Ovine KRTAP36-2: A New Keratin-Associated Protein Gene Related to Variation in Wool Yield
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Sheep Examined and the Collection of Wool Samples
2.2. PCR Amplification
2.3. Screening for Sequence Variation in the Amplicon and DNA Sequencing
2.4. DNA Sequence Analyses
2.5. Statistical Analyses of Associations
3. Results
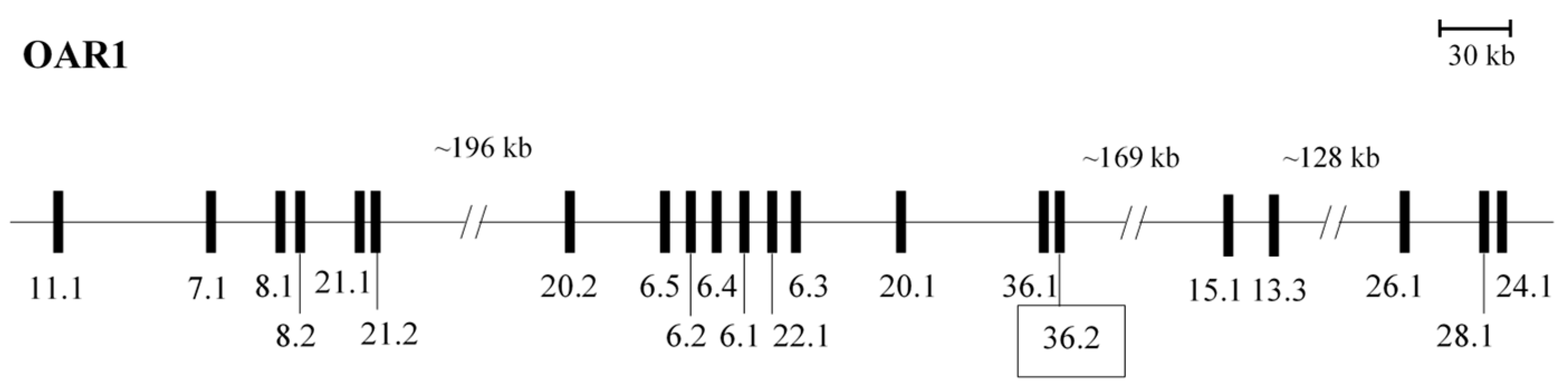
3.1. A Newly Identified Ovine HGT-KRTAP
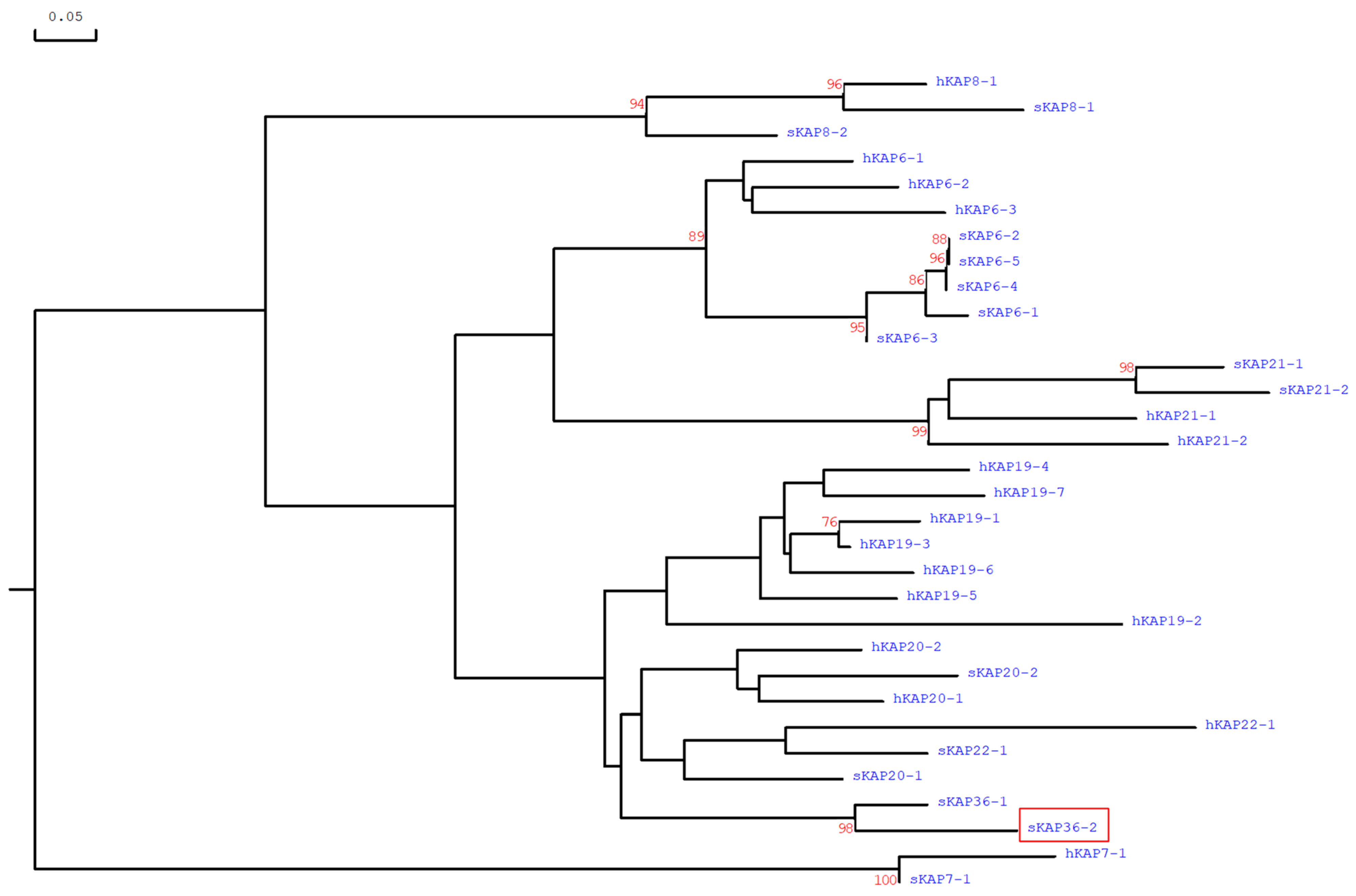
3.2. Nucleotide Sequence Variation in Ovine KRTAP36-2
3.3. Association of KRTAP36-2 Variation with Wool Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Powell, B.C.; Rogers, G.E. The role of keratin proteins and their genes in the growth, structure and properties of hair. EXS 1997, 78, 59–148. [Google Scholar]
- Yu, Z.; Wildermoth, J.E.; Wallace, O.A.; Gordon, S.W.; Maqbool, N.J.; Maclean, P.H.; Nixon, A.J.; Pearson, A.J. Annotation of sheep keratin intermediate filament genes and their patterns of expression. Exp. Dermatol. 2011, 20, 582–588. [Google Scholar] [CrossRef]
- Langbein, L.; Rogers, M.A.; Praetzel-Wunder, S.; Böckler, D.; Schirmacher, P.; Schweizer, J. Novel type I hair keratins K39 and K40 are the last to be expressed in differentiation of the hair: Completion of the human hair keratin catalog. J. Investig. Dermatol. 2007, 127, 1532–1535. [Google Scholar] [CrossRef]
- Rogers, M.A.; Schweizer, J. Human KAP genes, only the half of it? Extensive size polymorphisms in hair keratin-associated protein genes. J. Investig. Dermatol. 2005, 124, vii–ix. [Google Scholar] [CrossRef]
- Rogers, M.A.; Winter, H.; Langbein, L.; Wollschläger, A.; Praetzel-Wunder, S.; Jave-Suarez, L.F.; Schweizer, J. Characterization of human KAP24. 1, a cuticular hair keratin-associated protein with unusual amino-acid composition and repeat structure. J. Investig. Dermatol. 2007, 127, 1197–1204. [Google Scholar] [CrossRef]
- Rogers, M.A.; Langbein, L.; Praetzel Wunder, S.; Giehl, K. Characterization and expression analysis of the hair keratin associated protein KAP26.1. Brit. J. Dermatol. 2008, 159, 725–729. [Google Scholar] [CrossRef] [PubMed]
- Gong, H.; Zhou, H.; Forrest, R.H.; Li, S.; Wang, J.; Dyer, J.M.; Luo, Y.; Hickford, J.G.H. Wool keratin-associated protein genes in sheep- a review. Genes 2016, 7, 24. [Google Scholar] [CrossRef]
- Gong, H.; Zhou, H.; Wang, J.; Li, S.; Luo, Y.; Hickford, J.G. Characterisation of an ovine keratin associated protein (KAP) gene, which would produce a protein rich in glycine and tyrosine, but lacking in cysteine. Genes 2019, 10, 848. [Google Scholar] [CrossRef] [PubMed]
- Rogers, G.E. Biology of the wool follicle: An excursion into a unique tissue interaction system waiting to be re-discovered. Exp. Dermatol. 2006, 15, 931–949. [Google Scholar] [CrossRef]
- Gillespie, J.M. The proteins of hair and other hard a-keratins. In Cellular and Molecular Biology of Intermediate Filaments; Goldman, R.D., Steinert, P.M., Eds.; Plenum Press: New York, NY, USA, 1990; pp. 95–128. [Google Scholar]
- Li, S.W.; Ouyang, H.S.; Rogers, G.E.; Bawden, C.S. Characterization of the structural and molecular defects in fibres and follicles of the merino felting lustre mutant. Exp. Dermatol. 2009, 18, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Gong, H.; Zhou, H.; Tao, J.; Hickford, J.G. A nucleotide substitution in the ovine KAP20-2 gene leads to a premature stop codon that affects wool fibre curvature. Anim. Genet. 2018, 49, 357–358. [Google Scholar] [CrossRef]
- Ullah, F.; Jamal, S.M.; Zhou, H.; Hickford, J.G. Variation in ovine KRTAP8-1 affects mean staple length and opacity of wool fiber. Anim. Biotechnol. 2021, 34, 602–608. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Che, L.; Hickford, J.G.; Zhou, H.; Hao, Z.; Luo, Y.; Hu, J.; Liu, X.; Li, S. Identification of the caprine keratin-associated protein 20-2 (KAP20-2) gene and its effect on cashmere traits. Genes 2017, 8, 328. [Google Scholar] [CrossRef]
- Wang, J.; Hao, Z.; Zhou, H.; Luo, Y.; Hu, J.; Liu, X.; Li, S.; Hickford, J.G.H. A keratin-associated protein (KAP) gene that is associated with variation in cashmere goat fleece weight. Small Rumin. Res. 2018, 167, 104–109. [Google Scholar] [CrossRef]
- Zhou, H.; Gong, H.; Wang, J.; Luo, Y.; Li, S.; Tao, J.; Hickford, J.G.H. The complexity of the ovine and caprine keratin-associated protein genes. Int. J. Mol. Sci. 2021, 22, 12838. [Google Scholar] [CrossRef]
- Zhou, H.; Hickford, J.G.H.; Fang, Q. A two-step procedure for extracting genomic DNA from dried blood spots on filter paper for polymerase chain reaction amplification. Anal. Biochem. 2006, 354, 159–161. [Google Scholar] [CrossRef]
- Byun, S.O.; Fang, Q.; Zhou, H.; Hickford, J.G.H. An effective method for silver-staining DNA in large numbers of polyacrylamide gels. Anal. Biochem. 2009, 385, 174–175. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Gong, H.; Zhou, H.; Wang, J.; Liu, X.; Li, S.; Luo, Y.; Hickford, J.G.H. Variation in the ovine keratin-associated protein 15-1 gene affects wool yield. J. Agric. Sci. 2018, 156, 922–928. [Google Scholar] [CrossRef]
- Gong, H.; Zhou, H.; Bai, L.; Li, W.; Li, S.; Wang, J.; Luo, Y.; Hickford, J.G.H. Associations between variation in the ovine high glycine-tyrosine keratin-associated protein gene KRTAP20-1 and wool traits. J. Anim. Sci. 2019, 97, 587–595. [Google Scholar] [CrossRef]
- Li, S.; Zhou, H.; Gong, H.; Zhao, F.; Wang, J.; Liu, X.; Hu, J.; Luo, Y.; Hickford, J.G.H. Identification of the ovine keratin-associated protein 21-1 gene and its association with variation in wool traits. Animals 2019, 9, 450. [Google Scholar] [CrossRef]
- Li, S.; Zhou, H.; Gong, H.; Zhao, F.; Wang, J.; Liu, X.; Luo, Y.; Hickford, J.G.H. Identification of the ovine keratin-associated protein 22-1 (KAP22-1) gene and its effect on wool traits. Genes 2017, 8, 27. [Google Scholar] [CrossRef]
- McKittrick, J.; Chen, P.Y.; Bodde, S.; Yang, W.; Novitskaya, E.; Meyers, M. The structure, functions, and mechanical properties of keratin. JOM 2012, 64, 449–468. [Google Scholar] [CrossRef]
- Bradbury, J. The morphology and chemical structure of wool. In Macromolecular Chemistry-11; Eisenberg, H., Ed.; Elsevier: Amsterdam, The Netherlands, 1977; pp. 247–253. [Google Scholar]
- Wolner, B.S.; Gralla, J.D. TATA-flanking sequences influence the rate and stability of TATA-binding protein and TFIIB binding. J. Biol. Chem. 2001, 276, 6260–6266. [Google Scholar] [CrossRef] [PubMed]
- Babu, D.; Vannelli, S.; Fanelli, A.; Mellone, S.; Baffico, A.M.; Corrado, L.; Essa, W.A.; Grandone, A.; Bellone, S.; Monzani, A. Variants in the 5′ UTR reduce SHOX expression and contribute to SHOX haploinsufficiency. Eur. J. Hum. Genet. 2021, 29, 110–121. [Google Scholar] [CrossRef]
- Dvir, S.; Velten, L.; Sharon, E.; Zeevi, D.; Carey, L.B.; Weinberger, A.; Segal, E. Deciphering the rules by which 5′-UTR sequences affect protein expression in yeast. Proc. Natl. Acad. Sci. USA 2013, 110, E2792–E2801. [Google Scholar] [CrossRef]


| Wool Trait 1 | Variant Assessed | Absent | Present | P 3 | ||
|---|---|---|---|---|---|---|
| Mean ± SE 2 | n | Mean ± SE | n | |||
| GFW (kg) | A | 2.5 ± 0.16 | 38 | 2.4 ± 0.14 | 210 | 0.124 |
| B | 2.4 ± 0.15 | 109 | 2.4 ± 0.14 | 139 | 0.295 | |
| C | 2.4 ± 0.15 | 204 | 2.5 ± 0.17 | 44 | 0.255 | |
| CFW (kg) | A | 1.9 ± 0.15 | 38 | 1.8 ± 0.13 | 210 | 0.861 |
| B | 1.8 ± 0.12 | 109 | 1.8 ± 0.12 | 139 | 0.647 | |
| C | 1.8 ± 0.13 | 204 | 1.9 ± 0.15 | 44 | 0.575 | |
| YIELD (%) | A | 77.3 ± 2.44 | 38 | 74.2 ± 2.08 | 210 | 0.029 |
| B | 74.7 ± 2.21 | 109 | 74.1 ± 2.18 | 139 | 0.632 | |
| C | 74.9 ± 2.13 | 204 | 73.0 ± 2.43 | 44 | 0.233 | |
| MSL (mm) | A | 96.8 ± 4.86 | 38 | 97.6 ± 4.16 | 210 | 0.773 |
| B | 97.4 ± 4.33 | 109 | 97.6 ± 4.28 | 139 | 0.915 | |
| C | 97.4 ± 4.21 | 204 | 98.1 ± 4.75 | 44 | 0.805 | |
| MSS (N/ktex) | A | 26.3 ± 3.50 | 38 | 27.0 ± 3.00 | 210 | 0.717 |
| B | 27.1 ± 3.11 | 109 | 26.8 ± 3.08 | 139 | 0.848 | |
| C | 25.9 ± 2.99 | 204 | 29.2 ± 3.37 | 44 | 0.070 | |
| MFD (µm) | A | 18.5 ± 0.55 | 38 | 18.3 ± 0.48 | 210 | 0.494 |
| B | 18.2 ± 0.49 | 109 | 18.4 ± 0.48 | 139 | 0.322 | |
| C | 18.1 ± 0.52 | 204 | 18.6 ± 0.52 | 44 | 0.237 | |
| FDSD (µm) | A | 3.5 ± 0.22 | 38 | 3.5 ± 0.19 | 210 | 0.610 |
| B | 3.5 ± 0.18 | 109 | 3.5 ± 0.18 | 139 | 0.628 | |
| C | 3.5 ± 0.18 | 204 | 3.6 ± 0.20 | 44 | 0.386 | |
| CVFD (%) | A | 19.1 ± 0.97 | 38 | 19.0 ± 0.83 | 210 | 0.949 |
| B | 19.4 ± 0.87 | 109 | 18.9 ± 0.86 | 139 | 0.182 | |
| C | 19.1 ± 0.86 | 204 | 19.1 ± 0.95 | 44 | 0.997 | |
| PF (%) | A | 0.5 ± 0.51 | 38 | 0.7 ± 0.43 | 210 | 0.442 |
| B | 0.8 ± 0.45 | 109 | 0.6 ± 0.44 | 139 | 0.596 | |
| C | 0.6 ± 0.44 | 204 | 0.9 ± 0.50 | 44 | 0.428 | |
| MFC (°/mm) | A | 72.1 ± 6.80 | 38 | 78.0 ± 5.78 | 210 | 0.128 |
| B | 80.8 ± 5.99 | 109 | 74.9 ± 5.92 | 139 | 0.063 | |
| C | 76.9 ± 5.89 | 204 | 80.1 ± 6.66 | 44 | 0.447 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, H.; Li, W.; Bai, L.; Wang, J.; Luo, Y.; Li, S.; Hickford, J.G.H. Ovine KRTAP36-2: A New Keratin-Associated Protein Gene Related to Variation in Wool Yield. Genes 2023, 14, 2045. https://doi.org/10.3390/genes14112045
Zhou H, Li W, Bai L, Wang J, Luo Y, Li S, Hickford JGH. Ovine KRTAP36-2: A New Keratin-Associated Protein Gene Related to Variation in Wool Yield. Genes. 2023; 14(11):2045. https://doi.org/10.3390/genes14112045
Chicago/Turabian StyleZhou, Huitong, Wenhao Li, Lingrong Bai, Jiqing Wang, Yuzhu Luo, Shaobin Li, and Jonathan G. H. Hickford. 2023. "Ovine KRTAP36-2: A New Keratin-Associated Protein Gene Related to Variation in Wool Yield" Genes 14, no. 11: 2045. https://doi.org/10.3390/genes14112045
APA StyleZhou, H., Li, W., Bai, L., Wang, J., Luo, Y., Li, S., & Hickford, J. G. H. (2023). Ovine KRTAP36-2: A New Keratin-Associated Protein Gene Related to Variation in Wool Yield. Genes, 14(11), 2045. https://doi.org/10.3390/genes14112045








