Imidazolopiperazine (IPZ)-Induced Differential Transcriptomic Responses on Plasmodium falciparum Wild-Type and IPZ-Resistant Mutant Parasites
Abstract
1. Introduction
2. Materials and Methods
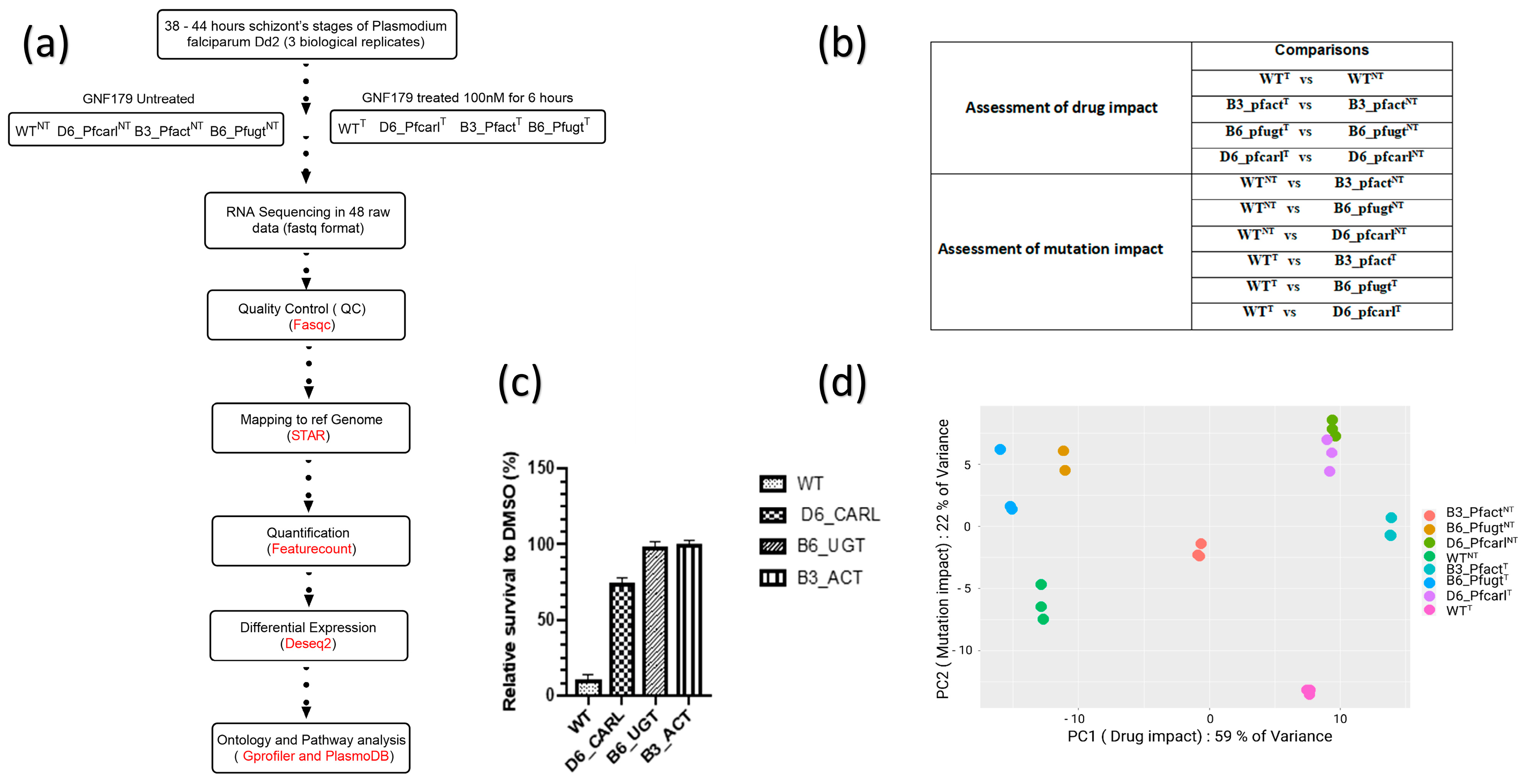
2.1. Drug Treatment for Transcriptomic Sample Preparation
2.2. mRNA Extraction and Sequencing
2.3. Bioinformatics Analysis
2.4. Comparison of Normalized Counts
3. Results
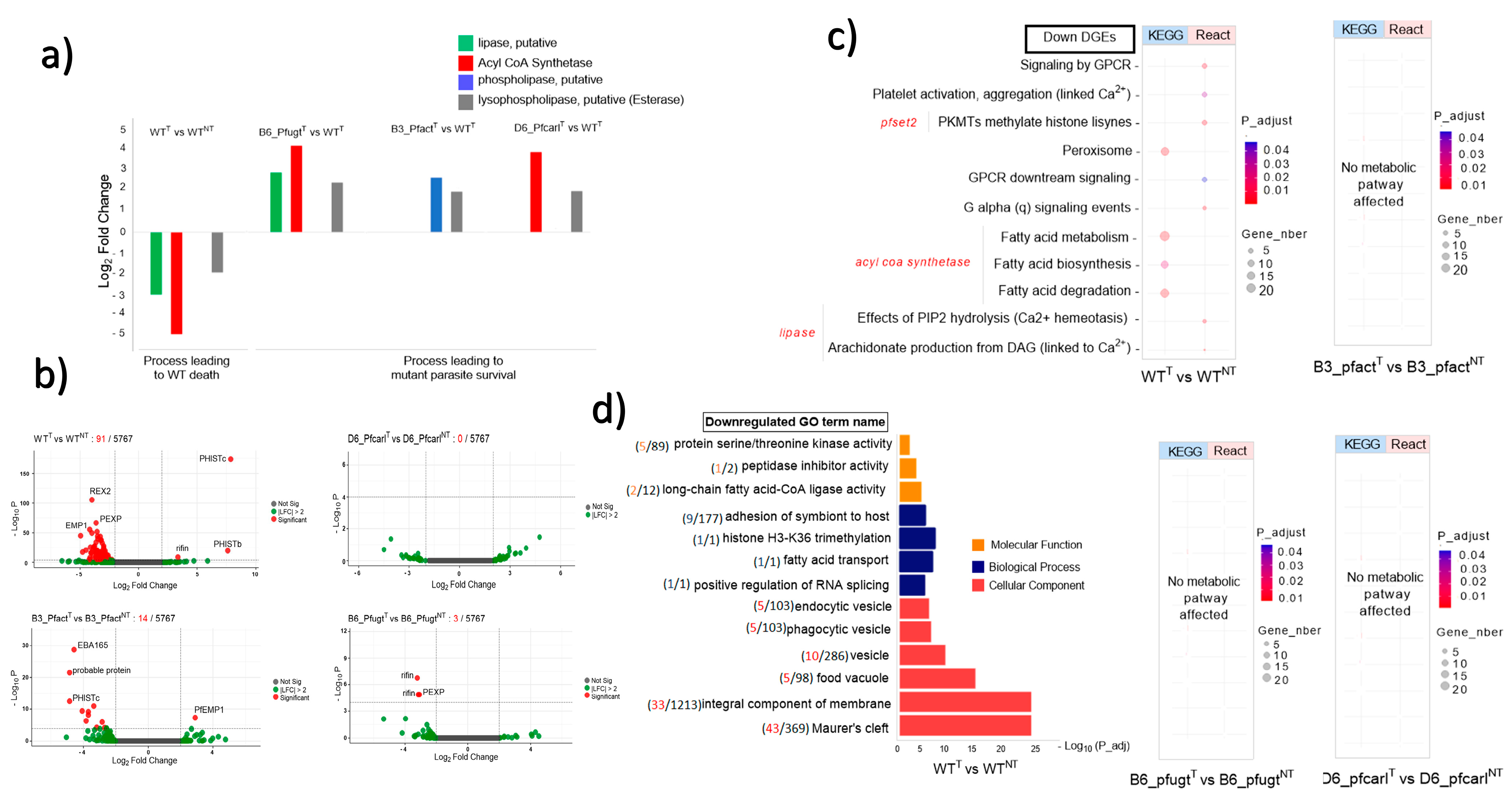
3.1. P. falciparum Wild-Type and Imidazolopiperazine-Resistant Mutant Parasites Displayed Differential Transcriptomic Response from GNF179 Exposure
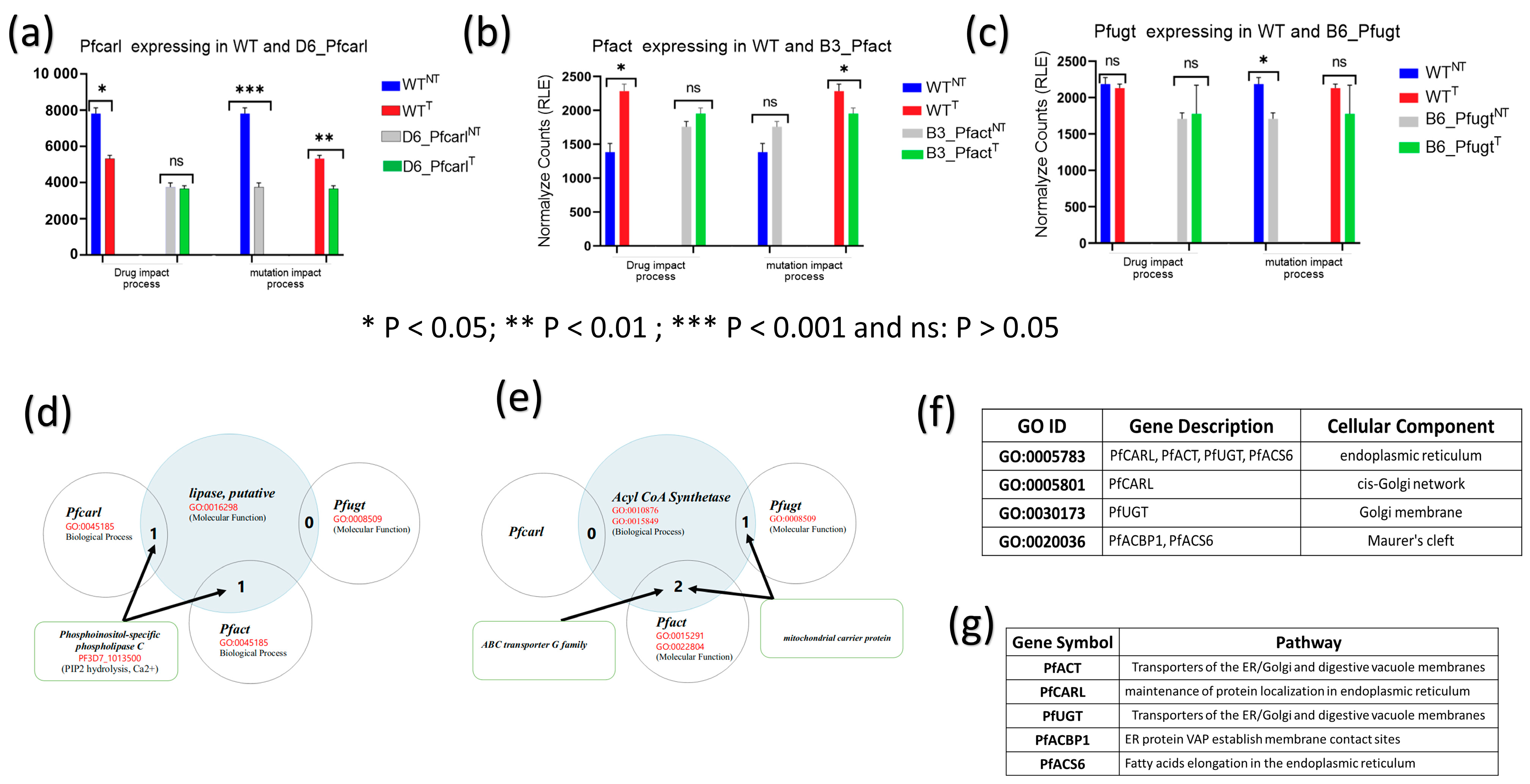
3.2. Imidazolopiperazine (IPZ) GNF179 Treatment Did Not Affect Pfcarl, Pfact, and Pfugt Genes Regulation Which Are Associated with Membrane Transport Targeted by IPZ
3.3. Key Uncommon Metabolic Pathways and Biological Process Disturbed by GNF179 between Wild and IPZ-Resistant Parasites in Treated and Untreated Conditions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. World Malaria Report 2022; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- White, N.J. Antimalarial drug resistance. J. Clin. Investig. 2004, 113, 1084–1092. [Google Scholar] [CrossRef] [PubMed]
- Dondorp, A.M.; Nosten, F.; Yi, P.; Das, D.; Phyo, A.P.; Tarning, J.; Lwin, K.M.; Ariey, F.; Hanpithakpong, W.; Lee, S.J.; et al. Artemisinin Resistance in Plasmodium falciparum Malaria. N. Engl. J. Med. 2009, 361, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.T.; Juliano, J.J.; Wongsrichanalai, C. Drug-Resistant Malaria: The Era of ACT. Curr. Infect. Dis. Rep. 2010, 12, 165–173. [Google Scholar] [CrossRef]
- Ouji, M.; Augereau, J.-M.; Paloque, L.; Benoit-Vical, F. Plasmodium falciparum resistance to artemisinin-based combination therapies: A sword of Damocles in the path toward malaria elimination. Parasite 2018, 25, 24. [Google Scholar] [CrossRef] [PubMed]
- Uwimana, A.; Legrand, E.; Stokes, B.H.; Ndikumana, J.-L.M.; Warsame, M.; Umulisa, N.; Ngamije, D.; Munyaneza, T.; Mazarati, J.-B.; Munguti, K.; et al. Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat. Med. 2020, 26, 1602–1608. [Google Scholar] [CrossRef]
- White, N.J.; Duong, T.T.; Uthaisin, C.; Nosten, F.; Phyo, A.P.; Hanboonkunupakarn, B.; Pukrittayakamee, S.; Jittamala, P.; Chuthasmit, K.; Cheung, M.S.; et al. Antimalarial Activity of KAF156 in Falciparum and Vivax Malaria. N. Engl. J. Med. 2016, 375, 1152–1160. [Google Scholar] [CrossRef] [PubMed]
- Meister, S.; Plouffe, D.M.; Kuhen, K.L.; Bonamy, G.M.C.; Wu, T.; Barnes, S.W.; Bopp, S.E.; Borboa, R.; Bright, A.T.; Che, J.; et al. Imaging of Plasmodium Liver Stages to Drive Next-Generation Antimalarial Drug Discovery. Science 2011, 334, 1372–1377. [Google Scholar] [CrossRef]
- Kuhen, K.L.; Chatterjee, A.K.; Rottmann, M.; Gagaring, K.; Borboa, R.; Buenviaje, J.; Chen, Z.; Francek, C.; Wu, T.; Nagle, A.; et al. KAF156 Is an antimalarial clinical candidate with potential for use in prophylaxis, treatment, and prevention of disease transmission. Antimicrob. Agents Chemother. 2014, 58, 5060–5067. [Google Scholar] [CrossRef]
- Dembele, L.; Gupta, D.K.; Lim, M.Y.-X.; Ang, X.; Selva, J.J.; Chotivanich, K.; Nguon, C.; Dondorp, A.M.; Bonamy, G.M.C.; Diagana, T.T.; et al. Imidazolopiperazines kill both rings and dormant rings in wild-type and K13 artemisinin-resistant Plasmodium falciparum In Vitro. Antimicrob. Agents Chemother. 2018, 62, 11. [Google Scholar] [CrossRef]
- Ouologuem, D.T.; Dembele, L.; Dara, A.; Kone, A.K.; Diallo, N.; Sangare, C.P.O.; Ballo, F.I.; Dao, F.; Goita, S.; Haidara, A.S.; et al. A Novel Ex Vivo Drug Assay for Assessing the Transmission-Blocking Activity of Compounds on Field-Isolated Plasmodium falciparum Gametocytes. Antimicrob. Agents Chemother. 2022, 66, e0100122. [Google Scholar] [CrossRef]
- LaMonte, G.; Lim, M.Y.-X.; Wree, M.; Reimer, C.; Nachon, M.; Corey, V.; Gedeck, P.; Plouffe, D.; Du, A.; Figueroa, N.; et al. Mutations in the Plasmodium falciparum Cyclic Amine Resistance Locus (PfCARL) Confer Multidrug Resistance. mBio 2016, 7, e00696-16. [Google Scholar] [CrossRef]
- Lim, M.Y.-X.; LaMonte, G.; Lee, M.C.S.; Reimer, C.; Tan, B.H.; Corey, V.; Tjahjadi, B.F.; Chua, A.; Nachon, M.; Wintjens, R.; et al. UDP-galactose and acetyl-CoA transporters as Plasmodium multidrug resistance genes. Nat. Microbiol. 2016, 1, 16166. [Google Scholar] [CrossRef] [PubMed]
- LaMonte, G.M.; Rocamora, F.; Marapana, D.S.; Gnädig, N.F.; Ottilie, S.; Luth, M.R.; Worgall, T.S.; Goldgof, G.M.; Mohunlal, R.; Kumar, T.R.S.; et al. Pan-active imidazolopiperazine antimalarials target the Plasmodium falciparum intracellular secretory pathway. Nat. Commun. 2020, 11, 1780. [Google Scholar] [CrossRef] [PubMed]
- Cowman, A.F.; Healer, J.; Marapana, D.; Marsh, K. Malaria: Biology and Disease. Cell 2016, 167, 610–624. [Google Scholar] [CrossRef]
- Jackson, K.E.; Klonis, N.; Ferguson, D.J.P.; Adisa, A.; Dogovski, C.; Tilley, L. Food vacuole-associated lipid bodies and heterogeneous lipid environments in the malaria parasite, Plasmodium falciparum. Mol. Microbiol. 2004, 54, 109–122. [Google Scholar] [CrossRef]
- Bunnik, E.M.; Chung, D.-W.D.; Hamilton, M.; Ponts, N.; Saraf, A.; Prudhomme, J.; Florens, L.; Le Roch, K.G. Polysome profiling reveals translational control of gene expression in the human malaria parasite Plasmodium falciparum. Genome Biol. 2013, 14, R128. [Google Scholar] [CrossRef]
- Staines, H.M.; Ashmore, S.; Felgate, H.; Moore, J.; Powell, T.; Ellory, J.C. Solute transport via the new permeability pathways in Plasmodium falciparum–infected human red blood cells is not consistent with a simple single-channel model. Blood 2006, 108, 3187–3194. [Google Scholar] [CrossRef][Green Version]
- McNamara, C.W.; Lee, M.C.S.; Lim, C.S.; Lim, S.H.; Roland, J.; Nagle, A.; Simon, O.; Yeung, B.K.S.; Chatterjee, A.K.; McCormack, S.L.; et al. Targeting Plasmodium PI(4)K to eliminate malaria. Nature 2013, 504, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Holz, G.G., Jr. Lipids and the malarial parasite. Bull. World Health Organ. 1977, 55, 237. [Google Scholar] [PubMed]
- Mikkelsen, R.B.; Kamber, M.; Wadwa, K.S.; Lin, P.S.; Schmidt-Ullrich, R. The role of lipids in Plasmodium falciparum invasion of erythrocytes: A coordinated biochemical and microscopic analysis. Proc. Natl. Acad. Sci. USA 1988, 85, 5956–5960. [Google Scholar] [CrossRef]
- Tran, P.N.; Brown, S.H.J.; Rug, M.; Ridgway, M.C.; Mitchell, T.W.; Maier, A.G. Changes in lipid composition during sexual development of the malaria parasite Plasmodium falciparum. Malar. J. 2016, 15, 73. [Google Scholar] [CrossRef] [PubMed]
- Flammersfeld, A.; Lang, C.; Flieger, A.; Pradel, G. Phospholipases during membrane dynamics in malaria parasites. Int. J. Med. Microbiol. 2018, 308, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Gulati, S.; Ekland, E.H.; Ruggles, K.V.; Chan, R.B.; Jayabalasingham, B.; Zhou, B.; Mantel, P.-Y.; Lee, M.C.; Spottiswoode, N.; Coburn-Flynn, O.; et al. Profiling the Essential Nature of Lipid Metabolism in Asexual Blood and Gametocyte Stages of Plasmodium falciparum. Cell Host Microbe 2015, 18, 371–381. [Google Scholar] [CrossRef]
- Pecenin, M.F.; Borges-Pereira, L.; Levano-Garcia, J.; Budu, A.; Alves, E.; Mikoshiba, K.; Thomas, A.; Garcia, C.R. Blocking IP 3 signal transduction pathways inhibits melatonin-induced Ca 2+ signals and impairs P. falciparum development and proliferation in erythrocytes. Cell Calcium 2018, 72, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Alves, E.; Bartlett, P.J.; Garcia, C.R.; Thomas, A.P. Melatonin and IP3-induced Ca2+ Release from Intracellular Stores in the Malaria Parasite Plasmodium falciparum within Infected Red Blood Cells. J. Biol. Chem. 2011, 286, 5905–5912. [Google Scholar] [CrossRef] [PubMed]
- Palacpac, N.M.Q.; Hiramine, Y.; Seto, S.; Hiramatsu, R.; Horii, T.; Mitamura, T. Evidence that Plasmodium falciparum diacylglycerol acyltransferase is essential for intraerythrocytic proliferation. Biochem. Biophys. Res. Commun. 2004, 321, 1062–1068. [Google Scholar] [CrossRef]
- Matesanz, F.; Téllez, M.-D.; Alcina, A. The Plasmodium falciparum fatty acyl-CoA synthetase family (PfACS) and differential stage-specific expression in infected erythrocytes. Mol. Biochem. Parasitol. 2003, 126, 109–112. [Google Scholar] [CrossRef]
- Mannaerts, G.P.; Van Veldhoven, P.P.; Casteels, M. Peroxisomal lipid degradation via beta- and alpha-oxidation in mammals. Cell Biochem. Biophys. 2000, 32, 73–87. [Google Scholar] [CrossRef] [PubMed]
- Zou, B.; Nagle, A.; Chatterjee, A.K.; Leong, S.Y.; Tan, L.J.; Sim, W.L.S.; Mishra, P.; Guntapalli, P.; Tully, D.C.; Lakshminarayana, S.B.; et al. Lead optimization of imidazopyrazines: A new class of antimalarial with activity on Plasmodium liver stages. ACS Med. Chem. Lett. 2014, 5, 947–950. [Google Scholar] [CrossRef]
- Lambros, C.; Vanderberg, J.P. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 1979, 65, 418–420. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J.; et al. Ensembl 2020. Nucleic Acids Res. 2020, 48, D682–D688. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. feature Counts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.-Y.; Dillies, M.-A. SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Reimand, J.; Kull, M.; Peterson, H.; Hansen, J.; Vilo, J. g:Profiler—A web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007, 35, W193–W200. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dembele, L.; Dara, A.; Maiga, M.; Maiga, F.O.; Cissoko, D.; Djimde, A.A. Imidazolopiperazine (IPZ)-Induced Differential Transcriptomic Responses on Plasmodium falciparum Wild-Type and IPZ-Resistant Mutant Parasites. Genes 2023, 14, 2124. https://doi.org/10.3390/genes14122124
Dembele L, Dara A, Maiga M, Maiga FO, Cissoko D, Djimde AA. Imidazolopiperazine (IPZ)-Induced Differential Transcriptomic Responses on Plasmodium falciparum Wild-Type and IPZ-Resistant Mutant Parasites. Genes. 2023; 14(12):2124. https://doi.org/10.3390/genes14122124
Chicago/Turabian StyleDembele, Laurent, Antoine Dara, Mohamed Maiga, Fatoumata O. Maiga, Djeneba Cissoko, and Abdoulaye A. Djimde. 2023. "Imidazolopiperazine (IPZ)-Induced Differential Transcriptomic Responses on Plasmodium falciparum Wild-Type and IPZ-Resistant Mutant Parasites" Genes 14, no. 12: 2124. https://doi.org/10.3390/genes14122124
APA StyleDembele, L., Dara, A., Maiga, M., Maiga, F. O., Cissoko, D., & Djimde, A. A. (2023). Imidazolopiperazine (IPZ)-Induced Differential Transcriptomic Responses on Plasmodium falciparum Wild-Type and IPZ-Resistant Mutant Parasites. Genes, 14(12), 2124. https://doi.org/10.3390/genes14122124





