Abstract
Mercury has high industrial utility and is present in many products, and environmental contamination and occupational exposure are widespread. There are numerous biological systems involved in the absorption, metabolism, and excretion of Hg, and it is possible that some systems may be impacted by genetic variation. If so, genotype may affect tissue concentrations of Hg and subsequent toxic effects. Genome-wide association testing was performed on blood Hg samples from pregnant women of the Avon Longitudinal Study of Parents and Children (n = 2893) and children of the Human Early Life Exposome (n = 1042). Directly-genotyped single-nucleotide polymorphisms (SNPs) were imputed to the Haplotype Reference Consortium r1.1 panel of whole genotypes and modelled againstlog-transformed Hg. Heritability was estimated using linkage disequilibrium score regression. The heritability of Hg was estimated as 24.0% (95% CI: 16.9% to 46.4%) in pregnant women, but could not be determined in children. There were 16 SNPs associated with Hg in pregnant women above a suggestive p-value threshold (p < 1 × 10−5), and 21 for children. However, no SNP passed this threshold in both studies, and none were genome-wide significant (p < 5 × 10−8). SNP-Hg associations were highly discordant between women and children, and this may reflect differences in metabolism, a gene–age interaction, or dose–response effects. Several suggestive variants had plausible links to Hg metabolism, such as rs146099921 in metal transporter SLC39A14, and two variants (rs28618224, rs7154700) in potassium voltage-gated channel genes. The findings would benefit from external validation, as suggestive results may contain both true associations and false positives.
1. Introduction
Environmental Hg concentrations have increased substantially over the past century, and this is largely attributed to human industrial activity [1]. Rising emissions from human activity is expected to lead to increased human exposure to Hg over the coming decades [2], and the health and economic costs are projected to be considerable [3]. There are three common forms of Hg found in the human environment—elemental, inorganic (I-Hg), and the organic compound methylmercury (MeHg) [4]—each of which is highly toxic [5].
Human exposure to environmental Hg is possible under a variety of circumstances, but in most populations occurs primarily through dietary consumption of I-Hg and MeHg. Emissions of elemental and I-Hg are released as a by-product of industrial processes such as coal burning, and these emissions are dispersed globally into the atmosphere, oceans, and soils [6,7]. Hg deposits in the ocean shallows can be methylated by microorganisms to form MeHg [8]. This organic compound tends to remain stable within organisms which leads to accumulation up the food chain in increasing concentrations, with the greatest concentrations of MeHg observed in long-living predatory fish such as swordfish, tuna, and king mackerel [9,10]. A major source of human exposure is the consumption of these fish [11], in addition to other potentially contaminated foods such as rice, cereals [12,13], and meat [14], and trace quantities are possible in other food products [11]. Apart from diet, exposure may occur in occupations which involve handling Hg [15], or among those who have silver amalgam fillings, smoke cigarettes, or use skin-whitening creams or traditional medicines which may be contaminated with Hg [16].
After absorption, I-Hg tends to bind to thiol-containing proteins, and is quickly transported by plasma proteins, such as albumin, out of circulation and into tissues throughout the body [17]. Most I-Hg is deposited into the kidneys, and only a small fraction of total exposure may be detected by blood measurements [18]. MeHg, on the other hand, binds to erythrocytes, potentially by binding to the erythrocyte membrane, or through other transport mechanisms such as those involving D-glucose or cysteine [17,19]. It is then distributed to all tissues, including the brain and placenta [20]. Most Hg measured in whole blood consists of MeHg, and therefore total blood Hg is commonly considered indicative of recent dietary MeHg exposure [21].
Hg toxicity is enabled or enhanced due to structural similarity to common amino acids [22], which facilitates binding with sulfhydryl group organic compounds, lipids, proteins, and enzymes. The toxic effects of Hg are therefore broad and can disrupt cellular functioning and tissue health throughout the body [23], leading to increased reactive oxygen species and oxidative stress [24,25], carcinogenesis [26], epigenetic changes [27], and cell death [28].
Health effects from high doses of exposure, such as from occupational accidents, can result in acute poisoning, and require immediate treatment to avoid organ failure, neurological impairment, and long-term harm [29,30,31]. However, for people who are not at risk of occupational exposure, there may still be long-term health risks from continuous dietary exposure. This is because Hg may be absorbed in greater quantities than can be excreted, leading to increasing concentrations of Hg accumulated in tissues. Hg is slow to clear from the body due to cycles of methylation and demethylation between the forms of Hg which alter metabolic pathways [32] and the tendency for Hg in the kidneys or intestines to be reabsorbed into circulation [18,33]. For these reasons, the biological half-life corresponding to an approximate halving of internal concentrations is 1–3 weeks for I-Hg [34], and several months for MeHg [35]. Environmental Hg exposure is associated with increased blood pressure [36], risk of heart disease [37], kidney disease [38], and a wide range of neurological symptoms [21,39]. Pregnant women are a particularly vulnerable population, because Hg can readily cross the placenta [40] and accumulate at a higher Hg-to-weight ratio in the developing infant. However, the evidence for an effect of mercury in the general population on detectable developmental impairment is currently mixed [41,42].
Considering the complex pathways of absorption, transport, tissue distribution, and excretion, it is likely that genetic variation may mediate the relationship between environmental Hg and human exposure. Studies have reported associations between single-nucleotide polymorphisms (SNPs) in genes related to Hg metabolism and internal levels of Hg, such as in the glutathione metal-binding detoxification system [43,44,45], the metallothionein metal transport family [46], lipid-transport protein apolipoprotein-E [47], and genes involved in iron homeostasis [48]. Hg interacts with other elements such as selenium [49], zinc [50,51], cadmium, and lead [52,53], and variants which alter concentrations of these elements may also impact Hg [54,55,56]. The utility of identifying SNP-Hg associations is twofold; first, it can enhance our understanding of the biological mechanisms of how Hg acts on the body, second, it may enable new methods to test the impact of Hg on health by using SNPs as randomised proxies of Hg exposure [57].
Prior studies have tested associations between Hg and SNPs in genes targeted for their theoretical relevance. However, genome-wide association testing may be advantageous in identifying a greater proportion of relevant SNPs. This approach is hypothesis-free and all available SNPs are tested, so variants can be identified not only in genes but also in non-coding regions of the genome [58,59]. This method has previously been used to identify novel genetic variants associated with blood concentrations of copper [55], iron [60], lead [54], manganese [61], selenium [55,56], and zinc [55].
The objective of this study was to assess the associations between SNPs and blood Hg concentrations in pregnant women and children using genome-wide association testing. Specific aims were (1) to estimate the heritability of blood Hg levels using linkage disequilibrium score (LDSC) regression, (2) to perform genome-wide association testing between imputed SNPs and blood Hg in two European populations, (3) to explore the function of strongly associated SNPs through in silico analyses, and (4) to compare associations between Hg and candidate variants identified from previous studies.
2. Materials and Methods
2.1. Overview
Genome-wide associations were estimated between SNPs and blood Hg concentrations in two separate European studies, one of pregnant women and one of children. Table 1 includes a brief summary of the characteristics of each study, with more details available in Supplementary Table S1.

Table 1.
Summary of included studies.
2.2. The Avon Longitudinal Study of Parents and Children (ALSPAC)
ALSPAC is a multi-generational birth cohort in the former Avon Health Authority area in the UK. All pregnant women living within this area with expected dates of delivery between 1 April 1991 and 31 December 1992 were invited to take part in the study. From 20,248 pregnancies identified as eligible, 14,541 were initially enrolled, which, after accounting for multiple pregnancies, resulted in 14,203 unique mothers. This was expanded with additional phases of recruitment to provide a total of 14,833 unique women in the study. Full details of the recruitment process and sample profile are described elsewhere [62,63]. Details of all the data that are available from the study are available in a fully searchable online data dictionary and variable search tool: http://www.bristol.ac.uk/alspac/researchers/our-data/ (accessed on 1 October 2023). Participant characteristics were representative of most UK women. However, women were predominantly of European ancestry, and due to the potential for population stratification, only European women were included in this GWAS analysis, which limits the generalizability of findings to populations with different ancestry [62].
Whole blood samples were taken from 4844 pregnant women during early antenatal care visits, with a median visit time of 11 weeks of gestation (IQR: 4 weeks). A vacutainer system was operated by midwives to draw the samples, which were stored at 4 °C for 1–4 days before being sent to the central Bristol laboratory. Samples were transported for up to 3 h at room temperature, and then stored at 4 °C until the time of analysis.
Whole blood Hg was measured using inductively coupled plasma dynamic reaction cell mass spectrometry (ICP-DRC-MS) at the Centers for Disease Control and Prevention (CDC), Bethesda, CDC method 3009.1. Quality control (QC) measures are described in earlier studies [55,64], which left 4131 measurements after exclusions. One sample was below the limit of detection for Hg (0.24 μg/L) and was assigned a value 0.7 times the lower limit of detection [65].
Blood samples for DNA analysis were taken during pregnancy from 10,015 women [66]. Samples were genotyped by Centre National de Génotypage (CNG) using the Illumina Human660W-Quad Array. Genotype annotation was performed using Illumina GenomeStudio [67], and aligned to GRCh37 with the software Burrows-Wheeler Aligner version 1. QC procedures were applied to the genotyped data using Plink v1.07 [68]. SNPs were excluded if they were missing from more than 5% of individuals, had a Hardy–Weinberg Equilibrium (HWE) p < 1.0 × 10−7, or a minor allele frequency (MAF) of less than 1%. Individuals were excluded if they were missing more than 5% of SNPs, had indeterminate X chromosome heterozygocity or extreme autosomal heterozygocity (>3 standard deviations from population mean), were population outliers using four HapMap populations as a reference, or had a cryptic relatedness estimate equivalent to first cousin or closer (identify by descent, IBD > 0.125) with another individual in the sample [69,70]. Directly genotyped SNPs were imputed to the Haplotype Reference Consortium (HRC r1.1) panel of approximately 31,000 phased whole genotypes. Phasing was performed using ShapeIt v2 [71] and imputation using Impute V3 on the Michigan Imputation Server [72]. SNPs were excluded following imputation where MAF <1% or imputation quality score (INFO) < 0.9.
2.3. The Human Early Life Exposome (HELIX)
HELIX comprises subcohorts of mother–child pairs from six European birth cohorts [73,74]. The cohorts enrolled approximately 32,000 pairs between 1999 and 2010 in the UK, France, Spain, Lithuania, Norway, and Greece [75] (Table 2). From these studies, 1301 children were included in the HELIX subcohort, which measured a variety of pre- and postnatal exposures, health outcomes, and genome-wide genotypes. The current study only included children with genetic data, Hg levels, and who were of European ancestry (determined from genome-wide genetic information) (n = 1042).

Table 2.
Locations of the study populations.
Child blood samples were collected during follow-up clinic visits between December 2013 and February 2016, when the children were aged 6 to 11 years old [76]. All cohorts followed the same procedures and analysis protocols. Whole blood was stored in EDTA vacutainers and analysed for trace element testing and DNA extraction at ALS Scandinavia (Sweden). Total Hg levels were measured using double focusing sector field inductively coupled plasma mass spectrometry (ICP-SFMS) as described elsewhere [77]. The limit of detection was 0.02 µg/L.
The Infinium Global Screening Array (GSA) (Illumina) was used for genome-wide genotyping at the Human Genomics Facility (HuGe-F), Erasmus MC (www.glimdna.org). GenomeStudio 2.0 software with the GenTrain2.0 algorithm was used for genotype calling, and annotation on GRCh37 using the GSAMD-24v1-0_20011747_A4 manifest. Samples were excluded if there was SNP missingness >3%, sex mismatch, heterozygosity (>4 SD), cryptic relatedness (Pi-hat > 0.185), or duplicates. SNPs were excluded if missing from >5% individuals, MAF < 1%, or HWE p < 1.0 × 10−6.
Genotype information was phased to HRC r1.1 using Eagle v2.4 and imputed using Minimac4 and the Michigan Imputation Server [78]. Post-imputation filtering was applied to exclude SNPs with low imputation quality (R2 < 0.9), allele frequency (MAF < 1%), or HWE p > 1.0 × 10−7.
2.4. SNP Heritability
Heritability refers to the amount of outcome variation which is attributable to genetic differences. We estimated Hg heritability for measured SNPs (h2g) by applying LD score regression [79,80] to summary statistics from each GWAS. In brief, this method involved taking SNP-level data and regressing standardised SNP-Hg associations on the sum of correlations between a SNP and those nearby, known as LD scores. The rationale behind this is that a high LD score increases the probability that a SNP is correlated with a true causal SNP of Hg. LD scores were taken from a reference panel computed from 1000 Genomes Project European data [81], which was accessed from https://data.broadinstitute.org/alkesgroup/LDSCORE (accessed on 1 October 2023) with the filename ‘eur_w_ld_chr’.
2.5. Genome-Wide Association Testing
Genome-wide association testing was performed to estimate the association between each SNP and a continuous Hg phenotype. In ALSPAC, the GWAS of women was conducted in SNPTEST version 2.5.2 using the frequentist option and “score” method of accounting for genotype uncertainty [82]. In HELIX, the GWAS of children was run using PLINK version 1.0 with the “--linear” option [68]. Each analysis was adjusted for age at the time when blood was taken and eigenvectors for the first 10 principal components estimated from GWAS data. The continuous linear models assumed an additive effect of SNPs and a normal distribution of phenotype residuals. The distribution of Hg had a strong right skew and if regressed in its raw form was unlikely to meet the latter assumption, and this could bias standard error and p-values. To address this, Hg measurements were log2-transformed to approximate a normal distribution.
Follow-up analysis was performed in R version 4.1.0 unless otherwise stated. Reference SNP IDs were missing from all ALSPAC results and some HELIX, and therefore chromosome and location were used to identify labels valid for GRCh37 using the “SNP locations for Homo sapiens (dbSNP Build 144)” reference table and “BSgenome” R packages [83,84]. Results were visualised with quantile–quantile (QQ) and Manhattan plots generated using the “qqman” package [85]. SNPs were classified as genome-wide significant if p < 5 × 10−8, and suggestively significant if p was between 1 × 10−5 and 5 × 10−8. Variants in linkage disequilibrium (R2 > 0.1 and 250 kb range) were grouped using the ld_clump function of the MRC IEU GWAS R package [86] and the most significant SNP kept. The strongest results from each GWAS were compared.
In addition to identifying suggestive and significant SNPs, summary statistics were extracted and reported for 13 variants of interest which were previously identified as (a) associated with Hg levels in candidate gene studies or (b) associated with metals which may interact with Hg levels in genome-wide association studies (Supplementary Table S2).
2.6. In Silico Functional Analysis
All variants with p < 1 × 10−5 were mapped to the nearest gene using the SNP2Gene function in Functional Mapping and Annotation of Genome-Wide Association Studies (FUMA) [87], and the results were verified in NCBI Sequence Viewer [88]. The potential biological mechanisms of how the variants may affect Hg were investigated using tools which aggregated prior genetic research. SNP-phenotype associations were explored using FUMA eQTL [87], LDtrait [89], and PhenoScanner [90,91]. Gene functions were explored in the GeneCards database [92], gene–tissue expression using the GTEx portal [93,94], and gene–phenotype associations in the Online Mendelian Inheritance in Man database [95].
3. Results
3.1. Study Characteristics
The derivation of the number of participants and SNPs included in each study is shown in Table 3. There were 2893 women included in this study, with a median age of 28.0 years (IQR: 6.0). The mean concentration of blood Hg was 2.09 μg/L (standard deviation, SD: 1.08) and median 1.89 μg/L (IQR: 1.16). The study included 1042 children with a median age of 8.0 years (IQR: 2.4) and 54.6% were male. As seen in Figure 1, Hg concentrations were lower for children, with mean Hg 1.35 μg/L and median 0.82 μg/L, and there was slightly more variance (IQR: 1.27). For improved readability, Figure 1 excludes seven samples where Hg > 8 μg/L, and complete histograms are available in Supplementary Figure S1.

Table 3.
Study size derivation.
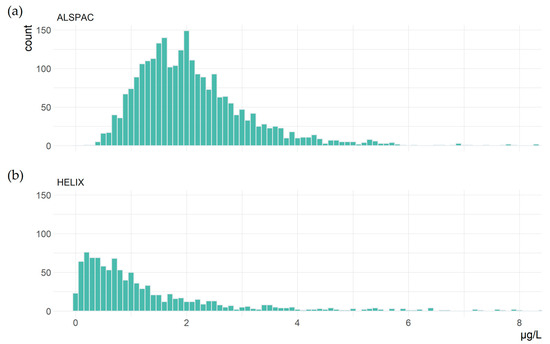
Figure 1.
Blood Hg concentrations in (a) 2890 women and (b) 1038 children aged 6–11 years old.
3.2. SNP Heritability
The SNP heritability (h2g) of Hg was calculated from summary statistics of the GWAS performed in this study. Data were available for 6,620,135 and 6,138,843 SNPs for women and children, respectively. Standardised effect estimates were regressed on LD scores of each SNP taken from the 1000 Genome Project Europeans reference panel. Merging summary statistics and the reference panel resulted in 1,137,154 and 965,135 SNPs for women and children available for LD score regression.
The estimated h2g for women was 24.0% (95% CI: 16.9% to 46.4%, p = 0.01). For children, the estimate was 4.8%, but the evidence for this was very weak (p = 0.85), and confidence intervals were very wide and overlapped with zero (95% CI: −45.7% to 55.4%).
3.3. Genome-Wide Association Testing
SNP-Hg associations are visualised in Manhattan plots in Supplementary Figure S2, and expected and observed p-values are compared in Supplementary Figure S3 using QQ plots. The plots did not show visible inflation or deflation of p-values, and this was reflected in lambda statistics of 1.01 for women and 1.00 for children.
No SNPs were found to be genome-wide-significant (p < 5 × 10−8) for women or children. At a lower threshold of p < 1 × 10−5, there were suggestive associations at 16 independent loci for women and 21 for children. The strongest association at each loci was selected, and summary statistics are presented in Table 4.

Table 4.
Summary statistics for the variants suggestively associated (p < 1 × 10−5) with blood Hg concentrations, pruned to the most significant SNP per independent genetic loci.
In total, there were 37 SNPs at independent genetic loci with suggestive associations in one of the studies. However, none of these were found to be associated in the alternative study even at a more relaxed threshold of p > 1 × 10−2. Not all SNPs could be compared between studies, due to presenting at low MAF (lower than the threshold for inclusion) in one of the cohorts.
3.4. In Silico Functional Analysis
The 37 SNPs with suggestively significant associations with blood Hg were mapped to their nearest genes using NCBI Sequence Viewer and FUMA SNP2Gene (Supplementary Table S3). Additionally, we identified variants strongly associated with the expression of other genes (p > 5 × 10−8) using Phenoscanner, and included those genes in the following analyses.
SNPs were most commonly associated with gene expression, histone modification, and methylation at genes or CpG sites close to the SNP locations (Supplementary Table S4). No SNPs or strongly correlated SNPs (r2 > 0.8) were found to have direct evidence of connections to Hg metabolism, but several were located inside genes with potential links to Hg.
In this study, the T allele of intronic variant rs146099921 was associated with −0.12 log Hg (p = 8.21 × 10−6) for women. This SNP is located in the gene Solute Carrier Family 39 Member 14 (SLC39A14), and within the gene is associated with DNA methylation at cg14348540 [96,97] and exon expression [98]. SLC39A14 (also referred to as ZIP14) is a metal transporter linked to cellular uptake of cadmium, iron, manganese, and zinc [99]. Mutations are associated with the impairment of manganese transport and homeostasis, leading to toxic accumulation [100,101,102]. There is evidence that the gene functions as a transporter of zinc [103], and mediates iron and cadmium uptake [104,105], and a detailed review of transport functions was identified [99]. According to data available in the GTEx Portal [94], SLC39A14 is expressed most highly in the liver, followed by the adipose tissue, the arteries, and pancreas.
There were further links between suggestive variants and genes with functions potentially affecting Hg levels, such as for rs17106291 (Solute Carrier Family 25 Member 21, SLC25A21) which transports dicarboxylates across the inner membranes of mitochondria [106]. Two variants (rs28618224, rs7154700) were in potassium voltage-gated channels genes, and two variants were near to genes affecting glutamate (rs361166) and phospholipid (rs115812569) transport.
3.5. Associations in Previous Candidate Variants
GWAS summary statistics were extracted for 14 SNPs which were identified a priori as variants of interest due to prior studies reporting associations with Hg or metals which interact with Hg (Supplementary Table S2). Associations with blood Hg are shown in Table 5. Most were not replicated in either women or children. Exceptions were the minor C allele of rs10636 which was associated with increased blood Hg in women (p = 0.01), and the minor C allele rs9936741 associated with lower Hg in children (p = 0.01). Neither association was replicated in the alternative study, and no association was found in variants previously reported as genome-wide-significant from GWAS of blood lead, selenium, or zinc.

Table 5.
SNP-Hg summary statistics for candidate variants identified in previous studies of Hg or other metal GWAS.
4. Discussion
Genome-wide association testing of blood Hg from British pregnant women and children across Europe did not identify strong associations with any imputed SNPs. Despite this, heritability from approximately 1 million SNPs was estimated to explain a considerable proportion of Hg variance in pregnant women (24.0%, 95% CI: 16.9 to 46.4). Considering that Hg is highly reactive with a wide range of molecules and exposure is affected by numerous biological processes, it is expected that a substantial component of its metabolism would be heritable and the finding is consistent with animal and plant studies [109,110] which also estimated there were large genetic components to Hg variation.
Although no variants passed genome-wide significance thresholds, there were 37 independent loci detected between the two studies at a lower threshold considered suggestive of an association. Surprisingly, no SNP was found to be suggestively associated in both women or children, even at a more relaxed threshold of p > 1 × 10−2. There are several possible reasons for this. First, there may be qualitative differences in Hg metabolism between pregnant women adults and children. It is possible that metabolic processes change with age or pregnancy, and although no human studies have explored or speculated on this, there is evidence in animals that rates of Hg absorption and excretion are different in early life compared to adulthood [111,112]. Secondly, the child GWAS was much smaller (1042 vs. 2893) and median blood Hg level was lower (0.82 vs. 1.89 μg/L), which may have led to a lack of statistical power, increased rates of false positives, and/or reduced SNP effects due to lower concentrations of Hg. We found indications of this in our heritability analyses, where there was insufficiant certainty to produce a meaningful estimate (h2g = 4.8%, 95% CI: −45.7% to 55.4%). A final reason for the heterogeneity may be if non-linear associations exist between SNPs and Hg, if for example a gene is only expressed above a certain threshold of Hg exposure.
The SNP rs146099921 was identified as the most biologically plausible of those with suggestive associations with Hg, located in the gene SLC39A14. While the variant is intronic, there is evidence it has an active effect through modification of SLC39A14 gene expression [98], methylation [96,97], and exon expression [98]. Most SLC39 genes are responsible for the cellular uptake of zinc [113], but studies suggest SLC39A14 is associated with multiple metals, including cadmium, iron, and manganese levels [99,100,104]. It is possible the gene impacts Hg levels indirectly through these metals, each of which may interact with Hg—for example, increased cellular zinc induces metallothionein synthesis which may promote removal of Hg [51]. Alternatively, SLC39A14 may directly affect the transport of Hg, but this does not appear to be reported in prior studies. In children, a suggestive association was found with rs17106291, which is located in SLC25A21, a transporter of C5-C7 oxodicarboxylates to mitochondria. Prior studies have linked other members of the SLC gene family to kidney uptake of I-Hg [114] and the intestinal transport of MeHg [17].
Several other suggestive SNPs were annotated to genes with possible connections to Hg metabolism. Associations were identified in SNPs located in potassium voltage-gated channels genes KCNH5 and KCNIP4 and the calcium ion channel gene TRPC4. These are potentially relevant to Hg, because expression of MeHg and I-Hg toxicity may be linked to inhibited potassium or calcium channels [51], although it is unclear what impact would be expected on blood Hg. Finally, there were variants located in proximity to genes affecting glutamate (GLS, GATB), relevant because Hg both inhibits glutamate uptake [115] and stimulates its release [116].
Associations reported in previous candidate gene studies of Hg were for the most part not replicated by this study. There were seven variants of interest to Hg metabolism, located in genes GCLM, GCLC, TF, MT1A, MT1M, and MT2A. In women, there was evidence of an association between blood HG and rs10636 (MT2A), and the direction of effect was consistent with that previously reported [46]. In children, rs9936741 (MT1M) was associated with Hg but in the opposite direction than a prior study [46], potentially due to the use of hair Hg in the original study. Both variants have a biologically plausible link to Hg: metallothionein generates proteins which bind to Hg toaid clearance [117]. Neither of the above associations were replicated in the alternative study, and the p-values of each association (p = 0.01) provide only tentative evidence because they were considerably below genome-wide thresholds. However, these variants were selected a priori and it therefore seemed more appropriate to apply standard observational thresholds. No other candidate SNP associations were replicated. This may be due to the smaller sample sizes used in prior studies which increased the likelihood of spurious results, or due to differences in tissues used to measure Hg. Finally, an epigenome-wide analysis of umbilical cord Hg identified associations in the genes GGH, MED31, and GRK1 and DNA methylation [118]. The direction of causality is unclear, but in this study no strong signals were found in variants near the reported CpG sites.
Limitations of this study were as follows. First, the lack of genome-wide significant SNPs and high heterogeneity between studies suggests one or more of the analyses may have been underpowered. By comparison, two larger GWAS reported one loci associated with blood lead levels (n = 5433) [54] and two with selenium levels (n = 9639) [56]. The required sample size is also a function of the trait variance, as demonstrated in an arsenic GWAS in Bangladesh which identified five independent loci from 1313 arsenic-exposed individuals [119]. The low power may be part of the reason for the discordance between ALSPAC and HELIX results, and in particular for SNPs with low MAF.
A second limitation is the use of blood Hg. This reflects relatively short-term exposure and is therefore subject to daily variation depending on diet, metabolism, and random noise [120,121]. In linear models, the measurement error between blood Hg and underlying exposure to Hg may lead to residual error and reduce study power. There are samples such as hair and nails which represent Hg exposure over a longer time-frame of several months, and this is something future studies should consider.
A third limitation is that the HELIX study comprised six subcohorts located in different countries. These populations had different profiles of Hg exposure and environmental variation, and while no heterogeneity was detected in the pooled analysis, it is possible this increased uncertainty in the GWAS estimates. Finally, this study was conducted on European ancestry populations, and populations with different ancestries are likely to have different genotype-Hg associations, thus our results may not be relevant to other populations.
5. Conclusions
In this GWAS of women and children, no SNPs were found to be associated with Hg above genome-wide significant thresholds. However, in women, SNP heritability was estimated to be around 24%, and some SNPs, in particular the variant rs146099921-located metal transport gene SLC39A14, were suggestively associated with blood Hg. Low correlations between results from pregnant women and children could reflect developmental changes in Hg metabolism, exposure levels or population heterogeneity.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes14122123/s1, Supplementary Table S1. Summary of study population, measurement methods, and GWAS characteristics. Supplementary Table S2. Previously identified variants. Supplementary Figure S1. Hg distributions. Supplementary Figure S2. GWAS Manhattan plots. Supplementary Figure S3. QQ plots. Supplementary Table S3. Gene mapping of selected results. Supplementary Table S4. Functional analysis of selected results.
Author Contributions
Conceptualization, K.D., P.Y. and S.J.L.; methodology, K.D. and S.J.L.; software, K.D.; validation K.D.; formal analysis, K.D. and M.B.; investigation, K.D.; resources, A.L.B., D.M., G.E., K.B.G., K.D. and R.G.; data curation, K.D. and M.B.; writing—original draft preparation, K.D.; writing—review and editing, A.L.B., C.M.T., D.M., G.E., K.B.G., M.B., M.L., P.Y., S.J.L., S.L. and R.G.; visualization, K.D.; supervision, S.J.L. and C.M.T.; project administration, K.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the following sources. K.D. is supported by a Ph.D. studentship from the MRC Integrative Epidemiology Unit at the University of Bristol (faculty matched place for MRC and Peter and Jean James Scholarship). C.M.T. is supported by an MRC Career Development Award (MR/T010010/1). S.J.L. and M.L. were funded by Generalitat Valenciana (GV/2021/111), Ministry of Universities (CAS21/00008 and NextGeneration EU), Instituto de Salud Carlos III (FIS-FEDER: 13/1944, 16/1288, 17/00663 and 19/1338; FIS-FSE: 17/00260; Miguel Servet-FSE: MSII20/0006). P.Y. is supported by the Medical Research Council Integrative Epidemiology Unit at the University of Bristol (MC_UU_00011/5). S.J.L. and P.Y. were supported by the NIHR Biomedical Research Centre at University Hospitals Bristol and Weston NHS Foundation Trust and the University of Bristol. ALSPAC: The UK Medical Research Council and Wellcome (Grant ref: 217065/Z/19/Z) and the University of Bristol provide core support for ALSPAC. This publication is the work of the authors and K.D. will serve as guarantor for the contents of this paper. Publication was supported by MRC IEU grant MC_UU_00011/1. A comprehensive list of grant funding is available on the ALSPAC website: http://www.bristol.ac.uk/alspac/external/documents/grant-acknowledgements.pdf; This research was specifically funded by Wellcome Trust Grant (WT088806) [genotyping] and NIHR (NF-SI-0611-10196) [blood samples]. The assays of the maternal blood samples were carried out at the Centers for Disease Control and Prevention with funding from NOAA, and the statistical analyses were carried out in Bristol with funding from NOAA and support from the Intramural Research Program of NIAAA, NIH. HELIX: The study has received funding from the European Community’s Seventh Framework Programme (FP7/2007-206) under grant agreement no 308333 (HELIX project) and the H2020-EU.3.1.2.—Preventing Disease Programme under grant agreement no 874583 (ATHLETE project). The genotyping was supported by the projects PI17/01225 and PI17/01935, funded by the Instituto de Salud Carlos III and co-funded by the European Union (ERDF, “A way to make Europe”) and the Centro Nacional de Genotipado-CEGEN (PRB2-ISCIII). BiB received core infrastructure funding from the Wellcome Trust (WT101597MA) and a joint grant from the UK Medical Research Council (MRC) and Economic and Social Science Research Council (ESRC) (MR/N024397/1). INMA data collections were supported by grants from the Instituto de Salud Carlos III (PI16/1288 and PI19/1338), CIBERESP, the Generalitat de Catalunya-CIRIT and the Generalitat Valenciana (CIAICO/2021/132). KANC was funded by the grant of the Lithuanian Agency for Science Innovation and Technology (6-04-2014_31V-66). The Norwegian Mother, Father and Child Cohort Study is supported by the Norwegian Ministry of Health and Care Services and the Ministry of Education and Research. The Rhea project was financially supported by European projects (EU FP6-2003-Food-3-NewGeneris, EU FP6. STREP Hiwate, EU FP7 ENV.2007.1.2.2.2. Project No 211250 Escape, EU FP7-2008-ENV-1.2.1.4 Envirogenomarkers, EU FP7-HEALTH-2009-single stage CHICOS, EU FP7 ENV.2008.1.2.1.6. Proposal No 226285 ENRIECO, EU-FP7-HEALTH-2012 Proposal No 308333 HELIX), and the Greek Ministry of Health (Program of Prevention of obesity and neurodevelopmental disorders in preschool children, in Heraklion district, Crete, Greece: 2011–2014; “Rhea Plus”: Primary Prevention Program of Environmental Risk Factors for Reproductive Health, and Child Health: 2012-15). ISGlobal acknowledges support from the Spanish Ministry of Science and Innovation through the “Centro de Excelencia Severo Ochoa 2019–2023” Program (CEX2018-000806-S), and support from the Generalitat de Catalunya through the CERCA Program.
Institutional Review Board Statement
Ethics approval for the study was obtained from the ALSPAC Ethics and Law Committee and the Local Research Ethics Committees. Consent for biological samples has been collected in accordance with the Human Tissue Act (2004). Informed consent for the use of data collected via questionnaires and clinics was obtained from participants following the recommendations of the ALSPAC Ethics and Law Committee at the time. In HELIX, local ethical committees approved the studies that were conducted according to the guidelines laid down in the Declaration of Helsinki. The ethical committees for each cohort were the following: BIB: Bradford Teaching Hospitals NHS Foundation Trust, EDEN: Agence nationale de sécurité du médicament et des produits de santé, INMA: Comité Ético de Inverticación Clínica Parc de Salut MAR, KANC: LIETUVOS BIOETIKOS KOMITETAS, MoBa: Regional komité for medisinsk og helsefaglig forskningsetikk, Rhea: Ethical committee of the general university hospital of Heraklion, Crete. Informed consent was obtained from a parent and/or legal guardian of all participants in the study. Participants did not receive any compensation.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
GWAS summary statistics for LD-clumped SNPs which were below a suggestive p-value threshold are reported in Table 2. Full summary statistics for European pregnant women are publicly available on the GWAS Catalog (https://www.ebi.ac.uk/gwas/ (accessed on 1 November 2023)), Study Accession: GCST90271315.
Acknowledgments
We are extremely grateful to all the families who took part in the ALSPAC and HELIX studies, the midwives for their help in recruiting them, and the ALSPAC and HELIX teams, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, and nurses.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gworek, B.; Dmuchowski, W.; Baczewska-Dąbrowska, A.H. Mercury in the terrestrial environment: A review. Environ. Sci. Eur. 2020, 32, 128. [Google Scholar] [CrossRef]
- Sunderland, E.M.; Selin, N.E. Future trends in environmental mercury concentrations: Implications for prevention strategies. Environ. Health 2013, 12, 2. [Google Scholar] [CrossRef]
- Zhang, Y.; Song, Z.; Huang, S.; Zhang, P.; Peng, Y.; Wu, P.; Gu, J.; Dutkiewicz, S.; Zhang, H.; Wu, S.; et al. Global health effects of future atmospheric mercury emissions. Nat. Commun. 2021, 12, 3035. [Google Scholar] [CrossRef]
- Broussard, L.A.; Hammett-Stabler, C.A.; Winecker, R.E.; Ropero-Miller, J.D. The Toxicology of Mercury. Lab. Med. 2002, 33, 614–625. [Google Scholar] [CrossRef]
- World Health Organization. Children’s Exposure to Mercury Compounds; World Health Organization: Geneva, Switzerland, 2010. [Google Scholar]
- Driscoll, C.T.; Mason, R.P.; Chan, H.M.; Jacob, D.J.; Pirrone, N. Mercury as a Global Pollutant: Sources, Pathways, and Effects. Environ. Sci. Technol. 2013, 47, 4967–4983. [Google Scholar] [CrossRef] [PubMed]
- Selin, N.E.; Jacob, D.J.; Park, R.J.; Yantosca, R.M.; Strode, S.; Jaeglé, L.; Jaffe, D. Chemical cycling and deposition of atmospheric mercury: Global constraints from observations. J. Geophys. Res. Atmos. 2007, 112, 1–14. [Google Scholar] [CrossRef]
- Parks, J.M.; Johs, A.; Podar, M.; Bridou, R.; Hurt, R.A., Jr.; Smith, S.D.; Tomanicek, S.J.; Qian, Y.; Brown, S.D.; Brandt, C.C. The genetic basis for bacterial mercury methylation. Science 2013, 339, 1332–1335. [Google Scholar] [CrossRef]
- Hammerschmidt, C.R.; Fitzgerald, W.F. Methylmercury in Freshwater Fish Linked to Atmospheric Mercury Deposition. Environ. Sci. Technol. 2006, 40, 7764–7770. [Google Scholar] [CrossRef]
- Médieu, A.; Point, D.; Itai, T.; Angot, H.; Buchanan, P.J.; Allain, V.; Fuller, L.; Griffiths, S.; Gillikin, D.P.; Sonke, J.E.; et al. Evidence that Pacific tuna mercury levels are driven by marine methylmercury production and anthropogenic inputs. Proc. Natl. Acad. Sci. USA 2022, 119, e2113032119. [Google Scholar] [CrossRef]
- Golding, J.; Steer, C.D.; Hibbeln, J.R.; Emmett, P.M.; Lowery, T.; Jones, R. Dietary Predictors of Maternal Prenatal Blood Mercury Levels in the ALSPAC Birth Cohort Study. Environ. Health Perspect. 2013, 121, 1214–1218. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, O.; Zamorano, P.; Garcia, O.; Bastías, J.M. Arsenic, cadmium, mercury, sodium, and potassium concentrations in common foods and estimated daily intake of the population in Valdivia (Chile) using a total diet study. Food Chem. Toxicol. 2017, 109, 1125–1134. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-S.; Cho, Y.-H.; Park, S.-O.; Kye, S.-H.; Kim, B.-H.; Hahm, T.-S.; Kim, M.; Lee, J.O.; Kim, C.-I. Dietary exposure of the Korean population to arsenic, cadmium, lead and mercury. J. Food Compos. Anal. 2006, 19, S31–S37. [Google Scholar] [CrossRef]
- Almerud, P.; Zamaratskaia, G.; Lindroos, A.K.; Bjermo, H.; Andersson, E.M.; Lundh, T.; Ankarberg, E.H.; Lignell, S. Cadmium, total mercury, and lead in blood and associations with diet, sociodemographic factors, and smoking in Swedish adolescents. Environ. Res. 2021, 197, 110991. [Google Scholar] [CrossRef]
- Berlin, M.; Zalups, R.K.; Fowler, B.A. Chapter 46–Mercury. In Handbook on the Toxicology of Metals, 4th ed.; Nordberg, G.F., Fowler, B.A., Nordberg, M., Eds.; Academic Press: San Diego, CA, USA, 2015; pp. 1013–1075. [Google Scholar]
- United Nations Environment Programme; World Health Organization. Guidance for Identifying Populations at Risk from Mercury Exposure; UNEP: Nairobi, Kenya, 2008. [Google Scholar]
- Bridges, C.C.; Zalups, R.K. Mechanisms involved in the transport of mercuric ions in target tissues. Arch. Toxicol. 2017, 91, 63–81. [Google Scholar] [CrossRef]
- Park, J.-D.; Zheng, W. Human Exposure and Health Effects of Inorganic and Elemental Mercury. J. Prev. Med. Public Health 2012, 45, 344–352. [Google Scholar] [CrossRef]
- Yin, Z.; Jiang, H.; Syversen, T.; Rocha, J.B.T.; Farina, M.; Aschner, M. The methylmercury-l-cysteine conjugate is a substrate for the L-type large neutral amino acid transporter. J. Neurochem. 2008, 107, 1083–1090. [Google Scholar] [CrossRef]
- Mortensen, M.E.; Caudill, S.P.; Caldwell, K.L.; Ward, C.D.; Jones, R.L. Total and methyl mercury in whole blood measured for the first time in the U.S. population: NHANES 2011–2012. Environ. Res. 2014, 134, 257–264. [Google Scholar] [CrossRef]
- Yokoo, E.M.; Valente, J.G.; Grattan, L.; Schmidt, S.L.; Platt, I.; Silbergeld, E.K. Low level methylmercury exposure affects neuropsychological function in adults. Environ. Health 2003, 2, 8. [Google Scholar] [CrossRef]
- Bridges, C.C.; Krasnikov, B.F.; Joshee, L.; Pinto, J.T.; Hallen, A.; Li, J.; Zalups, R.K.; Cooper, A.J. New insights into the metabolism of organomercury compounds: Mercury-containing cysteine S-conjugates are substrates of human glutamine transaminase K and potent inactivators of cystathionine γ-lyase. Arch. Biochem. Biophys. 2012, 517, 20–29. [Google Scholar] [CrossRef]
- Bernhoft, R.A. Mercury toxicity and treatment: A review of the literature. J. Environ. Public Health 2012, 2012, 460508. [Google Scholar] [CrossRef]
- Grotto, D.; Valentini, J.; Fillion, M.; Passos, C.J.S.; Garcia, S.C.; Mergler, D.; Barbosa, F., Jr. Mercury exposure and oxidative stress in communities of the Brazilian Amazon. Sci. Total Environ. 2010, 408, 806–811. [Google Scholar] [CrossRef]
- Syversen, T.; Kaur, P. The toxicology of mercury and its compounds. J. Trace Elem. Med. Biol. 2012, 26, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Zefferino, R.; Piccoli, C.; Ricciardi, N.; Scrima, R.; Capitanio, N. Possible Mechanisms of Mercury Toxicity and Cancer Promotion: Involvement of Gap Junction Intercellular Communications and Inflammatory Cytokines. Oxidative Med. Cell. Longev. 2017, 2017, 7028583. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Momtaz, S.; Abdollahi, M. The relationship between mercury exposure and epigenetic alterations regarding human health, risk assessment and diagnostic strategies. J. Trace Elements Med. Biol. 2019, 52, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Orr, S.E.; Barnes, M.C.; Joshee, L.; Uchakina, O.; McKallip, R.J.; Bridges, C.C. Potential mechanisms of cellular injury following exposure to a physiologically relevant species of inorganic mercury. Toxicol. Lett. 2019, 304, 13–20. [Google Scholar] [CrossRef]
- Ye, B.-J.; Kim, B.-G.; Jeon, M.-J.; Kim, S.-Y.; Kim, H.-C.; Jang, T.-W.; Chae, H.-J.; Choi, W.-J.; Ha, M.-N.; Hong, Y.-S. Evaluation of mercury exposure level, clinical diagnosis and treatment for mercury intoxication. Ann. Occup. Environ. Med. 2016, 28, 5. [Google Scholar] [CrossRef] [PubMed]
- Do, S.Y.; Lee, C.G.; Kim, J.Y.; Moon, Y.H.; Kim, M.S.; Bae, I.H.; Song, H.S. Cases of acute mercury poisoning by mercury vapor exposure during the demolition of a fluorescent lamp factory. Ann. Occup. Environ. Med. 2017, 29, 19. [Google Scholar] [CrossRef]
- Asano, S.; Eto, K.; Kurisaki, E.; Gunji, H.; Hiraiwa, K.; Sato, M.; Sato, H.; Hasuike, M.; Hagiwara, N.; Wakasa, H. Acute inorganic mercury vapor inhalation poisoning. Pathol. Int. 2000, 50, 169–174. [Google Scholar] [CrossRef]
- Clarkson, T.W. The three modern faces of mercury. Environ. Health Perspect. 2002, 110 (Suppl. 1), 11–23. [Google Scholar] [CrossRef]
- Bridges, C.C.; Zalups, R.K. Transport of Inorganic Mercury and Methylmercury in Target Tissues and Organs. J. Toxicol. Environ. Health Part B Crit. Rev. 2010, 13, 385–410. [Google Scholar] [CrossRef]
- Fisher, J.F.; World Health Organization. Elemental Mercury and Inorganic Mercury Compounds: Human Health Aspects; World Health Organization: Geneva, Switzerland, 2003. [Google Scholar]
- Jo, S.; Woo, H.D.; Kwon, H.-J.; Oh, S.-Y.; Park, J.-D.; Hong, Y.-S.; Pyo, H.; Park, K.S.; Ha, M.; Kim, H.; et al. Estimation of the Biological Half-Life of Methylmercury Using a Population Toxicokinetic Model. Int. J. Environ. Res. Public Health 2015, 12, 9054–9067. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.F.; Singh, K.; Chan, H.M. Mercury Exposure, Blood Pressure, and Hypertension: A Systematic Review and Dose–response Meta-analysis. Environ. Health Perspect. 2018, 126, 076002. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.F.; Lowe, M.; Chan, H.M. Mercury exposure, cardiovascular disease, and mortality: A systematic review and dose-response meta-analysis. Environ. Res. 2021, 193, 110538. [Google Scholar] [CrossRef]
- Bridges, C.C.; Zalups, R.K. The aging kidney and the nephrotoxic effects of mercury. J. Toxicol. Environ. Health Part B Crit. Rev. 2017, 20, 55–80. [Google Scholar] [CrossRef]
- Masley, S.C.; Masley, L.V.; Gualtieri, C. Effect of mercury levels and seafood intake on cognitive function in middle-aged adults. Integr. Med. 2012, 11, 32–40. [Google Scholar]
- National Research Council. Toxicological Effects of Methylmercury; The National Academies Press: Washington, DC, USA, 2000. [Google Scholar]
- Dack, K.; Fell, M.; Taylor, C.M.; Havdahl, A.; Lewis, S.J. Mercury and Prenatal Growth: A Systematic Review. Int. J. Environ. Res. Public Health 2021, 18, 7140. [Google Scholar] [CrossRef]
- Dack, K.; Fell, M.; Taylor, C.M.; Havdahl, A.; Lewis, S.J. Prenatal Mercury Exposure and Neurodevelopment up to the Age of 5 Years: A Systematic Review. Int. J. Environ. Res. Public Health 2022, 19, 1976. [Google Scholar] [CrossRef]
- Barcelos, G.R.M.; Grotto, D.; de Marco, K.C.; Valentini, J.; Lengert, A.v.H.; de Oliveira, A.S.; Garcia, S.C.; Braga, G.L.; Engström, K.S.; de Syllos Cólus, I.M.; et al. Polymorphisms in glutathione-related genes modify mercury concentrations and antioxidant status in subjects environmentally exposed to methylmercury. Sci. Total. Environ. 2013, 463–464, 319–325. [Google Scholar] [CrossRef]
- Engström, K.S.; Strömberg, U.; Lundh, T.; Johansson, I.; Vessby, B.; Hallmans, G.; Skerfving, S.; Broberg, K. Genetic Variation in Glutathione-Related Genes and Body Burden of Methylmercury. Environ. Health Perspect. 2008, 116, 734–739. [Google Scholar] [CrossRef]
- Gundacker, C.; Komarnicki, G.; Jagiello, P.; Gencikova, A.; Dahmen, N.; Wittmann, K.J.; Gencik, M. Glutathione-S-transferase polymorphism, metallothionein expression, and mercury levels among students in Austria. Sci. Total Environ. 2007, 385, 37–47. [Google Scholar] [CrossRef]
- Wang, Y.; Goodrich, J.M.; Gillespie, B.; Werner, R.; Basu, N.; Franzblau, A. An Investigation of Modifying Effects of Metallothionein Single-Nucleotide Polymorphisms on the Association between Mercury Exposure and Biomarker Levels. Environ. Health Perspect. 2012, 120, 530–534. [Google Scholar] [CrossRef]
- Ng, S.; Lin, C.-C.; Hwang, Y.-H.; Hsieh, W.-S.; Liao, H.-F.; Chen, P.-C. Mercury, APOE, and children’s neurodevelopment. NeuroToxicology 2013, 37, 85–92. [Google Scholar] [CrossRef]
- Julvez, J.; Smith, G.D.; Golding, J.; Ring, S.; Pourcain, B.S.; Gonzalez, J.R.; Grandjean, P. Prenatal Methylmercury Exposure and Genetic Predisposition to Cognitive Deficit at Age 8 Years. Epidemiology 2013, 24, 643–650. [Google Scholar] [CrossRef]
- Bjørklund, G.; Aaseth, J.; Ajsuvakova, O.P.; Nikonorov, A.A.; Skalny, A.V.; Skalnaya, M.G.; Tinkov, A.A. Molecular interaction between mercury and selenium in neurotoxicity. Coord. Chem. Rev. 2017, 332, 30–37. [Google Scholar] [CrossRef]
- Franciscato, C.; Moraes-Silva, L.; Duarte, F.; Oliveira, C.; Ineu, R.; Flores, E.; Dressler, V.; Peixoto, N.; Pereira, M. Delayed biochemical changes induced by mercury intoxication are prevented by zinc pre-exposure. Ecotoxicol. Environ. Saf. 2011, 74, 480–486. [Google Scholar] [CrossRef]
- Mesquita, M.; Pedroso, T.F.; Oliveira, C.S.; Oliveira, V.A.; Santos, R.F.D.; Bizzi, C.A.; Pereira, M.E. Effects of zinc against mercury toxicity in female rats 12 and 48 hours after HgCl2 exposure. EXCLI J. 2016, 15, 256–267. [Google Scholar] [CrossRef]
- Singh, N.; Gupta, V.K.; Kumar, A.; Sharma, B. Synergistic Effects of Heavy Metals and Pesticides in Living Systems. Front. Chem. 2017, 5, 70. [Google Scholar] [CrossRef]
- Hernandez, A.F.; Buha, A.; Constantin, C.; Wallace, D.R.; Sarigiannis, D.; Neagu, M.; Antonijevic, B.; Hayes, A.W.; Wilks, M.F.; Tsatsakis, A. Critical assessment and integration of separate lines of evidence for risk assessment of chemical mixtures. Arch. Toxicol. 2019, 93, 2741–2757. [Google Scholar] [CrossRef]
- Warrington, N.M.; Zhu, G.; Dy, V.; Heath, A.C.; Madden, P.A.; Hemani, G.; Kemp, J.P.; Mcmahon, G.; Pourcain, B.S.; Timpson, N.J.; et al. Genome-wide association study of blood lead shows multiple associations near ALAD. Hum. Mol. Genet. 2015, 24, 3871–3879. [Google Scholar] [CrossRef]
- Evans, D.M.; Zhu, G.; Dy, V.; Heath, A.C.; Madden, P.A.F.; Kemp, J.P.; McMahon, G.; St Pourcain, B.; Timpson, N.J.; Golding, J.; et al. Genome-wide association study identifies loci affecting blood copper, selenium and zinc. Hum. Mol. Genet. 2013, 22, 3998–4006. [Google Scholar] [CrossRef]
- Cornelis, M.C.; Fornage, M.; Foy, M.; Xun, P.; Gladyshev, V.N.; Morris, S.; Chasman, D.I.; Hu, F.B.; Rimm, E.B.; Kraft, P.; et al. Genome-wide association study of selenium concentrations. Hum. Mol. Genet. 2015, 24, 1469–1477. [Google Scholar] [CrossRef]
- Davies, N.M.; Holmes, M.V.; Smith, G.D. Reading Mendelian randomisation studies: A guide, glossary, and checklist for clinicians. BMJ 2018, 362, k601. [Google Scholar] [CrossRef]
- Duncan, L.E.; Ostacher, M.; Ballon, J. How genome-wide association studies (GWAS) made traditional candidate gene studies obsolete. Neuropsychopharmacology 2019, 44, 1518–1523. [Google Scholar] [CrossRef]
- Tam, V.; Patel, N.; Turcotte, M.; Bossé, Y.; Paré, G.; Meyre, D. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef]
- Bell, S.; Rigas, A.S.; Ferkingstad, E.; Allara, E.; Bjornsdottir, G.; Ramond, A.; Sørensen, E.; Halldorsson, G.H.; Paul, D.S.; Burgdorf, K.S.; et al. A genome-wide meta-analysis yields 46 new loci associating with biomarkers of iron homeostasis. Commun. Biol. 2021, 4, 156. [Google Scholar] [CrossRef]
- Ng, E.; Lind, P.M.; Lindgren, C.; Ingelsson, E.; Mahajan, A.; Morris, A.; Lind, L. Genome-wide association study of toxic metals and trace elements reveals novel associations. Hum. Mol. Genet. 2015, 24, 4739–4745. [Google Scholar] [CrossRef]
- Fraser, A.; Macdonald-Wallis, C.; Tilling, K.; Boyd, A.; Golding, J.; Smith, G.D.; Henderson, J.; Macleod, J.; Molloy, L.; Ness, A.; et al. Cohort Profile: The Avon Longitudinal Study of Parents and Children: ALSPAC mothers cohort. Eur. J. Endocrinol. 2012, 42, 97–110. [Google Scholar] [CrossRef]
- Boyd, A.; Golding, J.; Macleod, J.; A Lawlor, D.; Fraser, A.; Henderson, J.; Molloy, L.; Ness, A.; Ring, S.; Smith, G.D. Cohort Profile: The ‘Children of the 90s’—The index offspring of the Avon Longitudinal Study of Parents and Children. Eur. J. Endocrinol. 2012, 42 (Suppl. 3), 111–127. [Google Scholar] [CrossRef]
- Taylor, C.M.; Golding, J.; Hibbeln, J.; Emond, A.M. Environmental Factors Predicting Blood Lead Levels in Pregnant Women in the UK: The ALSPAC Study. PLoS ONE 2013, 8, e72371. [Google Scholar] [CrossRef]
- Hornung, R.W.; Reed, L.D. Estimation of Average Concentration in the Presence of Nondetectable Values. Appl. Occup. Environ. Hyg. 1990, 5, 46–51. [Google Scholar] [CrossRef]
- Pembrey, M. The Avon Longitudinal Study of Parents and Children (ALSPAC): A resource for genetic epidemiology. Eur. J. Endocrinol. 2004, 151 (Suppl. 3), U125–U129. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Steinthorsdottir, V.; McGinnis, R.; Williams, N.O.; Stefansdottir, L.; Thorleifsson, G.; Shooter, S.; Fadista, J.; Sigurdsson, J.K.; Auro, K.M.; Berezina, G.; et al. Genetic predisposition to hypertension is associated with preeclampsia in European and Central Asian women. Nat. Commun. 2020, 11, 5976. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Morris, T.T.; Davies, N.M.; Smith, G.D. Can education be personalised using pupils’ genetic data? eLife 2020, 9, e49962. [Google Scholar] [CrossRef] [PubMed]
- ALSPAC Team. ALSPAC OMICs Data Catalogue. Available online: https://alspac.github.io/omics_documentation/alspac_omics_data_catalogue.html (accessed on 16 October 2023).
- Delaneau, O.; Marchini, J.; Zagury, J.-F. A linear complexity phasing method for thousands of genomes. Nat. Methods 2012, 9, 179–181. [Google Scholar] [CrossRef]
- Das, S.; Forer, L.; Schönherr, S.; Sidore, C.; Locke, A.E.; Kwong, A.; Vrieze, S.I.; Chew, E.Y.; Levy, S.; McGue, M.; et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016, 48, 1284–1287. [Google Scholar] [CrossRef]
- Vrijheid, M.; Slama, R.; Robinson, O.; Chatzi, L.; Coen, M.; Hazel, P.v.D.; Thomsen, C.; Wright, J.; Athersuch, T.J.; Avellana, N.; et al. The Human Early-Life Exposome (HELIX): Project Rationale and Design. Environ. Health Perspect. 2014, 122, 535–544. [Google Scholar] [CrossRef]
- Maitre, L.; De Bont, J.; Casas, M.; Robinson, O.; Aasvang, G.M.; Agier, L.; Andrušaitytė, S.; Ballester, F.; Basagaña, X.; Borràs, E.; et al. Human Early Life Exposome (HELIX) study: A European population-based exposome cohort. BMJ Open 2018, 8, e021311. [Google Scholar] [CrossRef]
- Magnus, P.; Birke, C.; Vejrup, K.; Haugan, A.; Alsaker, E.; Daltveit, A.K.; Handal, M.; Haugen, M.; Høiseth, G.; Knudsen, G.P.; et al. Cohort Profile Update: The Norwegian Mother and Child Cohort Study (MoBa). Int. J. Epidemiol. 2016, 45, 382–388. [Google Scholar] [CrossRef]
- Haug, L.S.; Sakhi, A.K.; Cequier, E.; Casas, M.; Maitre, L.; Basagana, X.; Andrusaityte, S.; Chalkiadaki, G.; Chatzi, L.; Coen, M.; et al. In-utero and childhood chemical exposome in six European mother-child cohorts. Environ. Int. 2018, 121, 751–763. [Google Scholar] [CrossRef]
- Rodushkin, I.; Axelsson, M.D. Application of double focusing sector field ICP-MS for multielemental characterization of human hair and nails. Part I. Analytical methodology. Sci. Total. Environ. 2000, 250, 83–100. [Google Scholar] [CrossRef] [PubMed]
- Calvo-Serra, B.; Maitre, L.; E Lau, C.-H.; Siskos, A.P.; Gützkow, K.B.; Andrušaitytė, S.; Casas, M.; Cadiou, S.; Chatzi, L.; González, J.R.; et al. Urinary metabolite quantitative trait loci in children and their interaction with dietary factors. Hum. Mol. Genet. 2021, 29, 3830–3844. [Google Scholar] [CrossRef]
- Bulik-Sullivan, B.K.; Loh, P.R.; Finucane, H.K.; Ripke, S.; Yang, J.; Schizophrenia Working Group of the Psychiatric Genomics Consortium; Patterson, N.; Daly, M.J.; Price, A.L.; Neale, B.M. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015, 47, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Bulik-Sullivan, B.; Finucane, H.K.; Anttila, V.; Gusev, A.; Day, F.R.; Loh, P.-R.; Duncan, L.; Perry, J.R.B.; Patterson, N.; Robinson, E.B.; et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015, 47, 1236–1241. [Google Scholar] [CrossRef] [PubMed]
- Ni, G.; Moser, G.; Wray, N.R.; Lee, S.H.; Ripke, S.; Neale, B.M.; Corvin, A.; Walters, J.T.; Farh, K.-H.; Holmans, P.A.; et al. Estimation of Genetic Correlation via Linkage Disequilibrium Score Regression and Genomic Restricted Maximum Likelihood. Am. J. Hum. Genet. 2018, 102, 1185–1194. [Google Scholar] [CrossRef]
- Marchini, J.; Howie, B.; Myers, S.; McVean, G.; Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007, 39, 906–913. [Google Scholar] [CrossRef]
- Pagès, H.S. Hsapiens. dbSNPGRCh37: SNP Locations for Homo Sapiens (dbSNP Build 144). R Packag. version 0.99. 2017, Volume 20. Available online: https://bioconductor.org/packages/release/data/annotation/html/SNPlocs.Hsapiens.dbSNP144.GRCh37.html (accessed on 21 November 2023).
- Huber, W.; Carey, V.J.; Gentleman, R.; Anders, S.; Carlson, M.; Carvalho, B.S.; Bravo, H.C.; Davis, S.; Gatto, L.; Girke, T.; et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 2015, 12, 115–121. [Google Scholar] [CrossRef]
- Turner, S.D. qqman: An R package for visualizing GWAS results using Q-Q and manhattan plots. J. Open Source Softw. 2018, 3, 731. [Google Scholar] [CrossRef]
- Hemani, G.; Zheng, J.; Elsworth, B.; Wade, K.H.; Haberland, V.; Baird, D.; Laurin, C.; Burgess, S.; Bowden, J.; Langdon, R.; et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 2018, 7, e34408. [Google Scholar] [CrossRef]
- Watanabe, K.; Taskesen, E.; van Bochoven, A.; Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017, 8, 1826. [Google Scholar] [CrossRef]
- Rangwala, S.H.; Kuznetsov, A.; Ananiev, V.; Asztalos, A.; Borodin, E.; Evgeniev, V.; Joukov, V.; Lotov, V.; Pannu, R.; Rudnev, D.; et al. Accessing NCBI data using the NCBI Sequence Viewer and Genome Data Viewer (GDV). Genome Res. 2021, 31, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.-H.; Brown, D.W.; Machiela, M.J. LDtrait: An Online Tool for Identifying Published Phenotype Associations in Linkage Disequilibrium. Cancer Res. 2020, 80, 3443–3446. [Google Scholar] [CrossRef] [PubMed]
- Staley, J.R.; Blackshaw, J.; Kamat, M.A.; Ellis, S.; Surendran, P.; Sun, B.B.; Paul, D.S.; Freitag, D.; Burgess, S.; Danesh, J.; et al. PhenoScanner: A database of human genotype–phenotype associations. Bioinformatics 2016, 32, 3207–3209. [Google Scholar] [CrossRef] [PubMed]
- A Kamat, M.; A Blackshaw, J.; Young, R.; Surendran, P.; Burgess, S.; Danesh, J.; Butterworth, A.S.; Staley, J.R. PhenoScanner V2: An expanded tool for searching human genotype–phenotype associations. Bioinformatics 2019, 35, 4851–4853. [Google Scholar] [CrossRef] [PubMed]
- Safran, M.; Rosen, N.; Twik, M.; BarShir, R.; Stein, T.I.; Dahary, D.; Fishilevich, S.; Lancet, D. The GeneCards Suite. In Practical Guide to Life Science Databases; Abugessaisa, I., Kasukawa, T., Eds.; Springer: Singapore, 2021; pp. 27–56. [Google Scholar]
- Stanfill, A.G.; Cao, X. Enhancing Research through the Use of the Genotype-Tissue Expression (GTEx) Database. Biol. Res. Nurs. 2021, 23, 533–540. [Google Scholar] [CrossRef] [PubMed]
- Carithers, L.J.; Ardlie, K.; Barcus, M.; Branton, P.A.; Britton, A.; Buia, S.A.; Compton, C.C.; DeLuca, D.S.; Peter-Demchok, J.; Gelfand, E.T.; et al. A Novel Approach to High-Quality Postmortem Tissue Procurement: The GTEx Project. Biopreservation Biobanking 2015, 13, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Hamosh, A.; Scott, A.F.; Amberger, J.; Valle, D.; McKusick, V.A. Online Mendelian Inheritance in Man (OMIM). Hum. Mutat. 2000, 15, 57–61. [Google Scholar] [CrossRef]
- Gaunt, T.R.; Shihab, H.A.; Hemani, G.; Min, J.L.; Woodward, G.; Lyttleton, O.; Zheng, J.; Duggirala, A.; McArdle, W.L.; Ho, K.; et al. Systematic identification of genetic influences on methylation across the human life course. Genome Biol. 2016, 17, 61. [Google Scholar] [CrossRef]
- Bonder, M.J.; Luijk, R.; Zhernakova, D.; Moed, M.; Deelen, P.; Vermaat, M.; Van Iterson, M.M.; Van Dijk, F.; Van Galen, M.; Bot, J.; et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat. Genet. 2017, 49, 131–138. [Google Scholar] [CrossRef]
- Lappalainen, T.; Sammeth, M.; Friedländer, M.R.; ‘t Hoen, P.A.C.; Monlong, J.; Rivas, M.A.; Gonzàlez-Porta, M.; Kurbatova, N.; Griebel, T.; Ferreira, P.G.; et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 2013, 501, 506–511. [Google Scholar] [CrossRef]
- Aydemir, T.B.; Cousins, R.J. The Multiple Faces of the Metal Transporter ZIP14 (SLC39A14). J. Nutr. 2018, 148, 174–184. [Google Scholar] [CrossRef]
- Tuschl, K.; Meyer, E.; Valdivia, L.E.; Zhao, N.; Dadswell, C.; Abdul-Sada, A.; Hung, C.Y.; Simpson, M.A.; Chong, W.K.; Jacques, T.S.; et al. Mutations in SLC39A14 disrupt manganese homeostasis and cause childhood-onset parkinsonism–dystonia. Nat. Commun. 2016, 7, 11601. [Google Scholar] [CrossRef]
- Xin, Y.; Gao, H.; Wang, J.; Qiang, Y.; Imam, M.U.; Li, Y.; Wang, J.; Zhang, R.; Zhang, H.; Yu, Y.; et al. Manganese transporter Slc39a14 deficiency revealed its key role in maintaining manganese homeostasis in mice. Cell Discov. 2017, 3, 17025. [Google Scholar] [CrossRef] [PubMed]
- Jenkitkasemwong, S.; Akinyode, A.; Paulus, E.; Weiskirchen, R.; Hojyo, S.; Fukada, T.; Giraldo, G.; Schrier, J.; Garcia, A.; Janus, C.; et al. SLC39A14 deficiency alters manganese homeostasis and excretion resulting in brain manganese accumulation and motor deficits in mice. Proc. Natl. Acad. Sci. USA 2018, 115, E1769–E1778. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.; Morgan, H.; Johnson, A.; Nicholson, R. Structure–function analysis of a novel member of the LIV-1 subfamily of zinc transporters, ZIP14. FEBS Lett. 2005, 579, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Liuzzi, J.P.; Aydemir, F.; Nam, H.; Knutson, M.D.; Cousins, R.J. Zip14 (Slc39a14) mediates non-transferrin-bound iron uptake into cells. Proc. Natl. Acad. Sci. USA 2006, 103, 13612–13617. [Google Scholar] [CrossRef]
- Widhalm, R.; Ellinger, I.; Granitzer, S.; Forsthuber, M.; Bajtela, R.; Gelles, K.; Hartig, P.-Y.; Hengstschläger, M.; Zeisler, H.; Salzer, H.; et al. Human placental cell line HTR-8/SVneo accumulates cadmium by divalent metal transporters DMT1 and ZIP14. Metallomics 2020, 12, 1822–1833. [Google Scholar] [CrossRef]
- Boczonadi, V.; King, M.S.; Smith, A.C.; Olahova, M.; Bansagi, B.; Roos, A.; Eyassu, F.; Borchers, C.; Ramesh, V.; Lochmüller, H.; et al. Mitochondrial oxodicarboxylate carrier deficiency is associated with mitochondrial DNA depletion and spinal muscular atrophy–like disease. Anesthesia Analg. 2018, 20, 1224–1235. [Google Scholar] [CrossRef]
- Wahlberg, K.; Love, T.M.; Pineda, D.; Engström, K.; Watson, G.E.; Thurston, S.W.; Yeates, A.J.; Mulhern, M.S.; McSorley, E.M.; Strain, J.; et al. Maternal polymorphisms in glutathione-related genes are associated with maternal mercury concentrations and early child neurodevelopment in a population with a fish-rich diet. Environ. Int. 2018, 115, 142–149. [Google Scholar] [CrossRef]
- de Oliveira, A.S.; de Souza, M.F.; Lengert, A.v.H.; de Oliveira, M.T.; Camargo, R.B.d.O.G.; Braga, G.L.; Cólus, I.M.d.S.; Barbosa, F., Jr.; Barcelos, G.R.M. Genetic Polymorphisms in Glutathione (GSH-) Related Genes Affect the Plasmatic Hg/Whole Blood Hg Partitioning and the Distribution between Inorganic and Methylmercury Levels in Plasma Collected from a Fish-Eating Population. BioMed Res. Int. 2014, 2014, 940952. [Google Scholar] [CrossRef]
- Buck, K.A.; Varian-Ramos, C.W.; Cristol, D.A.; Swaddle, J.P. Blood Mercury Levels of Zebra Finches Are Heritable: Implications for the Evolution of Mercury Resistance. PLoS ONE 2016, 11, e0162440. [Google Scholar] [CrossRef]
- Paape, T.; Heiniger, B.; Domingo, M.S.; Clear, M.R.; Lucas, M.M.; Pueyo, J.J. Genome-Wide Association Study Reveals Complex Genetic Architecture of Cadmium and Mercury Accumulation and Tolerance Traits in Medicago truncatula. Front. Plant Sci. 2022, 12, 806949. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, K.; Yasutake, A. Sex and age differences in mercury distribution and excretion in methylmercury-administered mice. J. Toxicol. Environ. Health 1986, 18, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.T. The influence of age on the gastrointestinal absorption of mercuric chloride and methyl mercury chloride in the rat. Environ. Res. 1982, 27, 412–420. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.; Eide, D.J. The SLC39 family of zinc transporters. Mol. Asp. Med. 2013, 34, 612–619. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.M.; Dnyanmote, A.V.; Bush, K.T.; Wu, W.; Nigam, S.K. Deletion of Multispecific Organic Anion Transporter Oat1/Slc22a6 Protects against Mercury-induced Kidney Injury. J. Biol. Chem. 2011, 286, 26391–26395. [Google Scholar] [CrossRef] [PubMed]
- Borges, V.C.; Santos, F.W.; Rocha, J.B.T.; Nogueira, C.W. Heavy Metals Modulate Glutamatergic System in Human Platelets. Neurochem. Res. 2007, 32, 953–958. [Google Scholar] [CrossRef]
- Albrecht, J.; Matyja, E. Glutamate: A potential mediator of inorganic mercury neurotoxicity. Metab. Brain Dis. 1996, 11, 175–184. [Google Scholar] [CrossRef]
- Peixoto, N.; Serafim, A.; Flores, E.; Bebianno, M.; Pereira, M. Metallothionein, zinc, and mercury levels in tissues of young rats exposed to zinc and subsequently to mercury. Life Sci. 2007, 81, 1264–1271. [Google Scholar] [CrossRef]
- Lozano, M.; Yousefi, P.; Broberg, K.; Soler-Blasco, R.; Miyashita, C.; Pesce, G.; Kim, W.J.; Rahman, M.; Bakulski, K.M.; Haug, L.S.; et al. DNA methylation changes associated with prenatal mercury exposure: A meta-analysis of prospective cohort studies from PACE consortium. Environ. Res. 2022, 204, 112093. [Google Scholar] [CrossRef]
- Pierce, B.L.; Kibriya, M.G.; Tong, L.; Jasmine, F.; Argos, M.; Roy, S.; Paul-Brutus, R.; Rahaman, R.; Rakibuz-Zaman, M.; Parvez, F.; et al. Genome-Wide Association Study Identifies Chromosome 10q24.32 Variants Associated with Arsenic Metabolism and Toxicity Phenotypes in Bangladesh. PLoS Genet. 2012, 8, e1002522. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, M.F.H.; Grotto, D.; Barbosa, F., Jr. Inorganic and Methylmercury Levels in Plasma are Differentially Associated with Age, Gender, and Oxidative Stress Markers in a Population Exposed to Mercury through Fish Consumption. J. Toxicol. Environ. Health Part A 2014, 77, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.-G.; Jo, E.-M.; Kim, G.-Y.; Kim, D.-S.; Kim, Y.-M.; Kim, R.-B.; Suh, B.-S.; Hong, Y.-S. Analysis of Methylmercury Concentration in the Blood of Koreans by Using Cold Vapor Atomic Fluorescence Spectrophotometry. Ann. Lab. Med. 2012, 32, 31–37. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).