Chagas disease-vectoring kissing bugs (Reduviidae: Triatominae). Syst. Entomol. 2021, 46, 725–740. [Google Scholar] [CrossRef]
- Hughes-Schrader, S.; Schrader, F. The kinetochore of the Hemiptera. Chromosoma 1961, 12, 327–350. [Google Scholar] [CrossRef] [PubMed]
- Panzera, F.; Perez, R.; Panzera, Y.; Alvarez, F.; Scvortzoff, E.; Salvatella, R. Karyotype evolution in holocentric chromosomes of three related species of Triatomines (Hemiptera-Reduviidae). Chromosome Res. 1995, 3, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Panzera, Y.; Pita, S.; Ferreiro, M.J.; Ferrandis, I.; Lages, C.; Pérez, R.; Silva, A.E.; Guerra, M.; Panzera, F. High dynamics of rDNA cluster location in kissing bug holocentric chromosomes (Triatominae, Heteroptera). Cytogenet. Genome Res. 2012, 138, 56–67. [Google Scholar] [CrossRef]
- Bardella, V.B.; Pita, S.; Vanzela, A.L.L.; Galvão, C.; Panzera, F. Heterochromatin base pair composition and diversification in holocentric chromosomes of kissing bugs (Hemiptera, Reduviidae). Mem. Inst. Oswaldo Cruz 2016, 111, 614–624. [Google Scholar] [CrossRef] [PubMed]
- Pita, S.; Lorite, P.; Nattero, J.; Galvão, C.; Alevi, K.C.C.; Teves, S.C.; Azeredo-Oliveira, M.T.V.; Panzera, F. New arrangements on several species subcomplexes of Triatoma genus based on the chromosomal position of ribosomal genes (Hemiptera—Triatominae). Infect. Genet. Evol. 2016, 43, 225–231. [Google Scholar] [CrossRef]
- Pita, S.; Lorite, P.; Cuadrado, A.; Panzera, Y.; De Oliveira, J.; Alevi, K.C.C.; Rosa, J.A.; Freitas, S.P.C.; Gómez-Palacio, A.; Solari, A.; et al. High chromosomal mobility of rDNA clusters in holocentric chromosomes of Triatominae, vectors of Chagas disease (Hemiptera-Reduviidae). Med. Vet. Entomol. 2021, 36, 66–80. [Google Scholar] [CrossRef]
- Panzera, F.; Pita, S.; Lorite, P. Chromosome structure and evolution of Triatominae: A review. In Triatominae—The Biology of Chagas Disease Vectors; Entomology in Focus; Guarneri, A., Lorenzo, M., Eds.; Springer: New York, NY, USA, 2021; Volume 5, pp. 65–99. [Google Scholar]
- Panzera, F.; Ferrandis, I.; Ramsey, J.; Salazar-Schettino, P.M.; Cabrera, M.; Monroy, C.; Bargues, M.D.; Mas-Coma, S.; O’Connor, J.E.; Angulo, V.M.; et al. Genome size determination in Chagas disease transmitting bugs (hemiptera-triatominae) by flow cytometry. Am. J. Trop. Med. Hyg. 2007, 76, 516–521. [Google Scholar] [CrossRef]
- Sadílek, D.; Urfus, T.; Vilímová, J.; Hadrava, J.; Suda, J. Nuclear genome size in contrast to sex chromosome number variability in the human bed bug, Cimex lectularius (Heteroptera: Cimicidae). Cytometry A 2019, 95, 746–756. [Google Scholar] [CrossRef]
- Pita, S.; Panzera, F.; Sánchez, A.; Panzera, Y.; Palomeque, T.; Lorite, P. Distribution and evolution of repeated sequences in genomes of Triatominae (Hemiptera-Reduviidae) inferred from genomic in situ hybridization. PLoS ONE 2014, 9, e114298. [Google Scholar] [CrossRef] [PubMed]
- Pita, S.; Panzera, F.; Sánchez, A.; Palomeque, T.; Lorite, P. Chromosome painting in triatomine insects reveals shared sequences between X chromosomes and autosomes. J. Med. Entomol. 2017, 54, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Pita, S.; Lorite, P.; Vela, J.; Mora, P.; Palomeque, T.; Thi, K.P.; Panzera, F. Holocentric chromosome evolution in kissing bugs (Hemiptera: Reduviidae: Triatominae): Diversification of repeated sequences. Parasit. Vectors 2017, 10, 410. [Google Scholar] [CrossRef]
- Pita, S.; Panzera, F.; Mora, P.; Vela, J.; Cuadrado, Á.; Sánchez, A.; Palomeque, T.; Lorite, P. Comparative repeatome analysis on Triatoma infestans Andean and Non-Andean lineages, main vector of Chagas disease. PLoS ONE 2017, 12, e0181635. [Google Scholar] [CrossRef] [PubMed]
- Maumus, F.; Quesneville, H. Deep investigation of Arabidopsis thaliana junk DNA reveals a continuum between repetitive elements and genomic dark matter. PLoS ONE 2014, 9, e94101. [Google Scholar] [CrossRef] [PubMed]
- Mérel, V.; Boulesteix, M.; Fablet, M.; Vieira, C. Transposable elements in Drosophila. Mob. DNA 2020, 11, 23. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.N.; Feschotte, C. A field guide to eukaryotic transposable elements. Annu. Rev. Genet. 2020, 54, 539–561. [Google Scholar] [CrossRef]
- Palomeque, T.; Lorite, P. Satellite DNA in insects: A review. Heredity 2008, 100, 564–573. [Google Scholar] [CrossRef]
- Montiel, E.E.; Panzera, F.; Palomeque, T.; Lorite, P.; Pita, S. Satellitome analysis of Rhodnius prolixus, one of the main Chagas disease vector species. Int. J. Mol. Sci. 2021, 22, 6052. [Google Scholar] [CrossRef]
- Pezer, Ž.; Brajković, J.; Feliciello, I.; Ugarković, Đ. Transcription of satellite DNAs in insects. Prog. Mol. Subcell Biol. 2011, 51, 161–178. [Google Scholar] [CrossRef]
- Ferreira, D.; Meles, S.; Escudeiro, A.; Mendes-da-Silva, A.; Adega, F.; Chaves, R. Satellite non-coding RNAs: The emerging players in cells, cellular pathways and cancer. Chromosome Res. 2015, 23, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Ferree, P.M.; Prasad, S. How can satellite DNA divergence cause reproductive isolation? Let us count the chromosomal ways. Genet. Res. Int. 2012, 2012, e430136. [Google Scholar] [CrossRef] [PubMed]
- de Lima, L.G.; Ruiz-Ruano, F.J. In-depth satellitome analyses of 37 Drosophila species illuminate repetitive DNA evolution in the Drosophila genus. Genome Biol. Evol. 2022, 14, evac064. [Google Scholar] [CrossRef]
- Jagannathan, M.; Yamashita, Y.M. Defective Satellite DNA clustering into chromocenters underlies hybrid incompatibility in Drosophila. Mol. Bio. Evol. 2021, 38, 4977–4986. [Google Scholar] [CrossRef]
- Hughes, S.E.; Hawley, R.S. Heterochromatin: A rapidly evolving species barrier. PLoS Biol. 2009, 7, e1000233. [Google Scholar] [CrossRef] [PubMed]
- Mora, P.; Vela, J.; Ruiz-Ruano, F.J.; Ruiz-Mena, A.; Montiel, E.E.; Palomeque, T.; Lorite, P. Satellitome analysis in the ladybird beetle Hippodamia variegata (Coleoptera, Coccinellidae). Genes 2020, 11, 783. [Google Scholar] [CrossRef]
- Montiel, E.E.; Mora, P.; Rico-Porras, J.M.; Palomeque, T.; Lorite, P. Satellitome of the red palm weevil, Rhynchophorus ferrugineus (Coleoptera: Curculionidae), the most diverse among insects. Front. Ecol. Evol. 2022, 10, 826808. [Google Scholar] [CrossRef]
- Silva, B.S.M.L.; Heringer, P.; Dias, G.B.; Svartman, M.; Kuhn, G.C.S. De novo identification of satellite DNAs in the sequenced genomes of Drosophila virilis and D. americana using the RepeatExplorer and TAREAN pipelines. PLoS ONE 2019, 14, e0223466. [Google Scholar] [CrossRef]
- Bardella, V.B.; Milani, D.; Cabral-de-Mello, D.C. Analysis of Holhymenia histrio genome provides insight into the satDNA evolution in an insect with holocentric chromosomes. Chromosome Res. 2020, 28, 369–380. [Google Scholar] [CrossRef]
- Cabral-de-Mello, D.C.; Zrzavá, M.; Kubíčková, S.; Rendón, P.; Marec, F. The role of satellite DNAs in genome architecture and sex chromosome evolution in Crambidae moths. Front. Genet. 2021, 12, 661417. [Google Scholar] [CrossRef]
- Gasparotto, A.E.; Milani, D.; Martí, E.; Bardella, V.B.; Ferretti, A.B.S.M.; Hickmann, F.; Zrzavá, M.; Marec, F.; Cabral-de-Mello, D.C. A step forward in the genome characterization of the sugarcane borer, Diatraea saccharalis: Karyotype analysis, sex chromosome system and repetitive DNAs through a cytogenomic approach. Chromosoma 2022, 131, 253–267. [Google Scholar] [CrossRef] [PubMed]
- Pereira, J.A.; Milani, D.; Ferretti, A.B.S.M.; Bardella, V.B.; Cabral-de-Mello, D.C.; Lopes, D.M. The extensive amplification of heterochromatin in Melipona bees revealed by high throughput genomic and chromosomal analysis. Chromosoma 2021, 130, 251–262. [Google Scholar] [CrossRef]
- Ruiz-Ruano, F.J.; López-León, M.D.; Cabrero, J.; Camacho, J.P.M. High-throughput analysis of the satellitome illuminates satellite DNA evolution. Sci. Rep. 2016, 6, 28333. [Google Scholar] [CrossRef] [PubMed]
- Palacios-Gimenez, O.M.; Milani, D.; Song, H.; Marti, D.A.; López-León, M.D.; Ruiz-Ruano, F.J.; Camacho, J.P.M.; Cabral-de-Mello, D.C. Eight million years of satellite DNA evolution in grasshoppers of the genus Schistocerca illuminate the ins and outs of the library hypothesis. Genome Biol. Evol. 2020, 12, 88–102. [Google Scholar] [CrossRef]
- Camacho, J.P.M.; Cabrero, J.; López-León, M.D.; Martín-Peciña, M.; Perfectti, F.; Garrido-Ramos, M.A.; Ruiz-Ruano, F.J. Satellitome comparison of two oedipodine grasshoppers highlights the contingent nature of satellite DNA evolution. BMC Biol. 2022, 20, 36. [Google Scholar] [CrossRef]
- Tunjić-Cvitanić, M.; Pasantes, J.J.; García-Souto, D.; Cvitanić, T.; Plohl, M.; Šatović-Vukšić, E. Satellitome analysis of the pacific oyster Crassostrea gigas reveals new pattern of satellite DNA organization, highly scattered across the genome. Int. J. Mol. Sci. 2021, 22, 6798. [Google Scholar] [CrossRef]
- Boštjančić, L.L.; Bonassin, L.; Anušić, L.; Lovrenčić, L.; Besendorfer, V.; Maguire, I.; Grandjean, F.; Austin, C.M.; Greve, C.; Hamadou, A.B.; et al. The Pontastacus leptodactylus (Astacidae) repeatome provides insight into genome evolution and reveals remarkable diversity of satellite DNA. Front. Genet. 2021, 11, 611745. [Google Scholar] [CrossRef]
- Lower, S.S.; McGurk, M.P.; Clark, A.G.; Barbash, D.A. Satellite DNA evolution: Old ideas, new approaches. Curr. Opin. Genet. Dev. 2018, 49, 70–78. [Google Scholar] [CrossRef]
- Satovic, E.; Tunjić Cvitanić, M.; Plohl, M. Tools and databases for solving problems in detection and identification of repetitive DNA sequences. Period. Biol. 2020, 121–122, 7–14. [Google Scholar] [CrossRef]
- Novák, P.; Neumann, P.; Pech, J.; Steinhaisl, J.; Macas, J. RepeatExplorer: A galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 2013, 29, 792–793. [Google Scholar] [CrossRef]
- Novák, P.; Neumann, P.; Macas, J. Global Analysis of Repetitive DNA from unassembled sequence reads using RepeatExplorer2. Nat. Protoc. 2020, 15, 3745–3776. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Novák, P.; Ávila Robledillo, L.; Koblížková, A.; Vrbová, I.; Neumann, P.; Macas, J. TAREAN: A computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acids Res. 2017, 45, e111. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 1 November 2022).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; Available online: https://ggplot2.tidyverse.org (accessed on 24 November 2022).
- Negm, S.; Greenberg, A.; Larracuente, A.M.; Sproul, J.S. RepeatProfiler: A Pipeline for visualization and comparative analysis of repetitive DNA profiles. Mol. Ecol. Resour. 2021, 21, 969–981. [Google Scholar] [CrossRef]
- Palomeque, T.; Muñoz-López, M.; Carrillo, J.A.; Lorite, P. Characterization and evolutionary dynamics of a complex family of satellite DNA in the leaf beetle Chrysolina carnifex (Coleoptera, Chrysomelidae). Chromosome Res. 2005, 13, 795–807. [Google Scholar] [CrossRef]
- Pita, S.; Mora, P.; Vela, J.; Palomeque, T.; Sánchez, A.; Panzera, F.; Lorite, P. Comparative analysis of repetitive DNA between the main vectors of Chagas disease: Triatoma infestans and Rhodnius prolixus. Int. J. Mol. Sci. 2018, 19, 1277. [Google Scholar] [CrossRef]
- Castro, M.R.J.; Goubert, C.; Monteiro, F.A.; Vieira, C.; Carareto, C.M.A. Homology-free detection of transposable elements unveils their dynamics in three ecologically distinct Rhodnius species. Genes 2020, 11, 170. [Google Scholar] [CrossRef]
- Petitpierre, E.; Gatewood, J.M.; Schmid, C.W. Satellite DNA from the beetle Tenebrio molitor. Experientia 1988, 44, 498–499. [Google Scholar] [CrossRef]
- Marino, A.; Kizenko, A.; Wong, W.Y.; Ghiselli, F.; Simakov, O. Repeat age decomposition informs an ancient set of repeats associated with coleoid cephalopod divergence. Front Genet. 2022, 13, 793734. [Google Scholar] [CrossRef]
- Ugarković, Ð.; Podnar, M.; Plohl, M. Satellite DNA of the red flour beetle Tribolium castaneum—Comparative study of satellites from the genus Tribolium. Mol. Biol. Evol. 1996, 13, 1059–1066. [Google Scholar] [CrossRef]
- Huang, Y.C.; Lee, C.C.; Kao, C.Y.; Chang, N.C.; Lin, C.C.; Shoemaker, D.; Wang, J. Evolution of long centromeres in fire ants. BMC Evol. Biol. 2016, 16, 189. [Google Scholar] [CrossRef] [PubMed]
- Fry, K.; Salser, W. Nucleotide sequences of HS-alpha satellite DNA from kangaroo rat Dipodomys ordii and characterization of similar sequences in other rodents. Cell 1977, 12, 1069–1084. [Google Scholar] [CrossRef] [PubMed]
- Mestrović, N.; Plohl, M.; Mravinac, B.; Ugarković, Ð. Evolution of satellite DNAs from the genus Palorus--experimental evidence for the “library” hypothesis. Mol. Biol. Evol. 1998, 15, 1062–1068. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.H.; Grenier, J.K.; Barbash, D.A.; Clark, A.G. Correlated variation and population differentiation in satellite DNA abundance among lines of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2014, 111, 18793–18798. [Google Scholar] [CrossRef]
- Feliciello, I.; Akrap, I.; Brajković, J.; Zlatar, I.; Ugarković, Đ. Satellite DNA as a driver of population divergence in the red flour beetle Tribolium castaneum. Genome Biol. Evol. 2014, 7, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Haq, I.U.; Muhammad, M.; Yuan, H.; Ali, S.; Abbasi, A.; Asad, M.; Ashraf, H.J.; Khurshid, A.; Zhang, K.; Zhang, Q.; et al. Satellitome analysis and transposable elements comparison in geographically distant populations of Spodoptera frugiperda. Life 2022, 12, 521. [Google Scholar] [CrossRef]
- Ruiz-Ruano, F.J.; Castillo-Martínez, J.; Cabrero, J.; Gómez, R.; Camacho, J.P.M.; López-León, M.D. High-throughput analysis of satellite DNA in the grasshopper Pyrgomorpha conica reveals abundance of homologous and heterologous higher-order repeats. Chromosoma 2018, 127, 323–340. [Google Scholar] [CrossRef]
- Milani, D.; Bardella, V.B.; Ferretti, A.B.S.M.; Palacios-Gimenez, O.M.; de Melo, A.S.; Moura, R.C.; Loreto, V.; Song, H.; Cabral-de-Mello, D.C. Satellite DNAs unveil clues about the ancestry and composition of B chromosomes in three grasshopper species. Genes 2018, 9, 523. [Google Scholar] [CrossRef]
- Lorite, P.; Carrillo, J.A.; Aguilar, J.A.; Palomeque, T. Isolation and characterization of two families of satellite DNA with repetitive units of 135 bp and 2.5 kb in the ant Monomorium subopacum (Hymenoptera, Formicidae). Cytogenet. Genome Res. 2004, 105, 83–92. [Google Scholar] [CrossRef]
- Ruiz-Ruano, F.J.; Navarro-Domínguez, B.; Camacho, J.P.M.; Garrido-Ramos, M.A. Characterization of the satellitome in lower vascular plants: The case of the endangered fern Vandenboschia speciosa. Ann. Bot. 2019, 123, 587–599. [Google Scholar] [CrossRef]

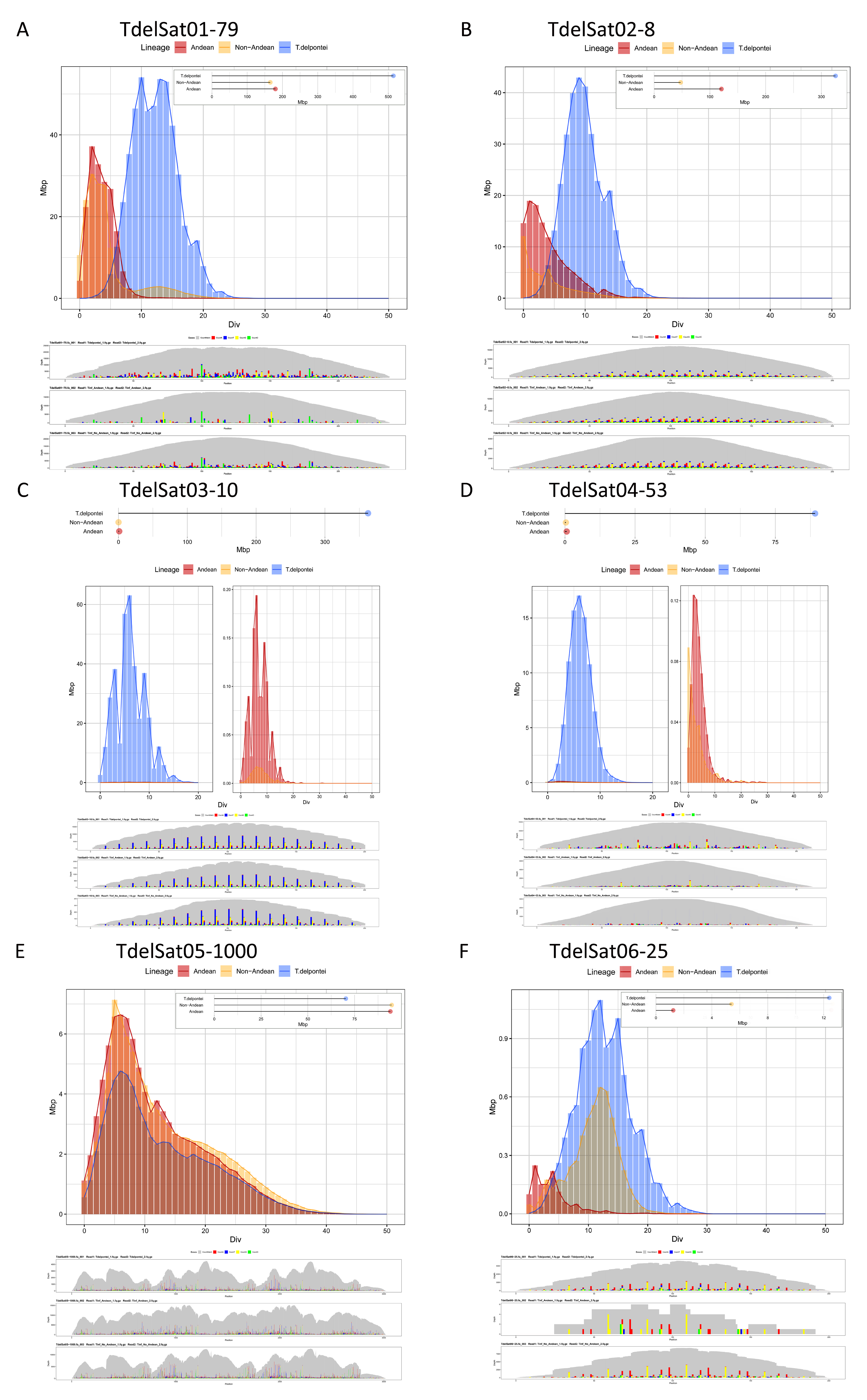
| SatDNA Family | Oligonucleotide * | Sequence |
|---|---|---|
| TdelSat01-79 | TinfSat02-79-F | 5′-TTGTAAGGTTCAAGAAAATCCC |
| TinfSat02-79-R | 5′-CTCACTCTTACGGTTGAAACGC | |
| TdelSat02-8 | (GATA)5 | 5′-GATAGATAGATAGATAGATA |
| TdelSat03-10 | TinfSat07-10 | 5′-RCATACTCGKRCATACTCGK |
| TdelSat04-53 | TinfSat10-53-F | 5′-CGGTTTTGGTTATACTATTTTTCCA |
| TinfSat10-53-R | 5′-GAAGGGGCAAACGTGTATT | |
| TdelSat05-1000 | TinfSat04-1000-F | 5′-GATATCGAAAATTTGACACG |
| TinfSat04-1000-R | 5′-ATGTATGTGAACAGCATAGC | |
| TdelSat06-25 | TinfSat09-113 | 5′-AGAATGTAKAACTTTG |
| TdelSat13-4 | (CATA)5 | 5′-CATACATACATACATACATA |
| TinfSat01-33 | TinfSat01-33 | 5′-TTTCCATAAGTCTATTACTTCGTAATTACTGCG |
| Name | Genome Abundance (%) | Similarity with T. infestans | Genome Abundance in T. infestans (%) | |
|---|---|---|---|---|
| Andean | Non-Andean | |||
| TdelSat01-79 | 18.158 | TinfSat02-79 | 9.300 | 11.127 |
| TdelSat02-8 (GATAGTTA) | 14.21 | TinfSat03-4 | 6.236 | 3.202 |
| TdelSat03-10 | 12.786 | TinfSat07-10 /36-10 | 0.066 | 0.01 |
| TdelSat04-53 | 3.146 | TinfSat10-53 | 0.033 | 0.024 |
| TdelSat05-1000 | 2.502 | TinfSat04-1000 | 4.927 | 6.473 |
| TdelSat06-25 | 0.437 | TinfSat09-113 | 0.064 | 0.358 |
| TdelSat07-334 | 0.410 | TinfSat52-334 | 0.028 | 0.150 |
| TdelSat08-247 | 0.303 | TinfSat43-242 | 0.697 | 0.697 |
| TdelSat09-181 | 0.173 | TinfSat06-181 | 0.204 | 0.129 |
| TdelSat10-315 | 0.117 | TinfSat37-314 | N.D. | 0.037 |
| TdelSat11-239 | 0.092 | TinfSat08-239 | 0.196 | 0.184 |
| TdelSat12-52 | 0.086 | TinfSat32-52 | 0.008 | 0.012 |
| TdelSat13-4 (CATA) | 0.056 | TinfSat05-4 | 2.627 | 0.734 |
| TdelSat14-94 | 0.053 | TinfSat45-94 | 0.318 | 0.342 |
| TdelSat15-93 | 0.053 | TinfSat49-94 | 0.033 | 0.045 |
| TdelSat16-31 | 0.053 | N.D. | ||
| TdelSat17-84 | 0.045 | TinfSat12-84 | 0.049 | 0.047 |
| TdelSat18-62 | 0.040 | TinfSat25-62 | 0.012 | 0.019 |
| TdelSat19-58 | 0.040 | TinfSat30-58 | 0.013 | 0.005 |
| TdelSat20-27 | 0.035 | R.P. | ||
| TdelSat21-33 | 0.034 | TinfSat60-33 | 0.014 | 0.015 |
| TdelSat22-104 | 0.033 | TinfSat19-104 | 0.020 | 0.002 |
| TdelSat23-112 | 0.031 | TinfSat42-112 | 0.014 | 0.017 |
| TdelSat24-147 | 0.030 | TinfSat14-147 | 0.031 | 0.032 |
| TdelSat25-138 | 0.028 | N.D. | ||
| TdelSat26-32 | 0.028 | N.D. | ||
| TdelSat27-57 | 0.026 | N.D. | ||
| TdelSat28-5 telomere | 0.024 | TinfSat39-5 | 0.021 | 0.011 |
| TdelSat29-137 | 0.023 | TinfSat51-137 | 0.032 | 0.041 |
| TdelSat30-38 | 0.022 | N.D. | ||
| TdelSat31-52 | 0.022 | R.P. | ||
| TdelSat32-49 | 0.022 | TinfSat16-49 | 0.015 | 0.004 |
| TdelSat33-46 | 0.022 | N.D. | ||
| TdelSat34-27 | 0.019 | R.P. | ||
| TdelSat35-188 | 0.018 | N.D. | ||
| TdelSat36-128 | 0.018 | TinfSat53-128 | 0.027 | 0.035 |
| TdelSat37-38 | 0.017 | R.P. | ||
| TdelSat38-147 | 0.016 | TinfSat13-147 | 0.025 | 0.014 |
| TdelSat39-18 | 0.016 | R.P. | ||
| TdelSat40-72 | 0.016 | R.P. | ||
| TdelSat41-49 | 0.015 | N.D. | ||
| TdelSat42-128 | 0.015 | N.D. | ||
| TdelSat43-111 | 0.015 | R.P. | ||
| TdelSat44-34 | 0.014 | R.P. | ||
| TdelSat45-102 | 0.014 | TinfSat18-102 | 0.014 | 0.006 |
| TdelSat46-114 | 0.014 | TinfSat67-114 | 0.008 | 0.007 |
| TdelSat47-25 | 0.014 | N.D. | ||
| TdelSat48-118 | 0.014 | TinfSat56-118 | 0.023 | 0.026 |
| TdelSat49-49 | 0.014 | R.P. | ||
| TdelSat50-101 | 0.013 | TinfSat96-101 | <0.001 | 0.01 |
| TdelSat51-46 | 0.013 | TinfSat28-46 | 0.008 | 0.009 |
| TdelSat52-88 | 0.013 | TinfSat64-88 | 0.009 | 0.003 |
| TdelSat53-48 | 0.013 | R.P. | ||
| TdelSat54-189 | 0.013 | R.P. | ||
| TdelSat55-51 | 0.012 | N.D. | ||
| TdelSat56-102 | 0.012 | N.D. | ||
| TdelSat57-271 | 0.012 | R.P. | ||
| TdelSat58-84 | 0.011 | N.D. | ||
| TdelSat59-63 | 0.011 | TinfSat50-63 | 0.032 | 0.011 |
| TdelSat60-99 | 0.011 | TinfSat15-99 | 0.013 | 0.011 |
| TdelSat61-315 | 0.010 | TinfSat38-315 | N.D. | 0.031 |
| TdelSat62-53 | 0.009 | N.D. | ||
| TdelSat63-51 | 0.009 | N.D. | ||
| TdelSat64-39 | 0.009 | R.P. | ||
| TdelSat65-71 | 0.008 | N.D. | ||
| TdelSat66-64 | 0.008 | TinfSat22-64 | 0.012 | 0.002 |
| TdelSat67-83 | 0.008 | TinfSat85-83 | 0.001 | 0.002 |
| TdelSat68-160 | 0.008 | TinfSat66-161 | 0.008 | 0.011 |
| TdelSat69-52 | 0.007 | N.D. | ||
| TdelSat70-86 | 0.007 | TinfSat47-87 | 0.133 | 0.135 |
| TdelSat71-102 | 0.007 | R.P. | ||
| TdelSat72-85 | 0.007 | R.P. | ||
| TdelSat73-372 | 0.007 | TinfSat33-372 | 0.015 | 0.017 |
| TdelSat74-137 | 0.007 | N.D. | ||
| TdelSat75-25 | 0.007 | R.P. | ||
| TdelSat76-96 | 0.007 | TinfSat58-96 | 0.016 | 0.010 |
| TdelSat77-102 | 0.007 | R.P. | ||
| TdelSat78-29 | 0.006 | R.P. | ||
| TdelSat79-26 | 0.006 | R.P. | ||
| TdelSat80-52 | 0.006 | R.P. | ||
| TdelSat81-185 | 0.006 | R.P. | ||
| TdelSat82-108 | 0.006 | R.P. | ||
| TdelSat83-66 | 0.006 | R.P. | ||
| TdelSat84-73 | 0.006 | R.P. | ||
| TdelSat85-49 | 0.006 | N.D. | ||
| TdelSat86-164 | 0.005 | TinfSat57-164 | 0.017 | 0.006 |
| TdelSat87-61 | 0.005 | N.D. | ||
| TdelSat88-87 | 0.005 | TinfSat55-89 | 0.026 | 0.031 |
| TdelSat89-86 | 0.005 | N.D. | ||
| TdelSat90-157 | 0.005 | R.P. | ||
| TdelSat91-23 | 0.005 | R.P. | ||
| TdelSat92-196 | 0.005 | N.D. | ||
| TdelSat93-57 | 0.004 | R.P. | ||
| TdelSat94-72 | 0.004 | TinfSat79-72 | 0.003 | 0.001 |
| TdelSat95-38 | 0.004 | R.P. | ||
| TdelSat96-58 | 0.004 | R.P. | ||
| TdelSat97-238 | 0.004 | R.P. | ||
| TdelSat98-104 | 0.004 | R.P. | ||
| TdelSat99-128 | 0.004 | N.D. | ||
| TdelSat100-99 | 0.004 | N.D. | ||
| TdelSat101-55 | 0.004 | N.D. | ||
| TdelSat102-39 | 0.004 | R.P. | ||
| TdelSat103-138 | 0.004 | R.P. | ||
| TdelSat104-78 | 0.004 | N.D. | ||
| TdelSat105-35 | 0.004 | R.P. | ||
| TdelSat106-44 | 0.004 | TinfSat27-47 | 0.008 | 0.008 |
| TdelSat107-57 | 0.004 | R.P. | ||
| TdelSat108-81 | 0.004 | R.P. | ||
| TdelSat109-142 | 0.004 | R.P. | ||
| TdelSat110-99 | 0.004 | R.P. | ||
| TdelSat111-7 | 0.004 | TinfSat95-7 | <0.001 | 0.005 |
| TdelSat112-113 | 0.004 | R.P. | ||
| TdelSat113-88 | 0.004 | N.D. | ||
| TdelSat114-56 | 0.004 | R.P. | ||
| TdelSat115-15 | 0.004 | N.D. | ||
| TdelSat116-66 | 0.003 | R.P. | ||
| TdelSat117-46 | 0.003 | N.D. | ||
| TdelSat118-207 | 0.003 | TinfSat82-207 | 0.002 | 0.002 |
| TdelSat119-348 | 0.003 | TinfSat78-348 | 0.004 | 0.004 |
| TdelSat120-101 | 0.003 | R.P. | ||
| TdelSat121-51 | 0.003 | R.P. | ||
| TdelSat122-43 | 0.003 | R.P. | ||
| TdelSat123-149 | 0.003 | R.P. | ||
| TdelSat124-69 | 0.003 | N.D. | ||
| TdelSat125-35 | 0.003 | R.P. | ||
| TdelSat126-46 | 0.003 | R.P. | ||
| TdelSat127-49 | 0.003 | R.P. | ||
| TdelSat128-66 | 0.003 | N.D. | ||
| TdelSat129-60 | 0.002 | R.P. | ||
| TdelSat130-105 | 0.002 | R.P. | ||
| TdelSat131-66 | 0.002 | R.P. | ||
| TdelSat132-39 | 0.002 | R.P. | ||
| TdelSat133-28 | 0.002 | R.P. | ||
| TdelSat134-32 | 0.002 | R.P. | ||
| TdelSat135-105 | 0.002 | R.P. | ||
| TdelSat136-61 | 0.002 | R.P. | ||
| TdelSat137-183 | 0.002 | N.D. | ||
| TdelSat138-63 | 0.002 | R.P. | ||
| TdelSat139-152 | 0.002 | N.D. | ||
| TdelSat140-68 | 0.002 | N.D. | ||
| TdelSat141-369 | 0.002 | R.P. | ||
| TdelSat142-23 | 0.002 | N.D. | ||
| TdelSat143-52 | 0.002 | N.D. | ||
| TdelSat144-121 | 0.002 | N.D. | ||
| TdelSat145-36 | 0.002 | R.P. | ||
| TdelSat146-62 | 0.002 | N.D. | ||
| TdelSat147-275 | 0.002 | R.P. | ||
| TdelSat148-53 | 0.001 | TinfSat26-53 | 0.011 | 0.002 |
| TdelSat149-26 | 0.001 | R.P. | ||
| TdelSat150-56 | 0.001 | N.D. | ||
| TdelSat151-149 | 0.001 | N.D. | ||
| TdelSat152-50 | 0.001 | R.P. | ||
| TdelSat153-75 | 0.001 | N.D. | ||
| TdelSat154-45 | 0.001 | R.P. | ||
| TdelSat155-89 | 0.001 | R.P. | ||
| TdelSat156-78 | 0.001 | R.P. | ||
| TdelSat157-51 | 0.001 | R.P. | ||
| TdelSat158-134 | 0.001 | R.P. | ||
| TdelSat159-241 | 0.001 | TinfSat63-237 | 0.010 | 0.002 |
| TdelSat160-49 | 0.001 | R.P. | ||
| TOTAL | 53.92 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mora, P.; Pita, S.; Montiel, E.E.; Rico-Porras, J.M.; Palomeque, T.; Panzera, F.; Lorite, P. Making the Genome Huge: The Case of Triatoma delpontei, a Triatominae Species with More than 50% of Its Genome Full of Satellite DNA. Genes 2023, 14, 371. https://doi.org/10.3390/genes14020371
Mora P, Pita S, Montiel EE, Rico-Porras JM, Palomeque T, Panzera F, Lorite P. Making the Genome Huge: The Case of Triatoma delpontei, a Triatominae Species with More than 50% of Its Genome Full of Satellite DNA. Genes. 2023; 14(2):371. https://doi.org/10.3390/genes14020371
Chicago/Turabian StyleMora, Pablo, Sebastián Pita, Eugenia E. Montiel, José M. Rico-Porras, Teresa Palomeque, Francisco Panzera, and Pedro Lorite. 2023. "Making the Genome Huge: The Case of Triatoma delpontei, a Triatominae Species with More than 50% of Its Genome Full of Satellite DNA" Genes 14, no. 2: 371. https://doi.org/10.3390/genes14020371
APA StyleMora, P., Pita, S., Montiel, E. E., Rico-Porras, J. M., Palomeque, T., Panzera, F., & Lorite, P. (2023). Making the Genome Huge: The Case of Triatoma delpontei, a Triatominae Species with More than 50% of Its Genome Full of Satellite DNA. Genes, 14(2), 371. https://doi.org/10.3390/genes14020371







