Analysis of Nucleotide Variations in Human G-Quadruplex Forming Regions Associated with Disease States
Abstract
1. Introduction
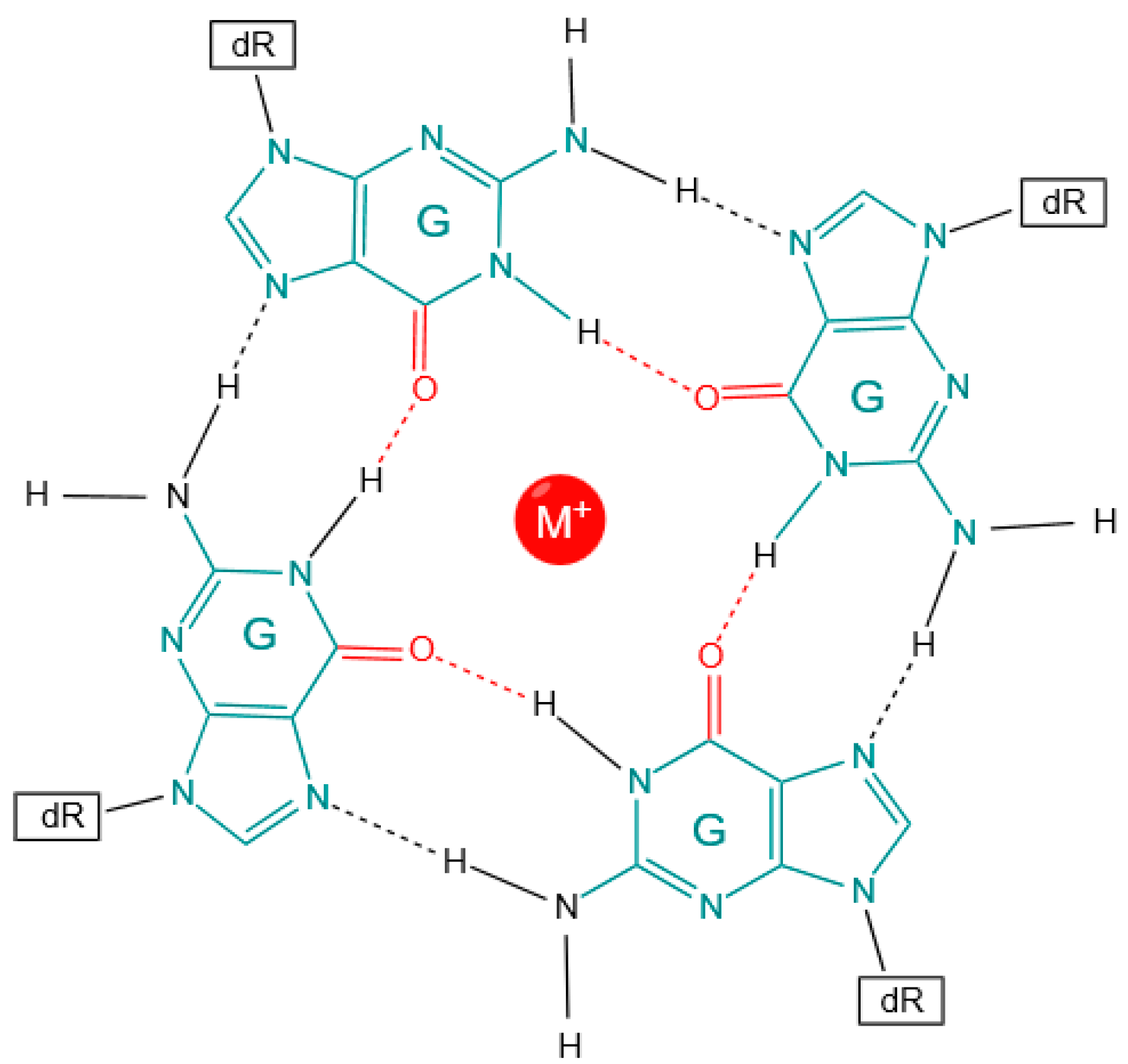
1.1. Functional Role of G4 Regions
1.2. Mutations within G4 Regions
1.3. Study Motivation
2. Materials and Methods
2.1. Putative and Validated G4 Identification
2.2. Somatic and Germline Variants in G4 Regions
2.3. Identification of SNPs Affecting G4 Formation
2.4. Functional and Transcription Factor Enrichment Analysis
3. Results
3.1. COSMIC Somatic Mutations
3.2. CLINVAR Germline Mutations
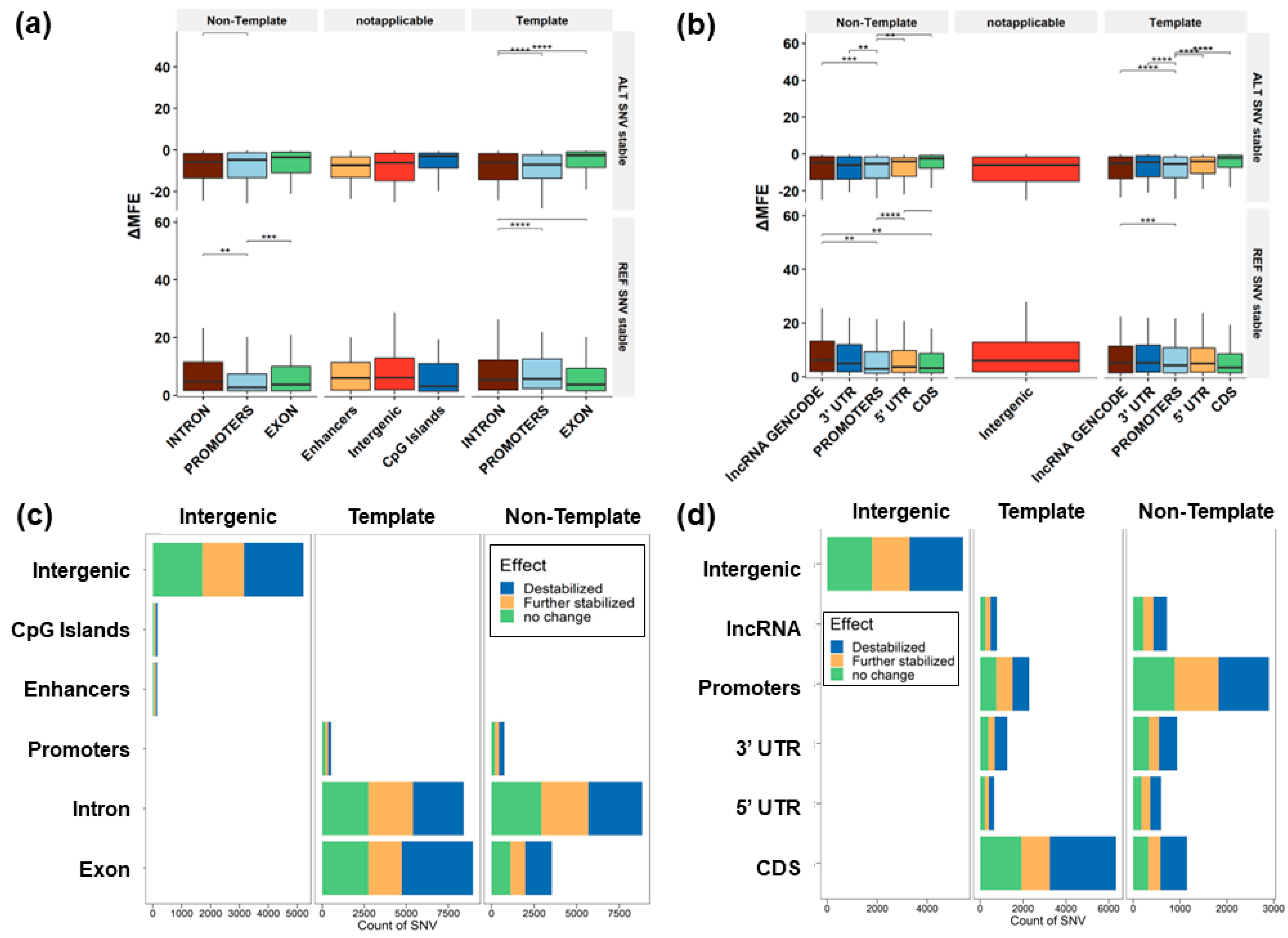
3.3. Predicted Change to G4 Stability
3.4. G4 Variants in Transcript Regions
3.5. G4 Variants in Gene Features
3.6. Enrichment Analysis
3.6.1. Gene Ontology
3.6.2. KEGG Metabolic Pathways
3.6.3. INTERPRO Protein Domains
3.6.4. Transcription Factors
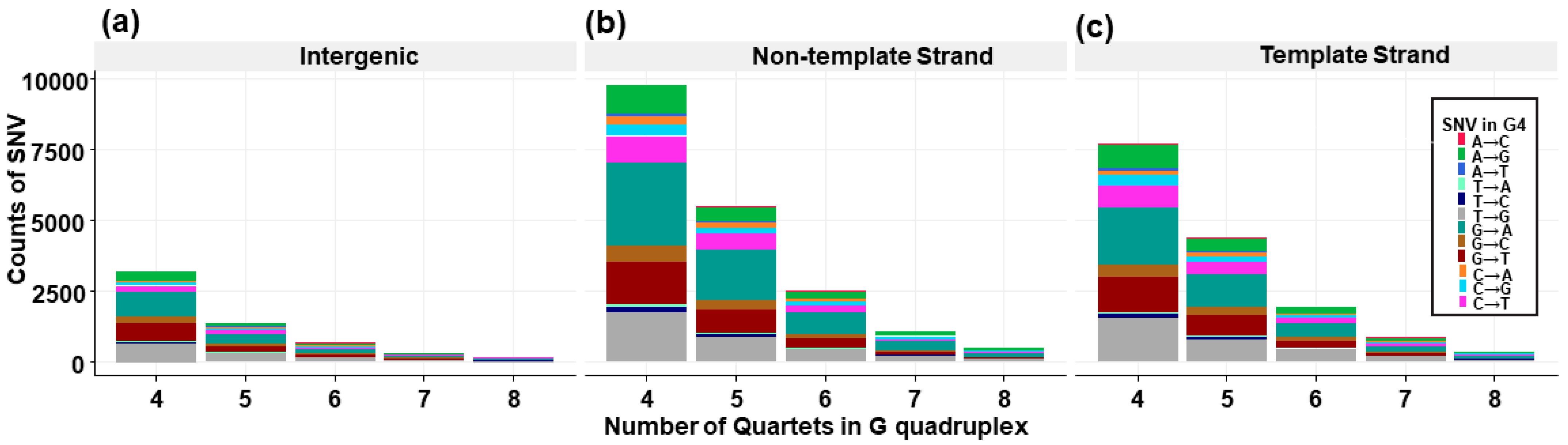
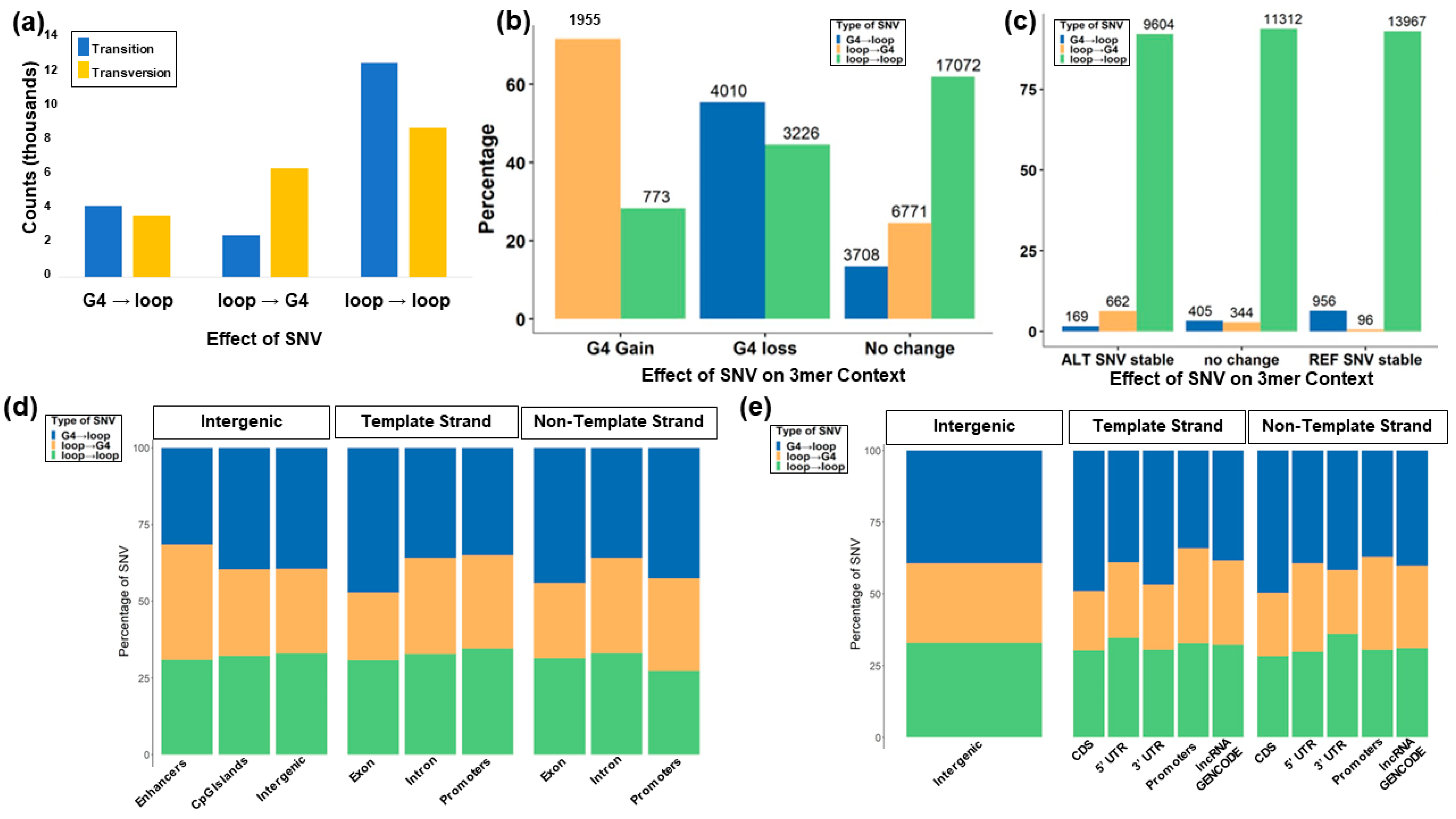
3.7. Trinucleotide Context Mutation in the G4 Sequence
3.8. Role of the Location of SNVs in G4s
4. Discussion
4.1. Molecular Mechanisms for Promoting Mutations in G4 Regions
4.2. TERT G4 Mutations
4.3. G4 Mutations Disrupting the Transcription Factor Binding
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sen, D.; Gilbert, W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 1988, 334, 364–366. [Google Scholar] [CrossRef]
- Sen, D.; Gilbert, W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature 1990, 344, 410–414. [Google Scholar] [CrossRef] [PubMed]
- Bugaut, A.; Balasubramanian, S. A sequence-independent study of the influence of short loop lengths on the stability and topology of intramolecular DNA G-quadruplexes. Biochemistry 2008, 47, 689–697. [Google Scholar] [CrossRef]
- Zhang, A.Y.; Bugaut, A.; Balasubramanian, S. A sequence-independent analysis of the loop length dependence of intramolecular RNA G-quadruplex stability and topology. Biochemistry 2011, 50, 7251–7258. [Google Scholar] [CrossRef] [PubMed]
- Sutyak, K.B.; Zavalij, P.Y.; Robinson, M.L.; Davis, J.T. Controlling molecularity and stability of hydrogen bonded G-quadruplexes by modulating the structure’s periphery. Chem. Commun. 2016, 52, 11112–11115. [Google Scholar] [CrossRef] [PubMed]
- Hänsel-Hertsch, R.; Beraldi, D.; Lensing, S.V.; Marsico, G.; Zyner, K.; Parry, A.; Di Antonio, M.; Pike, J.; Kimura, H.; Narita, M. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016, 48, 1267–1272. [Google Scholar] [CrossRef]
- Cheung, I.; Schertzer, M.; Rose, A.; Lansdorp, P.M. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat. Genet. 2002, 31, 405–409. [Google Scholar] [CrossRef]
- Dahan, D.; Tsirkas, I.; Dovrat, D.; Sparks, M.A.; Singh, S.P.; Galletto, R.; Aharoni, A. Pif1 is essential for efficient replisome progression through lagging strand G-quadruplex DNA secondary structures. Nucleic Acids Res. 2018, 46, 11847–11857. [Google Scholar] [CrossRef]
- Paeschke, K.; Capra, J.A.; Zakian, V.A. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 2011, 145, 678–691. [Google Scholar] [CrossRef]
- Rodriguez, R.; Miller, K.M.; Forment, J.V.; Bradshaw, C.R.; Nikan, M.; Britton, S.; Oelschlaegel, T.; Xhemalce, B.; Balasubramanian, S.; Jackson, S.P. Small-molecule–induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 2012, 8, 301–310. [Google Scholar] [CrossRef]
- Piazza, A.; Adrian, M.; Samazan, F.; Heddi, B.; Hamon, F.; Serero, A.; Lopes, J.; Teulade-Fichou, M.P.; Phan, A.T.; Nicolas, A. Short loop length and high thermal stability determine genomic instability induced by G-quadruplex-forming minisatellites. EMBO J. 2015, 34, 1718–1734. [Google Scholar] [CrossRef]
- London, T.B.; Barber, L.J.; Mosedale, G.; Kelly, G.P.; Balasubramanian, S.; Hickson, I.D.; Boulton, S.J.; Hiom, K. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J. Biol. Chem. 2008, 283, 36132–36139. [Google Scholar] [CrossRef] [PubMed]
- Ribeyre, C.; Lopes, J.; Boulé, J.-B.; Piazza, A.; Guédin, A.; Zakian, V.A.; Mergny, J.-L.; Nicolas, A. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009, 5, e1000475. [Google Scholar] [CrossRef] [PubMed]
- De Magis, A.; Manzo, S.G.; Russo, M.; Marinello, J.; Morigi, R.; Sordet, O.; Capranico, G. DNA damage and genome instability by G-quadruplex ligands are mediated by R loops in human cancer cells. Proc. Natl. Acad. Sci. USA 2019, 116, 816–825. [Google Scholar] [CrossRef] [PubMed]
- Madireddy, A.; Purushothaman, P.; Loosbroock, C.P.; Robertson, E.S.; Schildkraut, C.L.; Verma, S.C. G-quadruplex-interacting compounds alter latent DNA replication and episomal persistence of KSHV. Nucleic Acids Res. 2016, 44, 3675–3694. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Sung, K.; Joo, S.Y.; Jeong, J.-H.; Kim, S.K.; Lee, H. Dynamic interaction of BRCA2 with telomeric G-quadruplexes underlies telomere replication homeostasis. Nat. Commun. 2022, 2022, 3396. [Google Scholar] [CrossRef]
- Mei, Y.; Deng, Z.; Vladimirova, O.; Gulve, N.; Johnson, F.B.; Drosopoulos, W.C.; Schildkraut, C.L.; Lieberman, P.M. TERRA G-quadruplex RNA interaction with TRF2 GAR domain is required for telomere integrity. Sci. Rep. 2021, 11, 3509. [Google Scholar] [CrossRef]
- Zimmer, J.; Tacconi, E.M.; Folio, C.; Badie, S.; Porru, M.; Klare, K.; Tumiati, M.; Markkanen, E.; Halder, S.; Ryan, A. Targeting BRCA1 and BRCA2 deficiencies with G-quadruplex-interacting compounds. Mol. Cell 2016, 61, 449–460. [Google Scholar] [CrossRef]
- Siddiqui-Jain, A.; Grand, C.L.; Bearss, D.J.; Hurley, L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA 2002, 99, 11593–11598. [Google Scholar] [CrossRef]
- Gros, J.; Rosu, F.; Amrane, S.; De Cian, A.; Gabelica, V.; Lacroix, L.; Mergny, J.-L. Guanines are a quartet’s best friend: Impact of base substitutions on the kinetics and stability of tetramolecular quadruplexes. Nucleic Acids Res. 2007, 35, 3064–3075. [Google Scholar] [CrossRef]
- Didiot, M.-C.; Tian, Z.; Schaeffer, C.; Subramanian, M.; Mandel, J.-L.; Moine, H. The G-quartet containing FMRP binding site in FMR1 mRNA is a potent exonic splicing enhancer. Nucleic Acids Res. 2008, 36, 4902–4912. [Google Scholar] [CrossRef]
- Chaudhary, S.; Kaushik, M.; Ahmed, S.; Kukreti, R.; Kukreti, S. Structural switch from hairpin to duplex/antiparallel G-quadruplex at single-nucleotide polymorphism (SNP) site of human apolipoprotein E (APOE) gene coding region. ACS Omega 2018, 3, 3173–3182. [Google Scholar] [CrossRef]
- Bharti, S.K.; Sommers, J.A.; George, F.; Kuper, J.; Hamon, F.; Shin-ya, K.; Teulade-Fichou, M.-P.; Kisker, C.; Brosh, R.M. Specialization among iron-sulfur cluster helicases to resolve G-quadruplex DNA structures that threaten genomic stability. J. Biol. Chem. 2013, 288, 28217–28229. [Google Scholar] [CrossRef]
- Baral, A.; Kumar, P.; Halder, R.; Mani, P.; Yadav, V.K.; Singh, A.; Das, S.K.; Chowdhury, S. Quadruplex-single nucleotide polymorphisms (Quad-SNP) influence gene expression difference among individuals. Nucleic Acids Res. 2012, 40, 3800–3811. [Google Scholar] [CrossRef]
- Gong, J.-Y.; Wen, C.-J.; Tang, M.-L.; Duan, R.-F.; Chen, J.-N.; Zhang, J.-Y.; Zheng, K.-W.; He, Y.-D.; Hao, Y.-H.; Yu, Q. G-quadruplex structural variations in human genome associated with single-nucleotide variations and their impact on gene activity. Proc. Natl. Acad. Sci. USA 2021, 118, e2013230118. [Google Scholar] [CrossRef]
- Kuznetsova, A.A.; Fedorova, O.S.; Kuznetsov, N.A. Lesion recognition and cleavage of damage-containing quadruplexes and bulged structures by DNA glycosylases. Front. Cell Dev. Biol. 2020, 8, 595687. [Google Scholar] [CrossRef]
- Bedrat, A.; Lacroix, L.; Mergny, J.-L. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016, 44, 1746–1759. [Google Scholar] [CrossRef]
- Huppert, J.L.; Balasubramanian, S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005, 33, 2908–2916. [Google Scholar] [CrossRef]
- Fleming, A.M.; Burrows, C.J. G-quadruplex folds of the human telomere sequence alter the site reactivity and reaction pathway of guanine oxidation compared to duplex DNA. Chem. Res. Toxicol. 2013, 26, 593–607. [Google Scholar] [CrossRef]
- Lee, D.S.; Ghanem, L.R.; Barash, Y. Integrative analysis reveals RNA G-quadruplexes in UTRs are selectively constrained and enriched for functional associations. Nat. Commun. 2020, 11, 527. [Google Scholar] [CrossRef]
- Zhou, J.; Fleming, A.M.; Averill, A.M.; Burrows, C.J.; Wallace, S.S. The NEIL glycosylases remove oxidized guanine lesions from telomeric and promoter quadruplex DNA structures. Nucleic Acids Res. 2015, 43, 4039–4054. [Google Scholar] [CrossRef] [PubMed]
- Adrian, M.; Heddi, B.; Phan, A.T. NMR spectroscopy of G-quadruplexes. Methods 2012, 57, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Chambers, V.S.; Marsico, G.; Boutell, J.M.; Di Antonio, M.; Smith, G.P.; Balasubramanian, S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015, 33, 877–881. [Google Scholar] [CrossRef]
- Nakken, S.; Rognes, T.; Hovig, E. The disruptive positions in human G-quadruplex motifs are less polymorphic and more conserved than their neutral counterparts. Nucleic Acids Res. 2009, 37, 5749–5756. [Google Scholar] [CrossRef]
- Lim, K.W.; Alberti, P.; Guedin, A.; Lacroix, L.; Riou, J.-F.; Royle, N.J.; Mergny, J.-L.; Phan, A.T. Sequence variant (CTAGGG)n in the human telomere favors a G-quadruplex structure containing a G·C·G·C tetrad. Nucleic Acids Res. 2009, 37, 6239–6248. [Google Scholar] [CrossRef]
- Sagne, C.; Marcel, V.; Bota, M.; Martel-Planche, G.; Nobrega, A.; Palmero, E.I.; Perriaud, L.; Boniol, M.; Vagner, S.; Cox, D.G. Age at cancer onset in germline TP53 mutation carriers: Association with polymorphisms in predicted G-quadruplex structures. Carcinogenesis 2014, 35, 807–815. [Google Scholar] [CrossRef]
- Yeom, M.; Kim, I.-H.; Kim, J.-K.; Kang, K.; Eoff, R.L.; Guengerich, F.P.; Choi, J.-Y. Effects of twelve germline missense variations on DNA lesion and G-quadruplex bypass activities of human DNA polymerase REV1. Chem. Res. Toxicol. 2016, 29, 367–379. [Google Scholar] [CrossRef]
- Hänsel-Hertsch, R.; Simeone, A.; Shea, A.; Hui, W.W.; Zyner, K.G.; Marsico, G.; Rueda, O.M.; Bruna, A.; Martin, A.; Zhang, X. Landscape of G-quadruplex DNA structural regions in breast cancer. Nat. Genet. 2020, 52, 878–883. [Google Scholar] [CrossRef]
- Zhang, R.; Shu, H.; Wang, Y.; Tao, T.; Tu, J.; Wang, C.; Mergny, J.-L.; Sun, X. G-quadruplex structures are key modulators of somatic structural variants in cancers. Cancer Res. 2023, 83, 1234–1248. [Google Scholar] [CrossRef]
- Bamford, S.; Dawson, E.; Forbes, S.; Clements, J.; Pettett, R.; Dogan, A.; Flanagan, A.; Teague, J.; Futreal, P.A.; Stratton, M.R. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br. J. Cancer 2004, 91, 355–358. [Google Scholar] [CrossRef] [PubMed]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018, 46, D1062–D1067. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef] [PubMed]
- Shigemizu, D.; Fujimoto, A.; Akiyama, S.; Abe, T.; Nakano, K.; Boroevich, K.A.; Yamamoto, Y.; Furuta, M.; Kubo, M.; Nakagawa, H. A practical method to detect SNVs and indels from whole genome and exome sequencing data. Sci. Rep. 2013, 3, 2161. [Google Scholar] [CrossRef] [PubMed]
- Zook, J.M.; Chapman, B.; Wang, J.; Mittelman, D.; Hofmann, O.; Hide, W.; Salit, M. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat. Biotechnol. 2014, 32, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The vienna RNA websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef] [PubMed]
- Cavalcante, R.G.; Sartor, M.A. Annotatr: Genomic regions in context. Bioinformatics 2017, 33, 2381–2383. [Google Scholar] [CrossRef]
- Hammal, F.; de Langen, P.; Bergon, A.; Lopez, F.; Ballester, B. ReMap 2022: A database of Human, Mouse, Drosophila and Arabidopsis regulatory regions from an integrative analysis of DNA-binding sequencing experiments. Nucleic Acids Res. 2022, 50, D316–D325. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g: Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Coetzee, S.G.; Coetzee, G.A.; Hazelett, D.J. motifbreakR: An R/Bioconductor package for predicting variant effects at transcription factor binding sites. Bioinformatics 2015, 31, 3847–3849. [Google Scholar] [CrossRef]
- Sved, J.; Bird, A. The expected equilibrium of the CpG dinucleotide in vertebrate genomes under a mutation model. Proc. Natl. Acad. Sci. USA 1990, 87, 4692–4696. [Google Scholar] [CrossRef] [PubMed]
- Youk, J.; An, Y.; Park, S.; Lee, J.-K.; Ju, Y.S. The genome-wide landscape of C:G > T: A polymorphism at the CpG contexts in the human population. BMC Genom. 2020, 21, 270. [Google Scholar] [CrossRef]
- Beaudoin, J.-D.; Perreault, J.-P. 5′-UTR G-quadruplex structures acting as translational repressors. Nucleic Acids Res. 2010, 38, 7022–7036. [Google Scholar] [CrossRef] [PubMed]
- Bolduc, F.; Garant, J.-M.; Allard, F.; Perreault, J.-P. Irregular G-quadruplexes found in the untranslated regions of human mRNAs influence translation. J. Biol. Chem. 2016, 291, 21751–21760. [Google Scholar] [CrossRef]
- Capra, J.A.; Paeschke, K.; Singh, M.; Zakian, V.A. G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLoS Comput. Biol. 2010, 6, e1000861. [Google Scholar] [CrossRef]
- Cogoi, S.; Xodo, L.E. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006, 34, 2536–2549. [Google Scholar] [CrossRef] [PubMed]
- David, A.P.; Margarit, E.; Domizi, P.; Banchio, C.; Armas, P.; Calcaterra, N.B. G-quadruplexes as novel cis-elements controlling transcription during embryonic development. Nucleic Acids Res. 2016, 44, 4163–4173. [Google Scholar] [CrossRef]
- Endoh, T.; Kawasaki, Y.; Sugimoto, N. Suppression of gene expression by G-quadruplexes in open reading frames depends on G-quadruplex stability. Angew. Chem. Int. Ed. 2013, 52, 5522–5526. [Google Scholar] [CrossRef]
- Farhath, M.M.; Thompson, M.; Ray, S.; Sewell, A.; Balci, H.; Basu, S. G-Quadruplex-enabling sequence within the human tyrosine hydroxylase promoter differentially regulates transcription. Biochemistry 2015, 54, 5533–5545. [Google Scholar] [CrossRef]
- Fernando, H.; Sewitz, S.; Darot, J.; Tavare, S.; Huppert, J.L.; Balasubramanian, S. Genome-wide analysis of a G-quadruplex-specific single-chain antibody that regulates gene expression. Nucleic Acids Res. 2009, 37, 6716–6722. [Google Scholar] [CrossRef] [PubMed]
- Lago, S.; Nadai, M.; Cernilogar, F.M.; Kazerani, M.; Domíniguez Moreno, H.; Schotta, G.; Richter, S.N. Promoter G-quadruplexes and transcription factors cooperate to shape the cell type-specific transcriptome. Nat. Commun. 2021, 12, 3885. [Google Scholar] [CrossRef] [PubMed]
- Rezzoug, F.; Thomas, S.D.; Rouchka, E.C.; Miller, D.M. Discovery of a family of genomic sequences which interact specifically with the c-MYC promoter to regulate c-MYC expression. PLoS ONE 2016, 11, e0161588. [Google Scholar] [CrossRef]
- Shao, X.; Zhang, W.; Umar, M.I.; Wong, H.Y.; Seng, Z.; Xie, Y.; Zhang, Y.; Yang, L.; Kwok, C.K.; Deng, X. RNA G-quadruplex structures mediate gene regulation in bacteria. MBio 2020, 11, e02926-19. [Google Scholar] [CrossRef]
- Spiegel, J.; Cuesta, S.M.; Adhikari, S.; Hänsel-Hertsch, R.; Tannahill, D.; Balasubramanian, S. G-quadruplexes are transcription factor binding hubs in human chromatin. Genome Biol. 2021, 22, 117. [Google Scholar] [CrossRef]
- Belotserkovskii, B.P.; Soo Shin, J.H.; Hanawalt, P.C. Strong transcription blockage mediated by R-loop formation within a G-rich homopurine–homopyrimidine sequence localized in the vicinity of the promoter. Nucleic Acids Res. 2017, 45, 6589–6599. [Google Scholar] [CrossRef]
- Polak, P.; Arndt, P.F. Transcription induces strand-specific mutations at the 5′ end of human genes. Genome Res. 2008, 18, 1216–1223. [Google Scholar] [CrossRef]
- Park, J.W.; Han, Y.I.; Kim, S.W.; Kim, T.M.; Yeom, S.C.; Kang, J.; Park, J. 8-OxoG in GC-rich Sp1 binding sites enhances gene transcription in adipose tissue of juvenile mice. Sci. Rep. 2019, 9, 15618. [Google Scholar] [CrossRef]
- Cave, J.W.; Willis, D.E. G-quadruplex regulation of neural gene expression. FEBS J. 2022, 289, 3284–3303. [Google Scholar] [CrossRef]
- Westmark, C.J.; Malter, J.S. FMRP mediates mGluR5-dependent translation of amyloid precursor protein. PLoS Biol. 2007, 5, e52. [Google Scholar] [CrossRef]
- Huang, Z.-L.; Dai, J.; Luo, W.-H.; Wang, X.-G.; Tan, J.-H.; Chen, S.-B.; Huang, Z.-S. Identification of G-quadruplex-binding protein from the exploration of RGG motif/G-quadruplex interactions. J. Am. Chem. Soc. 2018, 140, 17945–17955. [Google Scholar] [CrossRef]
- Fürst, J.; Schedlbauer, A.; Gandini, R.; Garavaglia, M.L.; Saino, S.; Gschwentner, M.; Sarg, B.; Lindner, H.; Jakab, M.; Ritter, M. ICln159 Folds into a Pleckstrin Homology Domain-like Structure: Interaction with kinases and the splicing factor LSm4. J. Biol. Chem. 2005, 280, 31276–31282. [Google Scholar] [CrossRef]
- Gervais, V.; Lamour, V.; Jawhari, A.; Frindel, F.; Wasielewski, E.; Dubaele, S.; Egly, J.-M.; Thierry, J.-C.; Kieffer, B.; Poterszman, A. TFIIH contains a PH domain involved in DNA nucleotide excision repair. Nat. Struct. Mol. Biol. 2004, 11, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Srivastava, M.; Raghavan, S.C. GNG motifs can replace a GGG stretch during G-quadruplex formation in a context dependent manner. PLoS ONE 2016, 11, e0158794. [Google Scholar] [CrossRef] [PubMed]
- Neupane, A.; Chariker, J.H.; Rouchka, E.C. Structural and functional classification of G-quadruplex families within the human genome. Genes 2023, 14, 645. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Halder, K.; Halder, R.; Yadav, V.K.; Rawal, P.; Thakur, R.K.; Mohd, F.; Sharma, A.; Chowdhury, S. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J. Med. Chem. 2008, 51, 5641–5649. [Google Scholar] [CrossRef]
- Yadav, V.K.; Abraham, J.K.; Mani, P.; Kulshrestha, R.; Chowdhury, S. QuadBase: Genome-wide database of G4 DNA—Occurrence and conservation in human, chimpanzee, mouse and rat promoters and 146 microbes. Nucleic Acids Res. 2007, 36, D381–D385. [Google Scholar] [CrossRef] [PubMed]
- Pitié, M.; Boldron, C.; Pratviel, G. DNA oxidation by copper and manganese complexes. In Advances in Inorganic Chemistry; Elsevier: Amsterdam, The Netherlands, 2006; Volume 58, pp. 77–130. [Google Scholar]
- Bielskutė, S.; Plavec, J.; Podbevšek, P. Impact of oxidative lesions on the human telomeric G-quadruplex. J. Am. Chem. Soc. 2019, 141, 2594–2603. [Google Scholar] [CrossRef] [PubMed]
- Liguori, I.; Russo, G.; Curcio, F.; Bulli, G.; Aran, L.; Della-Morte, D.; Gargiulo, G.; Testa, G.; Cacciatore, F.; Bonaduce, D. Oxidative stress, aging, and diseases. Clin. Interv. Aging 2018, 13, 757. [Google Scholar] [CrossRef]
- Ishii, T.; Sekiguchi, M. Two ways of escaping from oxidative RNA damage: Selective degradation and cell death. DNA Repair 2019, 81, 102666. [Google Scholar] [CrossRef]
- Russo, M.T.; De Luca, G.; Degan, P.; Bignami, M. Different DNA repair strategies to combat the threat from 8-oxoguanine. Mutat. Res./Fundam. Mol. Mech. Mutagen. 2007, 614, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Holliday, R.; Grigg, G. DNA methylation and mutation. Mutat. Res./Fundam. Mol. Mech. Mutagen. 1993, 285, 61–67. [Google Scholar] [CrossRef]
- Banda, D.M.; Nuñez, N.N.; Burnside, M.A.; Bradshaw, K.M.; David, S.S. Repair of 8-oxoG: A mismatches by the MUTYH glycosylase: Mechanism, metals and medicine. Free Radic. Biol. Med. 2017, 107, 202–215. [Google Scholar] [CrossRef] [PubMed]
- van Loon, B.; Hübscher, U. An 8-oxo-guanine repair pathway coordinated by MUTYH glycosylase and DNA polymerase λ. Proc. Natl. Acad. Sci. USA 2009, 106, 18201–18206. [Google Scholar] [CrossRef]
- Viel, A.; Bruselles, A.; Meccia, E.; Fornasarig, M.; Quaia, M.; Canzonieri, V.; Policicchio, E.; Urso, E.D.; Agostini, M.; Genuardi, M. A specific mutational signature associated with DNA 8-oxoguanine persistence in MUTYH-defective colorectal cancer. EBioMedicine 2017, 20, 39–49. [Google Scholar] [CrossRef]
- Bellon, S.; Shikazono, N.; Cunniffe, S.; Lomax, M.; O’Neill, P. Processing of thymine glycol in a clustered DNA damage site: Mutagenic or cytotoxic. Nucleic Acids Res. 2009, 37, 4430–4440. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.M.; Zhou, J.; Wallace, S.S.; Burrows, C.J. A role for the fifth G-track in G-quadruplex forming oncogene promoter sequences during oxidative stress: Do these “spare tires” have an evolved function? ACS Cent. Sci. 2015, 1, 226–233. [Google Scholar] [CrossRef]
- Fleming, A.M.; Zhu, J.; Ding, Y.; Burrows, C.J. 8-Oxo-7, 8-dihydroguanine in the context of a gene promoter G-quadruplex is an on–off switch for transcription. ACS Chem. Biol. 2017, 12, 2417–2426. [Google Scholar] [CrossRef]
- Sun, D.; Liu, W.-J.; Guo, K.; Rusche, J.J.; Ebbinghaus, S.; Gokhale, V.; Hurley, L.H. The proximal promoter region of the human vascular endothelial growth factor gene has a G-quadruplex structure that can be targeted by G-quadruplex–interactive agents. Mol. Cancer Ther. 2008, 7, 880–889. [Google Scholar] [CrossRef]
- Monsen, R.C.; DeLeeuw, L.; Dean, W.L.; Gray, R.D.; Sabo, T.M.; Chakravarthy, S.; Chaires, J.B.; Trent, J.O. The hTERT core promoter forms three parallel G-quadruplexes. Nucleic Acids Res. 2020, 48, 5720–5734. [Google Scholar] [CrossRef]
- Palumbo, S.L.; Ebbinghaus, S.W.; Hurley, L.H. Formation of a unique end-to-end stacked pair of G-quadruplexes in the hTERT core promoter with implications for inhibition of telomerase by G-quadruplex-interactive ligands. J. Am. Chem. Soc. 2009, 131, 10878–10891. [Google Scholar] [CrossRef]
- Ramon, O.; Sauvaigo, S.; Gasparutto, D.; Faure, P.; Favier, A.; Cadet, J. Effects of 8-oxo-7, 8-dihydro-2′-deoxyguanosine on the binding of the transcription factor Sp1 to its cognate target DNA sequence (GC box). Free Radic. Res. 1999, 31, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Hailer-Morrison, M.K.; Kotler, J.M.; Martin, B.D.; Sugden, K.D. Oxidized guanine lesions as modulators of gene transcription. Altered p50 binding affinity and repair shielding by 7, 8-dihydro-8-oxo-2 ‘-deoxyguanosine lesions in the NF-κB promoter element. Biochemistry 2003, 42, 9761–9770. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.P.; Toomire, K.J.; Strauss, P.R. DNA modifications repaired by base excision repair are epigenetic. DNA Repair 2013, 12, 1152–1158. [Google Scholar] [CrossRef] [PubMed]
- Valinluck, V.; Tsai, H.-H.; Rogstad, D.K.; Burdzy, A.; Bird, A.; Sowers, L.C. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2). Nucleic Acids Res. 2004, 32, 4100–4108. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, N.; O’Shaughnessy, A.; Hendrich, B. Transcriptional repressors: Multifaceted regulators of gene expression. Development 2013, 140, 505–512. [Google Scholar] [CrossRef]
- Vidal, M.; Starowicz, K. Polycomb complexes PRC1 and their function in hematopoiesis. Exp. Hematol. 2017, 48, 12–31. [Google Scholar] [CrossRef] [PubMed]
- Beltran, M.; Tavares, M.; Justin, N.; Khandelwal, G.; Ambrose, J.; Foster, B.M.; Worlock, K.B.; Tvardovskiy, A.; Kunzelmann, S.; Herrero, J. G-tract RNA removes Polycomb repressive complex 2 from genes. Nat. Struct. Mol. Biol. 2019, 26, 899–909. [Google Scholar] [CrossRef]
- Wang, X.; Goodrich, K.J.; Gooding, A.R.; Naeem, H.; Archer, S.; Paucek, R.D.; Youmans, D.T.; Cech, T.R.; Davidovich, C. Targeting of polycomb repressive complex 2 to RNA by short repeats of consecutive guanines. Mol. Cell 2017, 65, 1056–1067.e1055. [Google Scholar] [CrossRef]
- Bak, S.T.; Sakellariou, D.; Pena-Diaz, J. The dual nature of mismatch repair as antimutator and mutator: For better or for worse. Front. Genet. 2014, 5, 287. [Google Scholar] [CrossRef]
- Carell, T.; Kurz, M.Q.; Müller, M.; Rossa, M.; Spada, F. Non-canonical bases in the genome: The regulatory information layer in DNA. Angew. Chem. Int. Ed. 2018, 57, 4296–4312. [Google Scholar] [CrossRef]
- Malfatti, M.C.; Antoniali, G.; Codrich, M.; Burra, S.; Mangiapane, G.; Dalla, E.; Tell, G. New perspectives in cancer biology from a study of canonical and non-canonical functions of base excision repair proteins with a focus on early steps. Mutagenesis 2020, 35, 129–149. [Google Scholar] [CrossRef]
- Saini, N.; Zhang, Y.; Usdin, K.; Lobachev, K.S. When secondary comes first–the importance of non-canonical DNA structures. Biochimie 2013, 95, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Bignami, M.; Mazzei, F. Base Excision Repair in Trinucleotide Repeat Expansion Disorders. In The Base Excision Repair Pathway: Molecular Mechanisms and Role in Disease Development and Therapeutic Design; World Scientific: Singapore, 2017; pp. 501–522. [Google Scholar]
- Dexheimer, T.S. DNA repair pathways and mechanisms. In DNA Repair of Cancer Stem Cells; Springer: Berlin/Heidelberg, Germany, 2013; pp. 19–32. [Google Scholar]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, J.-Q.; Chen, Z.; Zheng, K.-W.; Chen, C.-Y.; Hao, Y.-H.; Tan, Z. G-quadruplex formation at the 3′ end of telomere DNA inhibits its extension by telomerase, polymerase and unwinding by helicase. Nucleic Acids Res. 2011, 39, 6229–6237. [Google Scholar] [CrossRef]
- Lee, C.Y.; Park, K.S.; Park, H.G. A fluorescent G-quadruplex probe for the assay of base excision repair enzyme activity. Chem. Commun. 2015, 51, 13744–13747. [Google Scholar] [CrossRef] [PubMed]
- Leung, K.-H.; He, H.-Z.; Ma, V.P.-Y.; Zhong, H.-J.; Chan, D.S.-H.; Zhou, J.; Mergny, J.-L.; Leung, C.-H.; Ma, D.-L. Detection of base excision repair enzyme activity using a luminescent G-quadruplex selective switch-on probe. Chem. Commun. 2013, 49, 5630–5632. [Google Scholar] [CrossRef]
- Craddock, N.; Jones, L.; Jones, I.R.; Kirov, G.; Green, E.K.; Grozeva, D.; Moskvina, V.; Nikolov, I.; Hamshere, M.L.; Vukcevic, D. Strong genetic evidence for a selective influence of GABAA receptors on a component of the bipolar disorder phenotype. Mol. Psychiatry 2010, 15, 146–153. [Google Scholar] [CrossRef]
- Jin, Z.; Gao, F.; Flagg, T.; Deng, X. Tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone promotes functional cooperation of Bcl2 and c-Myc through phosphorylation in regulating cell survival and proliferation. J. Biol. Chem. 2004, 279, 40209–40219. [Google Scholar] [CrossRef]
- Mattila, P.M.; Röyttä, M.; Lönnberg, P.; Marjamäki, P.; Helenius, H.; Rinne, J.O. Choline acetyltransferase activity and striatal dopamine receptors in Parkinson’s disease in relation to cognitive impairment. Acta Neuropathol. 2001, 102, 160–166. [Google Scholar] [CrossRef]
- Tarabeux, J.; Kebir, O.; Gauthier, J.; Hamdan, F.; Xiong, L.; Piton, A.; Spiegelman, D.; Henrion, É.; Millet, B.; Fathalli, F. Rare mutations in N-methyl-D-aspartate glutamate receptors in autism spectrum disorders and schizophrenia. Transl. Psychiatry 2011, 1, e55. [Google Scholar] [CrossRef]
- Williams, N.M.; Bowen, T.; Spurlock, G.; Norton, N.; Williams, H.J.; Hoogendoorn, B.; Owen, M.J.; O’Donovan, M.C. Determination of the genomic structure and mutation screening in schizophrenic individuals for five subunits of the N-methyl-D-aspartate glutamate receptor. Mol. Psychiatry 2002, 7, 508–514. [Google Scholar] [CrossRef][Green Version]
- Yu, Y.; Lin, Y.; Takasaki, Y.; Wang, C.; Kimura, H.; Xing, J.; Ishizuka, K.; Toyama, M.; Kushima, I.; Mori, D. Rare loss of function mutations in N-methyl-D-aspartate glutamate receptors and their contributions to schizophrenia susceptibility. Transl. Psychiatry 2018, 8, 12. [Google Scholar] [CrossRef] [PubMed]
- Schofield, J.P.; Cowan, J.L.; Coldwell, M.J. G-quadruplexes mediate local translation in neurons. Biochem. Soc. Trans. 2015, 43, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, M.; Rage, F.; Tabet, R.; Flatter, E.; Mandel, J.L.; Moine, H. G–quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO Rep. 2011, 12, 697–704. [Google Scholar] [CrossRef] [PubMed]
- Kharel, P.; Balaratnam, S.; Beals, N.; Basu, S. The role of RNA G-quadruplexes in human diseases and therapeutic strategies. Wiley Interdiscip. Rev. RNA 2020, 11, e1568. [Google Scholar] [CrossRef] [PubMed]
- Lazniewska, J.; Milowska, K.; Zablocka, M.; Mignani, S.; Caminade, A.-M.; Majoral, J.-P.; Bryszewska, M.; Gabryelak, T. Mechanism of cationic phosphorus dendrimer toxicity against murine neural cell lines. Mol. Pharm. 2013, 10, 3484–3496. [Google Scholar] [CrossRef]
- Zeng, Y.; Kurokawa, Y.; Win-Shwe, T.-T.; Zeng, Q.; Hirano, S.; Zhang, Z.; Sone, H. Effects of PAMAM dendrimers with various surface functional groups and multiple generations on cytotoxicity and neuronal differentiation using human neural progenitor cells. J. Toxicol. Sci. 2016, 41, 351–370. [Google Scholar] [CrossRef]
- Grün, J.T.; Schwalbe, H. Folding dynamics of polymorphic G-quadruplex structures. Biopolymers 2022, 113, e23477. [Google Scholar] [CrossRef]
- Stevens, A.J.; de Jong, L.; Kennedy, M.A. The Dynamic Regulation of G-Quadruplex DNA Structures by Cytosine Methylation. Int. J. Mol. Sci. 2022, 23, 2407. [Google Scholar] [CrossRef]
- Lech, C.J.; Heddi, B.; Phan, A.T.n. Guanine base stacking in G-quadruplex nucleic acids. Nucleic Acids Res. 2013, 41, 2034–2046. [Google Scholar] [CrossRef] [PubMed]
- Illingworth, R.S.; Gruenewald-Schneider, U.; Webb, S.; Kerr, A.R.; James, K.D.; Turner, D.J.; Smith, C.; Harrison, D.J.; Andrews, R.; Bird, A.P. Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet. 2010, 6, e1001134. [Google Scholar] [CrossRef] [PubMed]
- Szeltner, Z.; Ferenc, G.; Juhász, T.; Kupihár, Z.; Váradi, Z.; Szüts, D.; Kovács, L. Probing telomeric-like G4 structures with full or partial 2′-deoxy-5-hydroxyuridine substitutions. Biochimie 2023, 214, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Dézé, O.; Laffleur, B.; Cogné, M. Roles of G4-DNA and G4-RNA in Class Switch Recombination and Additional Regulations in B-Lymphocytes. Molecules 2023, 28, 1159. [Google Scholar] [CrossRef]

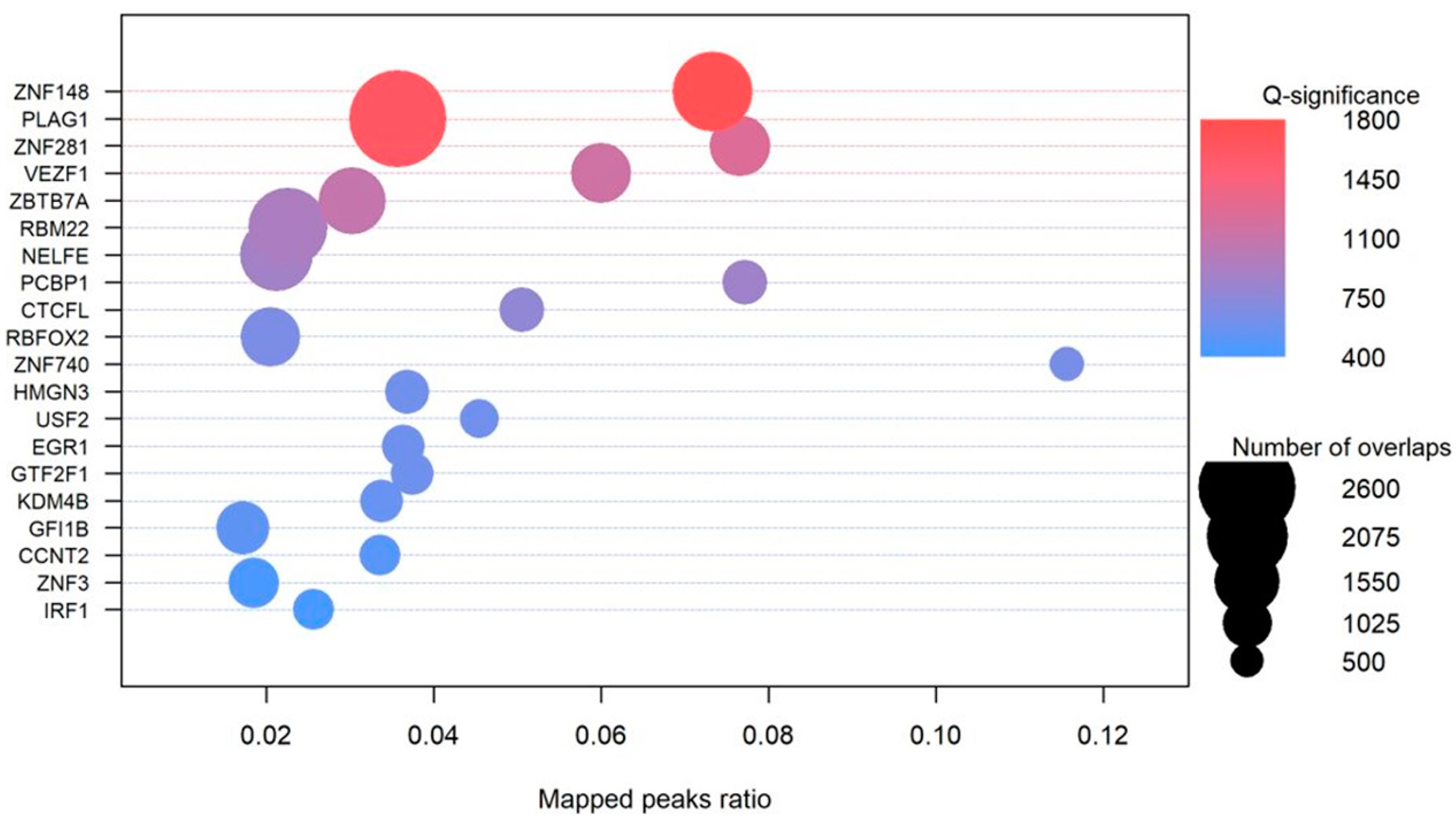

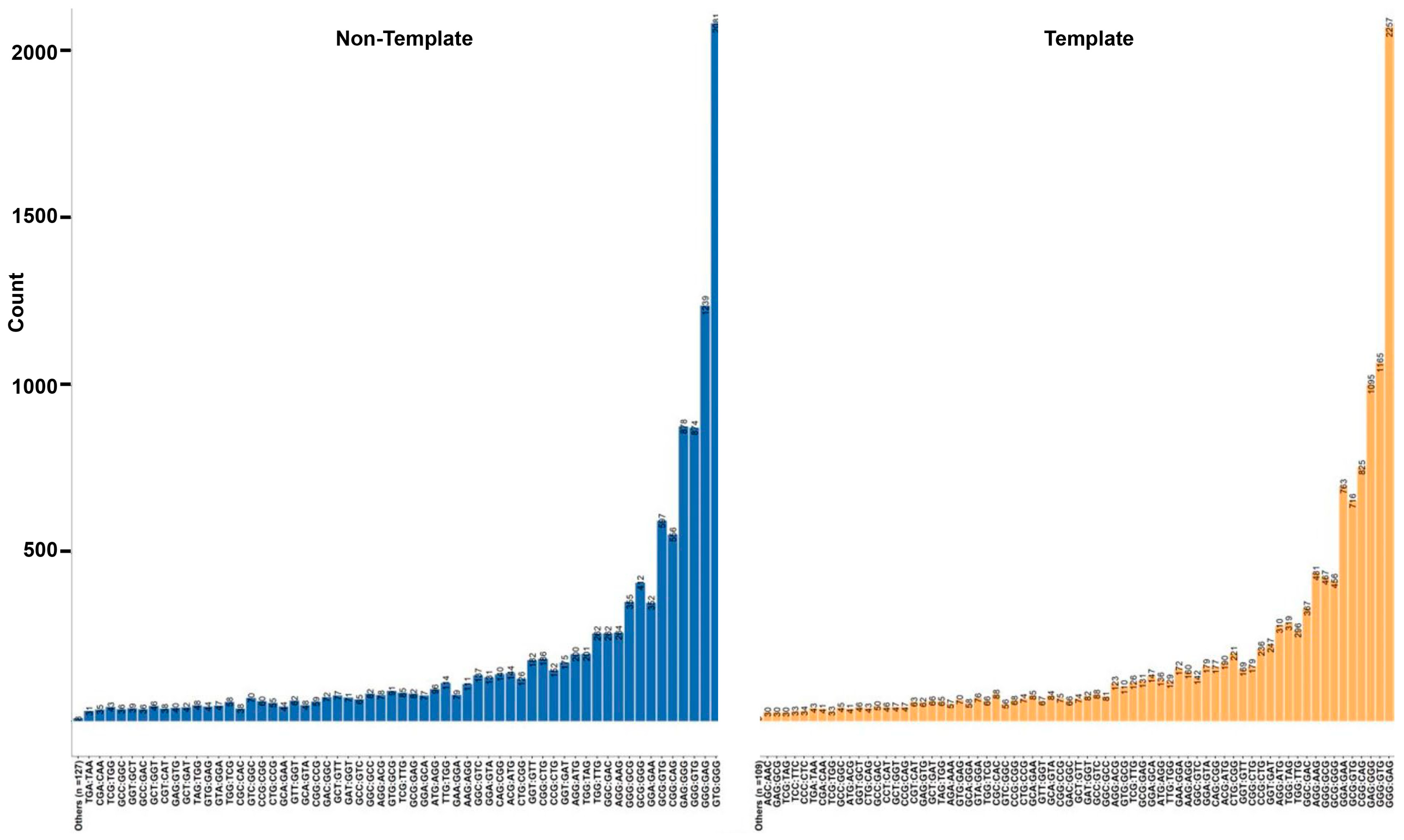
| Type of SNV | Effect of SNV on MFE | Freq | Percentage |
|---|---|---|---|
| Transition | Destabilized | 8600 | 22.93 |
| Transversion | Further stabilized | 6603 | 17.60 |
| Transition | No change | 6552 | 17.46 |
| Transversion | Destabilized | 6419 | 17.11 |
| Transversion | No change | 5509 | 14.68 |
| Transition | Further stabilized | 3832 | 10.21 |
| SNV | 3′ UTR | 5′ UTR | CDS | CpG Islands | Enhancers | Exon | Intergenic | Intron | lncRNA GENCODE | Promoter |
|---|---|---|---|---|---|---|---|---|---|---|
| G→A | 34.82 | 31.23 | 39.34 | 27.58 | 19.01 | 35.18 | 27.84 | 26.74 | 28.27 | 26.87 |
| G→T | 18.3 | 14.82 | 15.84 | 12.01 | 12.08 | 16.6 | 17.15 | 14.56 | 14.89 | 13.64 |
| C→T | 11.13 | 8.24 | 12.19 | 12.23 | 5.68 | 11.35 | 7.94 | 9.06 | 10.46 | 8.89 |
| T→G | 12.67 | 16.48 | 8.59 | 16.78 | 29.84 | 11.74 | 18.38 | 19.91 | 18.15 | 18.95 |
| A→G | 7.62 | 10.83 | 6.91 | 9.56 | 12.61 | 8 | 11.38 | 11.49 | 9.89 | 10.98 |
| G→C | 5.16 | 6.05 | 4.69 | 6.27 | 7.28 | 5.09 | 6.89 | 6.3 | 6.23 | 6.51 |
| C→G | 3.66 | 4.85 | 4.47 | 7.69 | 6.75 | 4.33 | 3.47 | 4.59 | 4.56 | 6.34 |
| C→A | 2.76 | 2.99 | 4.11 | 3.85 | 1.42 | 3.65 | 1.95 | 2.6 | 2.73 | 3.3 |
| T→C | 2.15 | 1.66 | 1.79 | 2 | 2.13 | 1.96 | 1.67 | 2.1 | 2.13 | 2.07 |
| T→A | 0.7 | 0.86 | 0.84 | 0.67 | 0.89 | 0.81 | 1.27 | 1 | 1.17 | 0.82 |
| A→T | 0.59 | 1.2 | 0.72 | 0.75 | 1.6 | 0.76 | 1.12 | 0.94 | 1 | 0.85 |
| A→C | 0.45 | 0.8 | 0.51 | 0.62 | 0.71 | 0.52 | 0.94 | 0.72 | 0.53 | 0.77 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neupane, A.; Chariker, J.H.; Rouchka, E.C. Analysis of Nucleotide Variations in Human G-Quadruplex Forming Regions Associated with Disease States. Genes 2023, 14, 2125. https://doi.org/10.3390/genes14122125
Neupane A, Chariker JH, Rouchka EC. Analysis of Nucleotide Variations in Human G-Quadruplex Forming Regions Associated with Disease States. Genes. 2023; 14(12):2125. https://doi.org/10.3390/genes14122125
Chicago/Turabian StyleNeupane, Aryan, Julia H. Chariker, and Eric C. Rouchka. 2023. "Analysis of Nucleotide Variations in Human G-Quadruplex Forming Regions Associated with Disease States" Genes 14, no. 12: 2125. https://doi.org/10.3390/genes14122125
APA StyleNeupane, A., Chariker, J. H., & Rouchka, E. C. (2023). Analysis of Nucleotide Variations in Human G-Quadruplex Forming Regions Associated with Disease States. Genes, 14(12), 2125. https://doi.org/10.3390/genes14122125






