The Role of IRX Homeobox Genes in Hematopoietic Progenitors and Leukemia
Abstract
1. Introduction
2. Homeobox Gene Codes
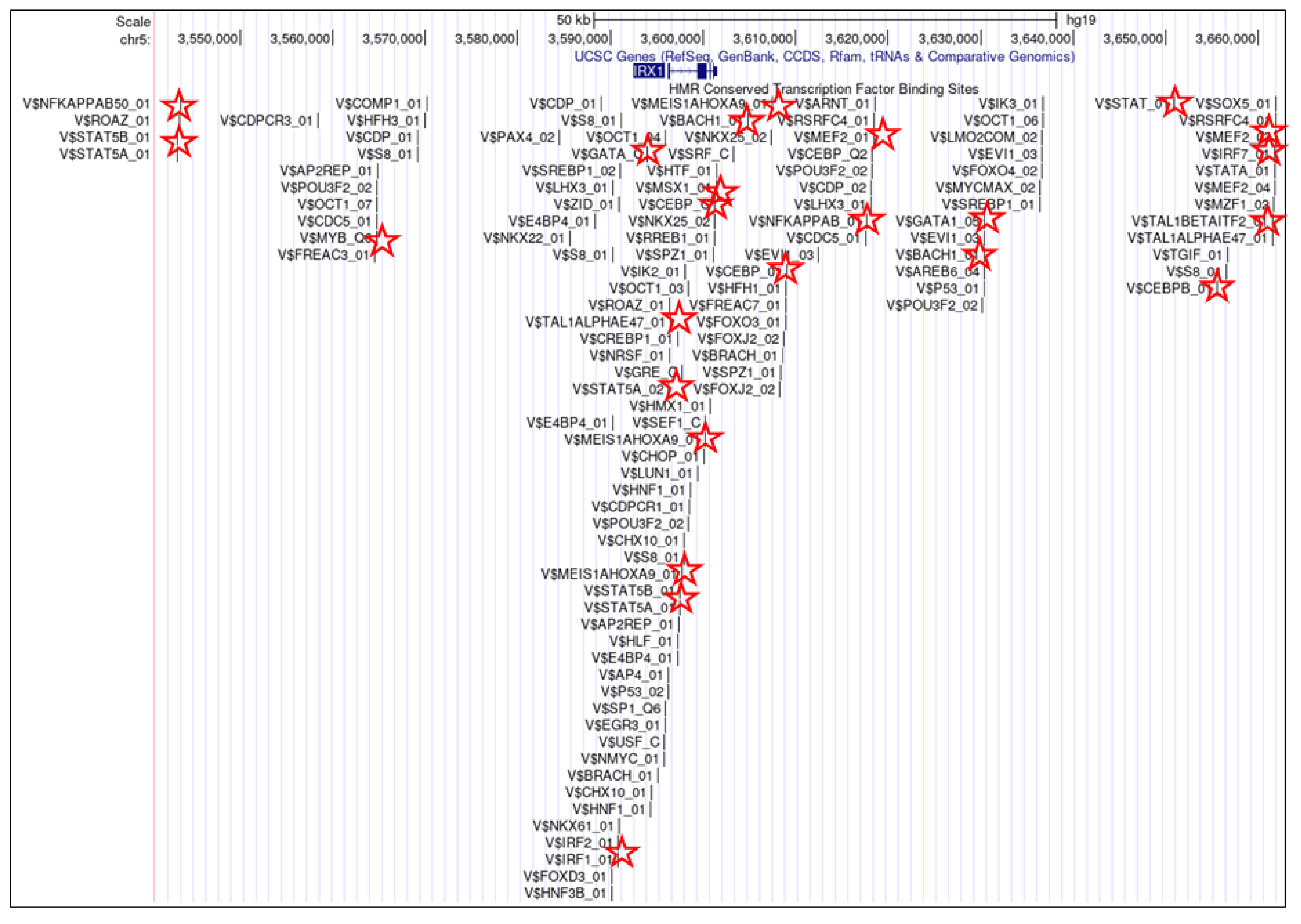
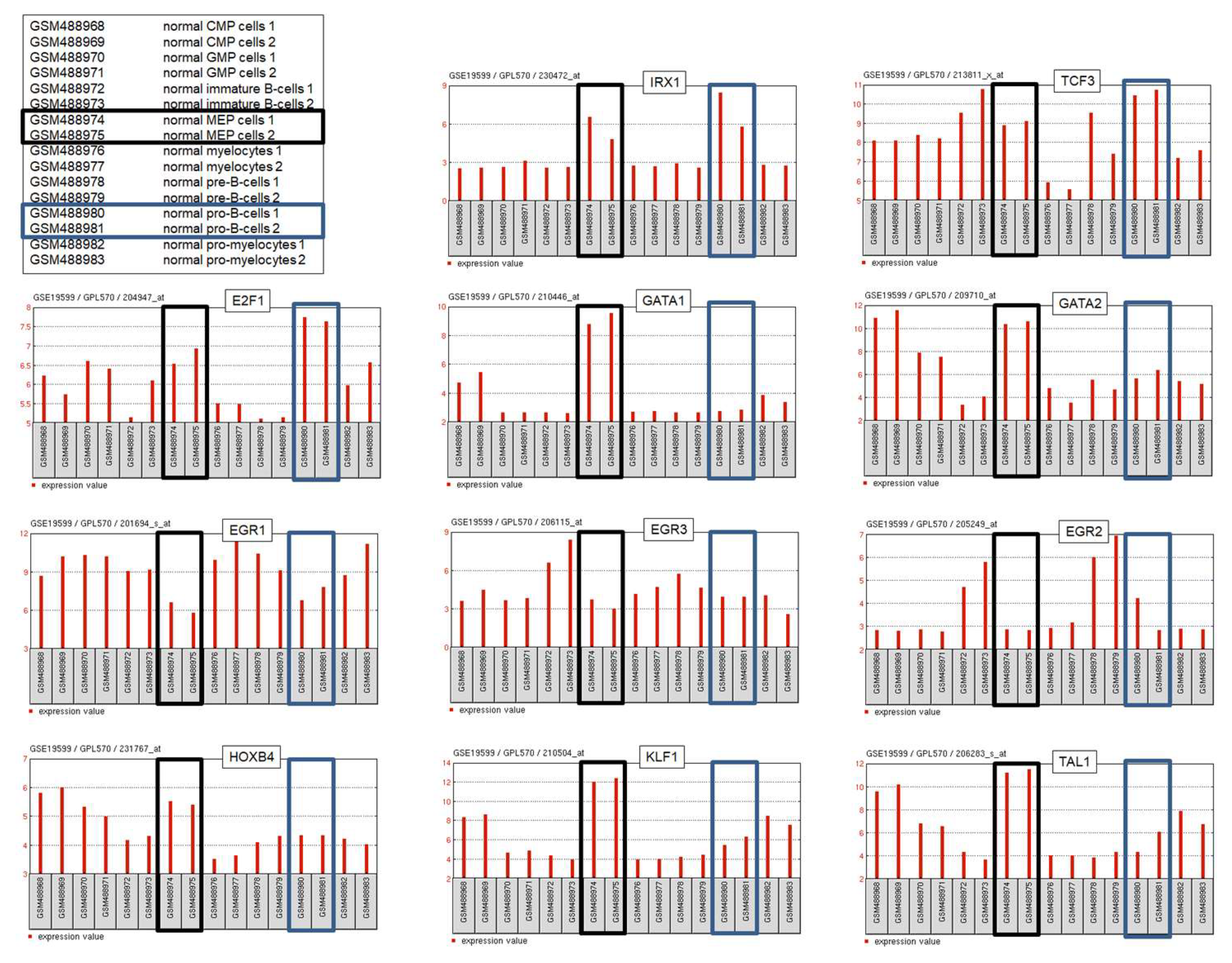
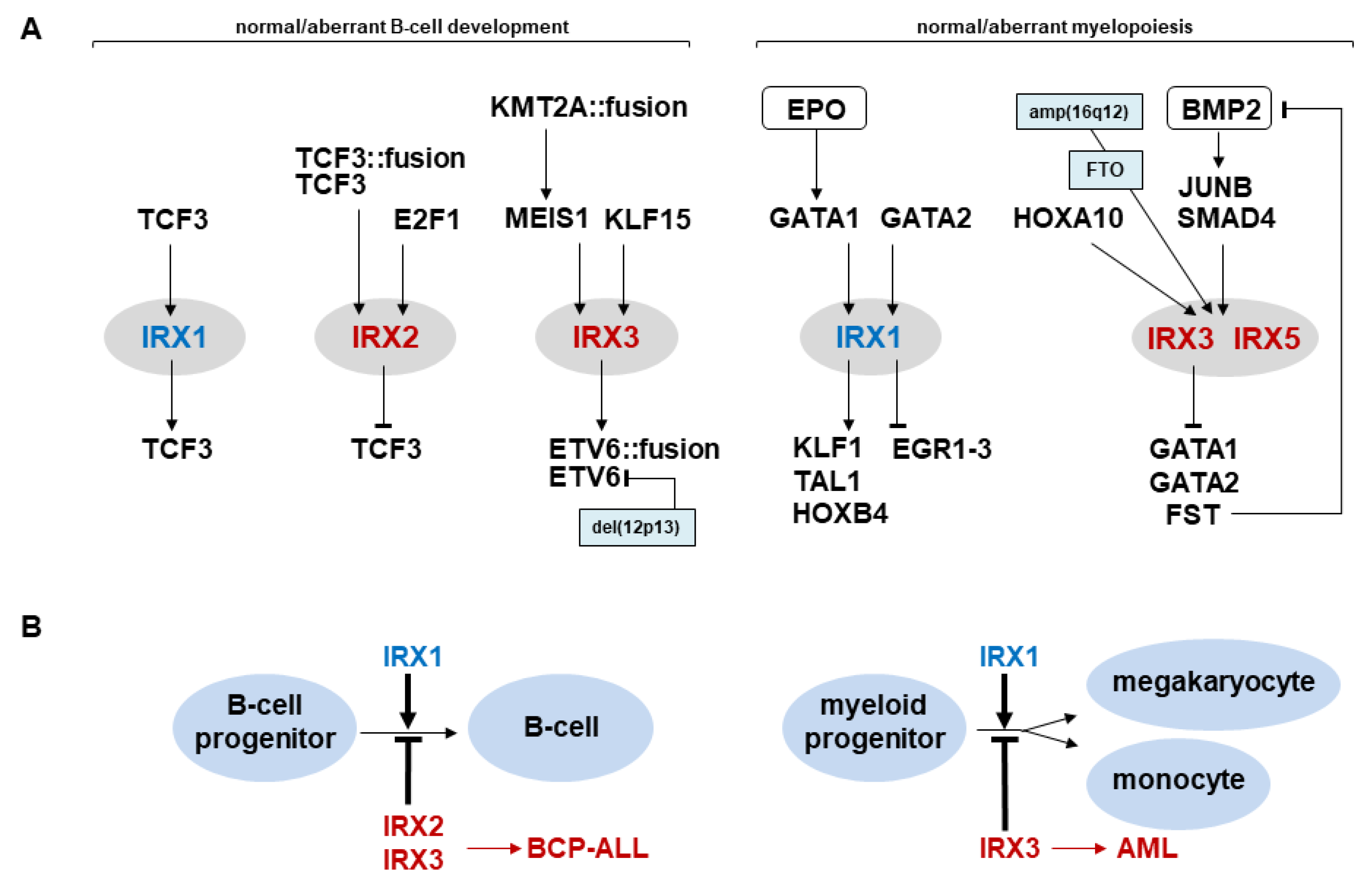
3. IRX1 Expression and Regulation in Hematopoiesis
3.1. IRX1 in Lymphopoiesis
3.2. IRX1 in Myelopoiesis
4. Aberrant IRX Gene Activities in Hematopoietic Malignancies
4.1. Deregulated IRX Genes in Acute Lymphoid Leukemia
4.2. Deregulated IRX Genes in Acute Myeloid Leukemia
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liggett, L.A.; Sankaran, V.G. Unraveling hematopoiesis through the lens of genomics. Cell 2020, 182, 1384–1400. [Google Scholar] [CrossRef] [PubMed]
- Kucinski, I.; Wilson, N.K.; Hannah, R.; Kinston, S.J.; Cauchy, P.; Lenaerts, A.; Grosschedl, R.; Göttgens, B. Interactions between lineage-associated transcription factors govern haematopoietic progenitor states. EMBO J. 2020, 39, e104983. [Google Scholar] [CrossRef] [PubMed]
- Somasundaram, R.; Prasad, M.A.; Ungerbäck, J.; Sigvardsson, M. Transcription factor networks in B-cell differentiation link development to acute lymphoid leukemia. Blood 2015, 126, 144–152. [Google Scholar] [CrossRef]
- Debili, N.; Coulombel, L.; Croisille, L.; Katz, A.; Guichard, J.; Breton-Gorius, J.; Vainchenker, W. Characterization of a bipotent erythro-megakaryocytic progenitor in human bone marrow. Blood 1996, 88, 1284–1296. [Google Scholar] [CrossRef] [PubMed]
- Xavier-Ferrucio, J.; Krause, D.S. Concise Review: Bipotent Megakaryocytic-Erythroid Progenitors: Concepts and Controversies. Stem Cells 2018, 36, 1138–1145. [Google Scholar] [CrossRef]
- Rothenberg, E.V. Transcriptional control of early T and B cell developmental choices. Annu. Rev. Immunol. 2014, 32, 283–321. [Google Scholar] [CrossRef]
- Rothenberg, E.V.; Kueh, H.Y.; Yui, M.A.; Zhang, J.A. Hematopoiesis and T-cell specification as a model developmental system. Immunol. Rev. 2016, 271, 72–97. [Google Scholar] [CrossRef]
- Belle, I.; Zhuang, Y. E proteins in lymphocyte development and lymphoid diseases. Curr. Top. Dev. Biol. 2014, 110, 153–187. [Google Scholar]
- Boller, S.; Grosschedl, R. The regulatory network of B-cell differentiation: A focused view of early B-cell factor 1 function. Immunol. Rev. 2014, 261, 102–115. [Google Scholar] [CrossRef]
- Fujiwara, T. GATA transcription factors: Basic principles and related human disorders. Tohoku J. Exp. Med. 2017, 242, 83–91. [Google Scholar] [CrossRef]
- Avram, D.; Califano, D. The multifaceted roles of Bcl11b in thymic and peripheral T cells: Impact on immune diseases. J. Immunol. 2014, 193, 2059–2065. [Google Scholar] [CrossRef] [PubMed]
- Kostrzewski, T.; Borg, A.J.; Meng, Y.; Filipovic, I.; Male, V.; Wack, A.; DiMaggio, P.A.; Brady, H.J.M. Multiple levels of control determine how E4bp4/Nfil3 regulates NK cell development. J. Immunol. 2018, 200, 1370–1381. [Google Scholar] [CrossRef] [PubMed]
- Tanriver, Y.; Diefenbach, A. Transcription factors controlling development and function of innate lymphoid cells. Int. Immunol. 2014, 26, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Hao, W.; Hu, W. Transcription factor PU.1 and immune cell differentiation (Review). Int. J. Mol. Med. 2020, 46, 1943–1950. [Google Scholar] [CrossRef] [PubMed]
- Vaquerizas, J.M.; Kummerfeld, S.K.; Teichmann, S.A.; Luscombe, N.M. A census of human transcription factors: Function, expression and evolution. Nat. Rev. Genet. 2009, 10, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Bürglin, T.R.; Affolter, M. Homeodomain proteins: An update. Chromosoma 2016, 125, 497–521. [Google Scholar] [CrossRef]
- Abate-Shen, C. Deregulated homeobox gene expression in cancer: Cause or consequence? Nat. Rev. Cancer 2002, 2, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Nagel, S.; Pommerenke, C.; Meyer, C.; Kaufmann, M.; MacLeod, R.A.F.; Drexler, H.G. NKL homeobox gene MSX1 acts like a tumor suppressor in NK-cell leukemia. Oncotarget 2017, 8, 66815–66832. [Google Scholar] [CrossRef]
- Gehring, W.J.; Müller, M.; Affolter, M.; Percival-Smith, A.; Billeter, M.; Qian, Y.Q.; Otting, G.; Wüthrich, K. The structure of the homeodomain and its functional implications. Trends Genet. 1990, 6, 323–329. [Google Scholar] [CrossRef]
- Holland, P.W.; Booth, H.A.; Bruford, E.A. Classification and nomenclature of all human homeobox genes. BMC Biol. 2007, 5, 47. [Google Scholar] [CrossRef]
- Mukherjee, K.; Bürglin, T.R. Comprehensive analysis of animal TALE homeobox genes: New conserved motifs and cases of accelerated evolution. J. Mol. Evol. 2007, 65, 137–153. [Google Scholar] [CrossRef] [PubMed]
- Hobert, O. Homeobox genes and the specification of neuronal identity. Nat. Rev. Neurosci. 2021, 22, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Hunt, P.; Gulisano, M.; Cook, M.; Sham, M.H.; Faiella, A.; Wilkinson, D.; Boncinelli, E.; Krumlauf, R. A distinct Hox code for the branchial region of the vertebrate head. Nature 1991, 353, 861–864. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K.E. How do genes regulate simple behaviours? Understanding how different neurons in the vertebrate spinal cord are genetically specified. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2006, 361, 45–66. [Google Scholar] [CrossRef] [PubMed]
- Parker, H.J.; Pushel, I.; Krumlauf, R. Coupling the roles of Hox genes to regulatory networks patterning cranial neural crest. Dev. Biol. 2018, 444 (Suppl. 1), S67–S78. [Google Scholar] [CrossRef] [PubMed]
- Depew, M.J.; Simpson, C.A.; Morasso, M.; Rubenstein, J.L. Reassessing the Dlx code: The genetic regulation of branchial arch skeletal pattern and development. J. Anat. 2005, 207, 501–561. [Google Scholar] [CrossRef]
- Streit, A. The cranial sensory nervous system: Specification of sensory progenitors and placodes. In StemBook; Harvard Stem Cell Institute: Cambridge, MA, USA, 2008. [Google Scholar]
- Nagel, S. NKL-code in normal and aberrant hematopoiesis. Cancers 2021, 13, 1961. [Google Scholar] [CrossRef]
- Nagel, S.; Pommerenke, C.; Meyer, C.; MacLeod, R.A.F.; Drexler, H.G. Establishment of the TALE-code reveals aberrantly activated homeobox gene PBX1 in Hodgkin lymphoma. PLoS ONE 2021, 16, e0246603. [Google Scholar] [CrossRef]
- Nagel, S.; Pommerenke, C.; Meyer, C.; MacLeod, R.A.F. The hematopoietic TALE-code shows normal activity of IRX1 in myeloid progenitors and reveals ectopic expression of IRX3 and IRX5 in acute myeloid leukemia. Int. J. Mol. Sci. 2022, 23, 3192. [Google Scholar] [CrossRef]
- Nagel, S.; Meyer, C. Normal and aberrant TALE-class homeobox gene activities in pro-B-cells and B-cell precursor acute lymphoblastic leukemia. Int. J. Mol. Sci. 2022, 23, 11874. [Google Scholar] [CrossRef]
- Nagel, S. The NKL- and TALE-codes represent hematopoietic gene signatures to evaluate deregulated homeobox genes in Hodgkin lymphoma. Hemato 2022, 3, 122–130. [Google Scholar] [CrossRef]
- Bosse, A.; Zülch, A.; Becker, M.B.; Torres, M.; Gómez-Skarmeta, J.L.; Modolell, J.; Gruss, P. Identification of the vertebrate Iroquois homeobox gene family with overlapping expression during early development of the nervous system. Mech. Dev. 1997, 69, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Bruneau, B.G.; Bao, Z.Z.; Tanaka, M.; Schott, J.J.; Izumo, S.; Cepko, C.L.; Seidman, J.G.; Seidman, C.E. Cardiac expression of the ventricle-specific homeobox gene Irx4 is modulated by Nkx2-5 and dHand. Dev. Biol. 2000, 217, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Cohen, D.R.; Cheng, C.W.; Cheng, S.H.; Hui, C.C. Expression of two novel mouse Iroquois homeobox genes during neurogenesis. Mech. Dev. 2000, 91, 317–321. [Google Scholar] [CrossRef]
- Gomez-Skarmeta, J.L.; Diez del Corral, R.; de la Calle-Mustienes, E.; Ferré-Marcó, D.; Modolell, J. Araucan and caupolican, two members of the novel iroquois complex, encode homeoproteins that control proneural and vein-forming genes. Cell 1996, 85, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Leyns, L.; Gómez-Skarmeta, J.L.; Dambly-Chaudière, C. iroquois: A prepattern gene that controls the formation of bristles on the thorax of Drosophila. Mech. Dev. 1996, 59, 63–72. [Google Scholar] [CrossRef]
- Uhlen, M.; Karlsson, M.J.; Zhong, W.; Tebani, A.; Pou, C.; Mikes, J.; Lakshmikanth, T.; Forsström, B.; Edfors, F.; Odeberg, J.; et al. A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science 2019, 366, eaax9198. [Google Scholar] [CrossRef]
- Alarcón, P.; Rodríguez-Seguel, E.; Fernández-González, A.; Rubio, R.; Gómez-Skarmeta, J.L. A dual requirement for Iroquois genes during Xenopus kidney development. Development 2008, 135, 3197–3207. [Google Scholar] [CrossRef]
- Freese, N.H.; Lam, B.A.; Staton, M.; Scott, A.; Chapman, S.C. A novel gain-of-function mutation of the proneural IRX1 and IRX2 genes disrupts axis elongation in the Araucana rumpless chicken. PLoS ONE 2014, 9, e112364. [Google Scholar] [CrossRef] [PubMed]
- Küster, M.M.; Schneider, M.A.; Richter, A.M.; Richtmann, S.; Winter, H.; Kriegsmann, M.; Pullamsetti, S.S.; Stiewe, T.; Savai, R.; Muley, T.; et al. Epigenetic inactivation of the tumor suppressor IRX1 occurs frequently in lung adenocarcinoma and its silencing is associated with impaired prognosis. Cancers 2020, 12, 3528. [Google Scholar] [CrossRef]
- Lu, J.; Song, G.; Tang, Q.; Zou, C.; Han, F.; Zhao, Z.; Yong, B.; Yin, J.; Xu, H.; Xie, X.; et al. IRX1 hypomethylation promotes osteosarcoma metastasis via induction of CXCL14/NF-κB signaling. J. Clin. Investig. 2015, 125, 1839–1856. [Google Scholar] [CrossRef] [PubMed]
- Lentjes, M.H.; Niessen, H.E.; Akiyama, Y.; de Bruïne, A.P.; Melotte, V.; van Engeland, M. The emerging role of GATA transcription factors in development and disease. Expert Rev. Mol. Med. 2016, 18, e3. [Google Scholar] [CrossRef] [PubMed]
- Kühn, A.; Löscher, D.; Marschalek, R. The IRX1/HOXA connection: Insights into a novel t(4;11)- specific cancer mechanism. Oncotarget 2016, 7, 35341–35352. [Google Scholar] [CrossRef] [PubMed]
- Hunger, S.P.; Mullighan, C.G. Acute lymphoblastic leukemia in children. N. Engl. J. Med. 2015, 373, 1541–1552. [Google Scholar] [CrossRef] [PubMed]
- Lilljebjörn, H.; Fioretos, T. New oncogenic subtypes in pediatric B-cell precursor acute lymphoblastic leukemia. Blood 2017, 130, 1395–1401. [Google Scholar] [CrossRef]
- Hirabayashi, S.; Ohki, K.; Nakabayashi, K.; Ichikawa, H.; Momozawa, Y.; Okamura, K.; Yaguchi, A.; Terada, K.; Saito, Y.; Yoshimi, A.; et al. ZNF384-related fusion genes define a subgroup of childhood B-cell precursor acute lymphoblastic leukemia with a characteristic immunotype. Haematologica 2017, 102, 118–129. [Google Scholar] [CrossRef]
- Li, J.F.; Dai, Y.T.; Lilljebjörn, H.; Shen, S.H.; Cui, B.W.; Bai, L.; Liu, Y.F.; Qian, M.X.; Kubota, Y.; Kiyoi, H.; et al. Transcriptional landscape of B cell precursor acute lymphoblastic leukemia based on an international study of 1223 cases. Proc. Natl. Acad. Sci. USA 2018, 115, E11711–E11720. [Google Scholar] [CrossRef]
- Brady, S.W.; Roberts, K.G.; Gu, Z.; Shi, L.; Pounds, S.; Pei, D.; Cheng, C.; Dai, Y.; Devidas, M.; Qu, C.; et al. The genomic landscape of pediatric acute lymphoblastic leukemia. Nat. Genet. 2022, 54, 1376–1389. [Google Scholar] [CrossRef]
- Somerville, T.D.D.; Simeoni, F.; Chadwick, J.A.; Williams, E.L.; Spencer, G.J.; Boros, K.; Wirth, C.; Tholouli, E.; Byers, R.J.; Somervaille, T.C.P. Derepression of the Iroquois homeodomain transcription factor gene IRX3 confers differentiation block in acute leukemia. Cell Rep. 2018, 22, 638–652. [Google Scholar] [CrossRef]
- Rasighaemi, P.; Ward, A.C. ETV6 and ETV7: Siblings in hematopoiesis and its disruption in disease. Crit. Rev. Oncol. Hematol. 2017, 116, 106–115. [Google Scholar] [CrossRef]
- O’Neil, J.; Billa, M.; Oikemus, S.; Kelliher, M. The DNA binding activity of TAL-1 is not required to induce leukemia/lymphoma in mice. Oncogene 2001, 20, 3897–3905. [Google Scholar] [CrossRef] [PubMed]
- Palomero, T.; Odom, D.T.; O’Neil, J.; Ferrando, A.A.; Margolin, A.; Neuberg, D.S.; Winter, S.S.; Larson, R.S.; Li, W.; Liu, X.S.; et al. Transcriptional regulatory networks downstream of TAL1/SCL in T-cell acute lymphoblastic leukemia. Blood 2006, 108, 986–992. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Wilson, C.S.; Harvey, R.C.; Chen, I.M.; Murphy, M.H.; Atlas, S.R.; Bedrick, E.J.; Devidas, M.; Carroll, A.J.; Robinson, B.W.; et al. Gene expression profiles predictive of outcome and age in infant acute lymphoblastic leukemia: A Children’s Oncology Group study. Blood 2012, 119, 1872–1881. [Google Scholar] [CrossRef] [PubMed]
- Symeonidou, V.; Ottersbach, K. HOXA9/IRX1 expression pattern defines two subgroups of infant MLL-AF4-driven acute lymphoblastic leukemia. Exp. Hematol. 2021, 93, 38–43. [Google Scholar] [CrossRef]
- Somerville, T.D.; Wiseman, D.H.; Spencer, G.J.; Huang, X.; Lynch, J.T.; Leong, H.S.; Williams, E.L.; Cheesman, E.; Somervaille, T.C. Frequent derepression of the mesenchymal transcription factor gene FOXC1 in acute myeloid leukemia. Cancer Cell 2015, 28, 329–342. [Google Scholar] [CrossRef]
- Claussnitzer, M.; Dankel, S.N.; Kim, K.H.; Quon, G.; Meuleman, W.; Haugen, C.; Glunk, V.; Sousa, I.S.; Beaudry, J.L.; Puviindran, V.; et al. FTO obesity variant circuitry and adipocyte browning in humans. N. Engl. J. Med. 2015, 373, 895–907. [Google Scholar] [CrossRef]
- Rask-Andersen, M.; Almén, M.S.; Schiöth, H.B. Scrutinizing the FTO locus: Compelling evidence for a complex, long-range regulatory context. Hum. Genet. 2015, 134, 1183–1193. [Google Scholar] [CrossRef]
- Deng, X.; Su, R.; Stanford, S.; Chen, J. Critical enzymatic functions of FTO in obesity and cancer. Front. Endocrinol. 2018, 9, 396. [Google Scholar] [CrossRef]
- Li, Z.; Weng, H.; Su, R.; Weng, X.; Zuo, Z.; Li, C.; Huang, H.; Nachtergaele, S.; Dong, L.; Hu, C.; et al. FTO plays an oncogenic role in acute myeloid leukemia as a N6-methyladenosine RNA demethylase. Cancer Cell. 2017, 31, 127–141. [Google Scholar] [CrossRef]
- Li, B.; An, W.; Wang, H.; Baslan, T.; Mowla, S.; Krishnan, A.; Xiao, W.; Koche, R.P.; Liu, Y.; Cai, S.F.; et al. BMP2/SMAD pathway activation in JAK2/p53-mutant megakaryocyte/erythroid progenitors promotes leukemic transformation. Blood 2022, 139, 3630–3646. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nagel, S. The Role of IRX Homeobox Genes in Hematopoietic Progenitors and Leukemia. Genes 2023, 14, 297. https://doi.org/10.3390/genes14020297
Nagel S. The Role of IRX Homeobox Genes in Hematopoietic Progenitors and Leukemia. Genes. 2023; 14(2):297. https://doi.org/10.3390/genes14020297
Chicago/Turabian StyleNagel, Stefan. 2023. "The Role of IRX Homeobox Genes in Hematopoietic Progenitors and Leukemia" Genes 14, no. 2: 297. https://doi.org/10.3390/genes14020297
APA StyleNagel, S. (2023). The Role of IRX Homeobox Genes in Hematopoietic Progenitors and Leukemia. Genes, 14(2), 297. https://doi.org/10.3390/genes14020297





