Genome-Wide Association Study of the Reproductive Traits of the Dazu Black Goat (Capra hircus) Using Whole-Genome Resequencing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Phenotypic and Genotypic Data
- (1)
- Udder depth (UD) is the vertical distance from the root of the left udder to the top of the teat.
- (2)
- Teat diameter (TD) is the teat diameter of the left udder.
- (3)
- Teat length (TL) is the teat length of the left udder.
- (4)
- Supernumerary teats (SNTs) were observed and recorded with the naked eye.
2.3. Statistical Analysis
3. Results
3.1. Phenotypic Data Analysis
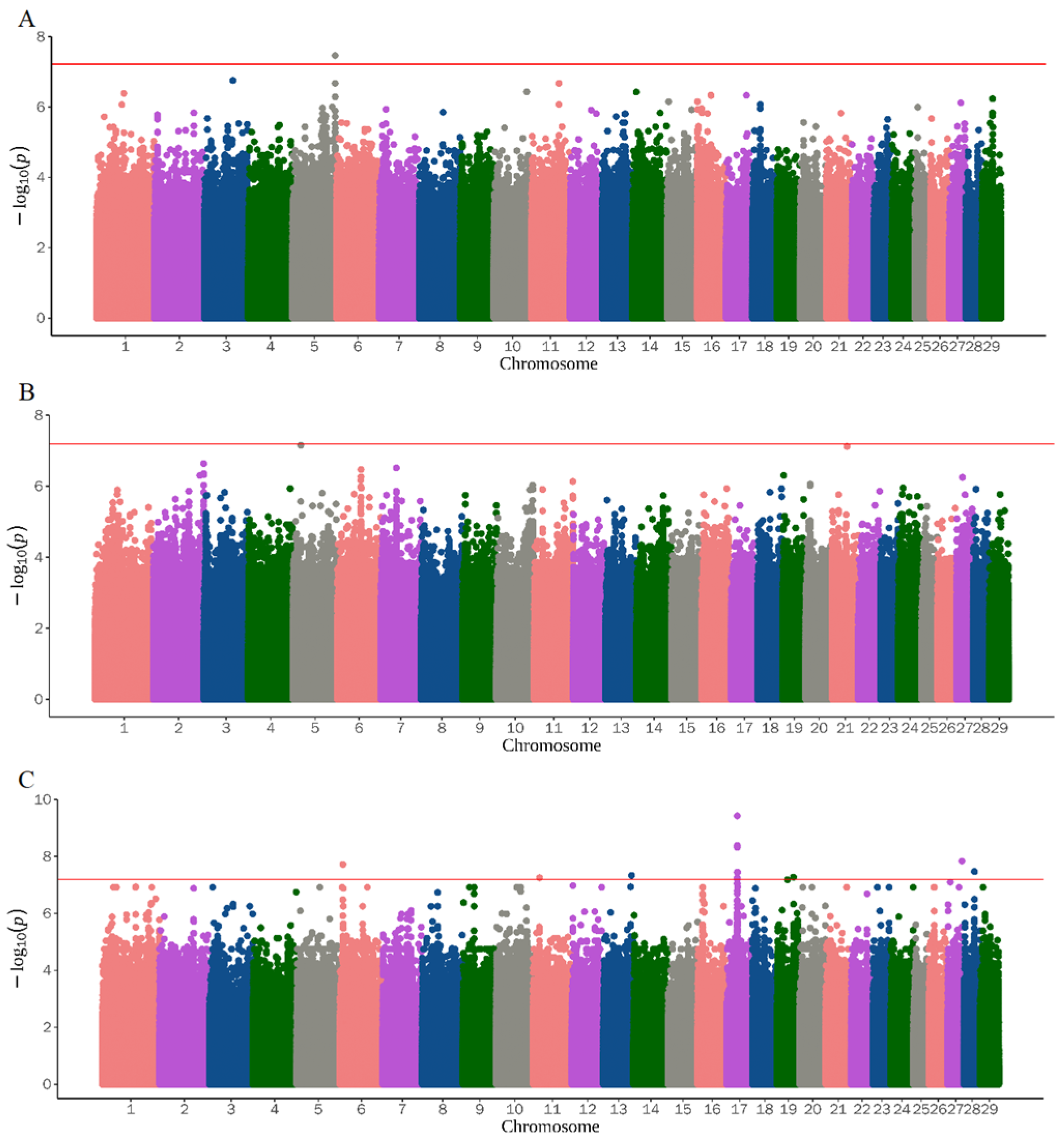
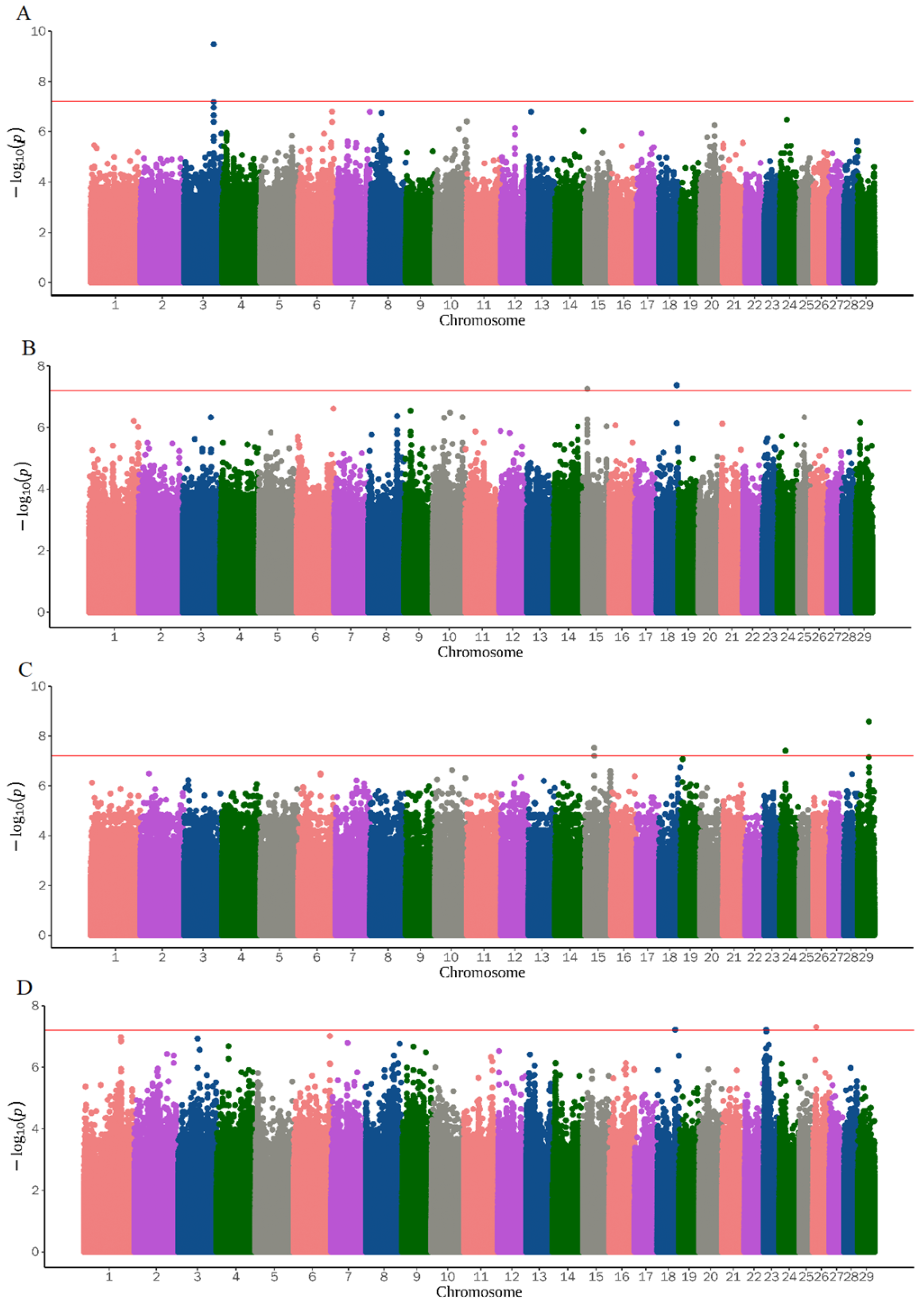
3.2. Genome-Wide Association Study
3.3. GO Enrichment and KEGG Analysis
4. Discussion
4.1. Kidding Traits
4.2. Udder Traits
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, Z.; Yang, T.; Qiao, L.; He, Q.; Dai, Z. Reproductive Characteristics of Dazu Black Goats, a Newly Discovered Chinese Indigenous Breed Resource with High Litter Sizes. Pak. J. Zool. 2019, 51, 399–403. [Google Scholar] [CrossRef]
- Lai, F.-N.; Zhai, H.-L.; Cheng, M.; Ma, J.-Y.; Cheng, S.-F.; Ge, W.; Zhang, G.-L.; Wang, J.-J.; Zhang, R.-Q.; Wang, X.; et al. Whole-genome scanning for the litter size trait associated genes and SNPs under selection in dairy goat (Capra hircus). Sci. Rep. 2016, 6, 38096. [Google Scholar] [CrossRef] [PubMed]
- Odubote, I.K. Genetic parameters for litter size at birth and kidding interval in the West African Dwarf goats. Small Rumin. Res. 1996, 20, 261–265. [Google Scholar] [CrossRef]
- Bangar, Y.C.; Magotra, A.; Yadav, A.S.; Chauhan, A. Estimation of genetic parameters for early reproduction traits in Beetal goat. Zygote 2022, 30, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Guang-Xin, E.; Zhu, Y.-B.; Basang, W.-D.; Na, R.-S.; Han, Y.-G.; Zeng, Y. Comparative and selection sweep analysis of CNV was associated to litter size in Dazu black goats. Anim. Biotechnol. 2021, 32, 792–797. [Google Scholar] [CrossRef]
- Islam, R.; Liu, X.; Gebreselassie, G.; Abied, A.; Ma, Q.; Ma, Y. Genome-wide association analysis reveals the genetic locus for high reproduction trait in Chinese Arbas Cashmere goat. Genes Genom. 2020, 42, 893–899. [Google Scholar] [CrossRef]
- Wang, J.-J.; Zhang, T.; Chen, Q.-M.; Zhang, R.-Q.; Li, L.; Cheng, S.-F.; Shen, W.; Lei, C.-Z. Genomic Signatures of Selection Associated With Litter Size Trait in Jining Gray Goat. Front. Genet. 2020, 11, 286. [Google Scholar] [CrossRef]
- Visscher, P.M.; Wray, N.R.; Zhang, Q.; Sklar, P.; McCarthy, M.I.; Brown, M.A.; Yang, J. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Hum. Genet. 2017, 101, 5–22. [Google Scholar] [CrossRef]
- Xu, S.-S.; Gao, L.; Xie, X.-L.; Ren, Y.-L.; Shen, Z.-Q.; Wang, F.; Shen, M.; Eyporsdottir, E.; Hallsson, J.H.; Kiseleva, T.; et al. Genome-Wide Association Analyses Highlight the Potential for Different Genetic Mechanisms for Litter Size Among Sheep Breeds. Front. Genet. 2018, 9, 118. [Google Scholar] [CrossRef]
- Guo, J.; Jiang, R.; Mao, A.; Liu, G.E.; Zhan, S.; Li, L.; Zhong, T.; Wang, L.; Cao, J.; Chen, Y.; et al. Genome-wide association study reveals 14 new SNPs and confirms two structural variants highly associated with the horned/polled phenotype in goats. BMC Genom. 2021, 22, 769. [Google Scholar] [CrossRef]
- Moaeen-ud-Din, M.; Muner, R.D.; Khan, M.S. Genome wide association study identifies novel candidate genes for growth and body conformation traits in goats. Sci. Rep. 2022, 12, 9891. [Google Scholar] [CrossRef] [PubMed]
- Scholtens, M.; Jiang, A.; Smith, A.; Littlejohn, M.; Lehnert, K.; Snell, R.; Lopez-Villalobos, N.; Garrick, D.; Blair, H. Genome-wide association studies of lactation yields of milk, fat, protein and somatic cell score in New Zealand dairy goats. J. Anim. Sci. Biotechnol. 2020, 11, 55. [Google Scholar] [CrossRef] [PubMed]
- Islam, R.; Li, Y.; Liu, X.; Berihulay, H.; Abied, A.; Gebreselassie, G.; Ma, Q.; Ma, Y. Genome-Wide Runs of Homozygosity, Effective Population Size, and Detection of Positive Selection Signatures in Six Chinese Goat Breeds. Genes 2019, 10, 938. [Google Scholar] [CrossRef] [PubMed]
- Devani, K.; Plastow, G.; Orsel, K.; Valente, T.S. Genome-wide association study for mammary structure in Canadian Angus cows. PLoS ONE 2020, 15, e0237818. [Google Scholar] [CrossRef]
- Abdoli, R.; Mirhoseini, S.Z.; Hossein-Zadeh, N.G.; Zamani, P.; Moradi, M.H.; Ferdosi, M.H.; Gondro, C. Genome-wide association study of first lambing age and lambing interval in sheep. Small Rumin. Res. 2019, 178, 43–45. [Google Scholar] [CrossRef]
- Gu, B.W.; Sun, R.F.; Fang, X.Q.; Zhang, J.P.; Zhao, Z.Q.; Huang, D.L.; Zhao, Y.P.; Zhao, Y.J. Genome-Wide Association Study of Body Conformation Traits by Whole Genome Sequencing in Dazu Black Goats. Animals 2022, 12, 548. [Google Scholar] [CrossRef]
- Browning, B.L.; Zhou, Y.; Browning, S.R. A One-Penny Imputed Genome from Next-Generation Reference Panels. Am. J. Hum. Genet. 2018, 103, 338–348. [Google Scholar] [CrossRef]
- Yu, J.M.; Pressoir, G.; Briggs, W.H.; Bi, I.V.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. Omics-A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- E, G.-X.; Duan, X.-H.; Zhang, J.-H.; Huang, Y.-F.; Zhao, Y.-J.; Na, R.-S.; Zhao, Z.-Q.; Ma, Y.-H.; Chu, M.-X.; Basang, W.-D.; et al. Genome-wide selection signatures analysis of litter size in Dazu black goats using single-nucleotide polymorphism. 3 Biotech 2019, 9, 336. [Google Scholar] [CrossRef]
- Hu, Z.; Niu, G.; Ren, J.; Wang, X.; Chen, L.; Hong, R.; Ke, C. TAFA5 promotes proliferation and migration in gastric cancer. Mol. Med. Rep. 2019, 20, 4477–4488. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, S.J.; Christensen, M.T.; Vrang, N.; Larsen, L.K. The putative neuropeptide TAFA5 is expressed in the hypothalamic paraventricular nucleus and is regulated by dehydration. Brain Res. 2008, 1199, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Bundzikova, J.; Pirnik, Z.; Zelena, D.; Mikkelsen, J.D.; Kiss, A. Response of Substances Co-Expressed in Hypothalamic Magnocellular Neurons to Osmotic Challenges in Normal and Brattleboro Rats. Cell. Mol. Neurobiol. 2008, 28, 1033–1047. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Wang, A.; Feng, L.; Wang, Y.; Zhang, H.; Zhang, I.; Bany, B.M.; Ma, L. Heparan Sulfate Proteoglycan Sulfation Regulates Uterine Differentiation and Signaling During Embryo Implantation. Endocrinology 2018, 159, 2459–2472. [Google Scholar] [CrossRef]
- Pallerla, S.R.; Lawrence, R.; Lewejohann, L.; Pan, Y.; Fischer, T.; Schlomann, U.; Zhang, X.; Esko, J.D.; Grobe, K. Altered heparan sulfate structure in mice with deleted NDST3 gene function. J. Biol. Chem. 2008, 283, 16885–16894. [Google Scholar] [CrossRef]
- Orlowski, J.; Grinstein, S. Diversity of the mammalian sodium/proton exchanger SLC9 gene family. Pflug. Arch.-Eur. J. Physiol. 2004, 447, 549–565. [Google Scholar] [CrossRef] [PubMed]
- Oberheide, K.; Puchkov, D.; Jentsch, T.J. Loss of the Na+/H+ exchanger NHE8 causes male infertility in mice by disrupting acrosome formation. J. Biol. Chem. 2017, 292, 10845–10854. [Google Scholar] [CrossRef]
- Freebern, E.; Santos, D.J.A.; Fang, L.; Jiang, J.; Parker Gaddis, K.L.; Liu, G.E.; VanRaden, P.M.; Maltecca, C.; Cole, J.B.; Ma, L. GWAS and fine-mapping of livability and six disease traits in Holstein cattle. BMC Genom. 2020, 21, 41. [Google Scholar] [CrossRef]
- Kumar, P.; Meizel, S. Identification and spatial distribution of glycine receptor subunits in human sperm. Reproduction 2008, 136, 387–390. [Google Scholar] [CrossRef]
- Tahir, M.S.; Porto-Neto, L.R.; Gondro, C.; Shittu, O.B.; Wockner, K.; Andre, T.W.L.; Smith, H.R.; Gouveia, G.C.; Kour, J.; Fortes, M.R.S. Meta-Analysis of Heifer Traits Identified Reproductive Pathways in Bos indicus Cattle. Genes 2021, 12, 768. [Google Scholar] [CrossRef] [PubMed]
- Iremonger, K.J.; Constantin, S.; Liu, X.; Herbison, A.E. Glutamate regulation of GnRH neuron excitability. Brain Res. 2010, 1364, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Spergel, D.J.; Krsmanovic, L.Z.; Stojilkovic, S.S.; Catt, K.J. Glutamate modulates Ca2+ (I) and gonadotropin-releasing-hormone secretion in immortalized hypothalamic GT1-7 neurons. Neuroendocrinology 1994, 59, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Legare, C.; Akintayo, A.; Blondin, P.; Calvo, E.; Sullivan, R. Impact of male fertility status on the transcriptome of the bovine epididymis. Mol. Hum. Reprod. 2017, 23, 355–369. [Google Scholar] [CrossRef]
- Sun, X.; Jiang, J.; Wang, G.; Zhou, P.; Li, J.; Chen, C.; Liu, L.; Li, N.; Xia, Y.; Ren, H. Genome-wide association analysis of nine reproduction and morphological traits in three goat breeds from Southern China. Anim. Biosci. 2023, 36, 191–199. [Google Scholar] [CrossRef]
- Mahmoudi, P.; Rashidi, A.; Nazari-Ghadikolaei, A.; Rostamzadeh, J.; Razmkabir, M.; Huson, H.J. Genome-wide association study reveals novel candidate genes for litter size in Markhoz goats. Front. Vet. Sci. 2022, 9, 1045589. [Google Scholar] [CrossRef]
- Jena, S.; Malik, D.S.; Kaswan, S.; Sharma, A.; Kashyap, N.; Singh, U. Relationship of udder morphometry with milk yield and body condition traits in Beetal goats. Indian J. Anim. Sci. 2019, 89, 204–208. [Google Scholar] [CrossRef]
- Erduran, H.; Dag, B. Determination of factors affecting milk yield, composition and udder morphometry of Hair and cross-bred dairy goats in a semi-intensive system. J. Dairy Res. 2021, 88, 265–269. [Google Scholar] [CrossRef]
- Hardwick, L.J.A.; Phythian, C.J.; Fowden, A.L.; Hughes, K. Size of supernumerary teats in sheep correlates with complexity of the anatomy and microenvironment. J. Anat. 2020, 236, 954–962. [Google Scholar] [CrossRef]
- Ghaffarilaleh, V.; Javanmard, A.; Saberivand, A.; Asadzadeh, N.; Masoudi, R.; Barfourooshi, H.J.; Rashidi, A.; Eghbalsaied, S. Variation and frequency of supernumerary teats, litter size, histological features and the fibroblast growth factor 2 (FGF-2) gene expression pattern in goats. Theriogenology 2022, 179, 141–148. [Google Scholar] [CrossRef]
- Martin, P.; Palhiere, I.; Tosser-Klopp, G.; Rupp, R. Heritability and genome-wide association mapping for supernumerary teats in French Alpine and Saanen dairy goats. J. Dairy Sci. 2016, 99, 8891–8900. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.X.; Xu, C.H.; Gao, T.Y.; Sun, Y. Polymorphisms of the ATP1A1 gene associated with mastitis in dairy cattle. Genet. Mol. Res. 2012, 11, 651–660. [Google Scholar] [CrossRef] [PubMed]
- Barish, S.; Barakat, T.S.; Michel, B.C.; Mashtalir, N.; Phillips, J.B.; Valencia, A.M.; Ugur, B.; Wegner, J.; Scott, T.M.; Bostwick, B.; et al. BICRA, a SWI/SNF Complex Member, Is Associated with BAF-Disorder Related Phenotypes in Humans and Model Organisms. Am. J. Hum. Genet. 2020, 107, 1096–1112. [Google Scholar] [CrossRef]
- Li, Z.; Ju, X.; Silveira, P.A.; Abadir, E.; Hsu, W.-H.; Hart, D.N.J.; Clark, G.J. CD83: Activation Marker for Antigen Presenting Cells and Its Therapeutic Potential. Front. Immunol. 2019, 10, 1312. [Google Scholar] [CrossRef] [PubMed]
- Batool, A.; Liu, H.; Liu, Y.-X.; Chen, S.-R. CD83, a Novel MAPK Signaling Pathway Interactor, Determines Ovarian Cancer Cell Fate. Cancers 2020, 12, 2269. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.K.; Lin, J.; Ye, L.L.; Zhang, Q.; Chen, J.Q.; Yang, Q.; Yu, Q.H. Modulation of Mammary Gland Development and Milk Production by Growth Hormone Expression in GH Transgenic Goats. Front. Physiol. 2016, 7, 278. [Google Scholar] [CrossRef]
- Spencer, T.E.; Johnson, G.A.; Burghardt, R.C.; Bazer, F.W. Progesterone and placental hormone actions on the uterus: Insights from domestic animals. Biol. Reprod. 2004, 71, 2–10. [Google Scholar] [CrossRef]
- Lei, J.H.; Lee, M.H.; Miao, K.; Huang, Z.B.; Yao, Z.C.; Zhang, A.P.; Xu, J.; Zhao, M.; Huang, Z.N.; Zhang, X.; et al. Activation of FGFR2 Signaling Suppresses BRCA1 and Drives Triple-Negative Mammary Tumorigenesis That is Sensitive to Immunotherapy. Adv. Sci. 2021, 8, 2100974. [Google Scholar] [CrossRef]
- Filant, J.; DeMayo, F.J.; Pru, J.K.; Lydon, J.P.; Spencer, T.E. Fibroblast Growth Factor Receptor Two (FGFR2) Regulates Uterine Epithelial Integrity and Fertility in Mice. Biol. Reprod. 2014, 90, 7. [Google Scholar] [CrossRef]
- Jiang, C.; Moorthy, B.T.; Patel, D.M.; Kumar, A.; Morgan, W.M.; Alfonso, B.; Huang, J.; Lampidis, T.J.; Isom, D.G.; Barrientos, A.; et al. Regulation of Mitochondrial Respiratory Chain Complex Levels, Organization, and Function by Arginyltransferase 1. Front. Cell Dev. Biol. 2020, 8, 603688. [Google Scholar] [CrossRef]
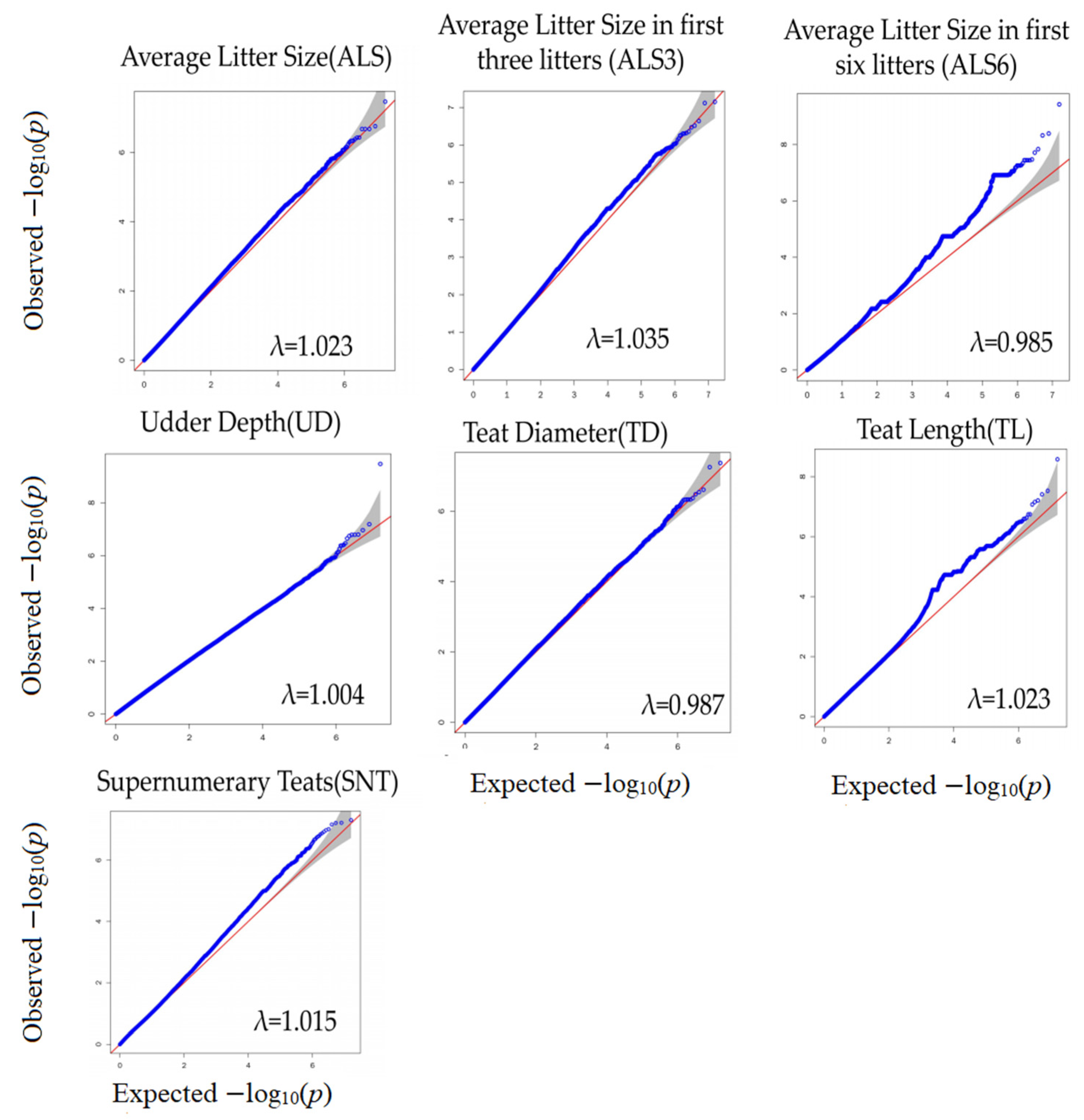

| Trait | Max | Min | Mean | Var | Std. Dev | CV |
|---|---|---|---|---|---|---|
| ALS | 3 | 1 | 1.91 | 0.17 | 0.41 | 21.4% |
| ALS3 | 3 | 1 | 1.81 | 0.26 | 0.51 | 28.2% |
| ALS6 | 2.83 | 1.17 | 1.83 | 0.13 | 0.35 | 19.3% |
| UD | 23.6 cm | 10.6 cm | 17.27 cm | 8.36 | 2.89 | 16.7% |
| TD | 26 mm | 9.5 mm | 15.6 mm | 10.87 | 3.29 | 21.1% |
| TL | 39.8 mm | 21.1 mm | 29.9 mm | 22.43 | 4.74 | 15.8% |
| SNT | 2 | 0 | 0.525 | 0.78 | 0.89 | 168.6% |
| Trait | Chr | Position | SNP | Gene | MAF | Effect | p-Value |
|---|---|---|---|---|---|---|---|
| ALS | 5 | 116147132 | rs644062850 | TBC1D22A, TAFA5 | 0.017 | 1.011881 | 3.43 × 10−8 |
| ALS6 | 17 | 28987976 | rs671071009 | NDST3, TRAM1L1 | 0.090 | 0.7715029 | 3.74 × 10−10 |
| 17 | 28985790 | rs639337328 | ENSCHIG00000026285, ENSCHIG00000009886 | 0.073 | 0.7564855 | 4.06 × 10−9 | |
| 17 | 28993318 | None | ENSCHIG00000026285, ENSCHIG00000009886 | 0.063 | 0.7777119 | 4.74 × 10−9 | |
| 27 | 39690010 | rs658833319 | ENSCHIG00000026285, ENSCHIG00000009886 | 0.087 | 0.5117511 | 1.45 × 10−8 | |
| 6 | 7982288 | rs650366329 | SLC9A8 | 0.047 | 0.5783803 | 1.93 × 10−8 | |
| 28 | 28949854 | rs642166030 | GLRB | 0.063 | 0.5344838 | 3.39 × 10−8 | |
| 17 | 28984146 | rs655588910 | GLRB | 0.077 | 0.9069677 | 3.59 × 10−8 | |
| 17 | 29975255 | rs640960816 | GLRB | 0.067 | 0.9069677 | 3.59 × 10−8 | |
| 17 | 29975258 | rs656987736 | GLRB | 0.067 | 0.9069677 | 3.59 × 10−8 | |
| 17 | 29975313 | rs663645959 | GLRB | 0.767 | 0.9069677 | 3.59 × 10−8 | |
| 13 | 77438831 | rs640662512 | GLRB | 0.120 | 0.6405018 | 4.60 × 10−8 | |
| 19 | 45379378 | rs672124332 | GRIA2, GASK1B | 0.157 | 0.4522095 | 5.28 × 10−8 | |
| 17 | 28986352 | rs672162117 | GRIA2, GASK1B | 0.037 | 0.7584963 | 5.53 × 10−8 | |
| 11 | 17646846 | rs651860215 | GRIA2, GASK1B | 0.067 | 0.889428 | 5.57 × 10−8 | |
| 11 | 17660782 | rs661293290 | ENSCHIG00000027099, ENSCHIG00000019825 | 0.080 | 0.889428 | 5.57 × 10−8 | |
| 11 | 17779280 | rs661424069 | ENSCHIG00000002625, ENSCHIG00000020975 | 0.073 | 0.889428 | 5.57 × 10−8 | |
| 17 | 29002516 | None | RHOBTB1, ENSCHIG00000019367 | 0.093 | 0.4836778 | 6.34 × 10−8 | |
| UD | 3 | 93754926 | rs639924456 | ATP1A1, ENSCHIG00000022779 | 0.493 | 2.180614 | 3.37 × 10−10 |
| TD | 18 | 62626266 | None | LRRC4C | 0.040 | 6.440032 | 4.25 × 10−8 |
| 15 | 13770235 | rs654987322 | ENSCHIG00000000547 | 0.320 | 2.478602 | 5.57 × 10−8 | |
| TL | 29 | 34789189 | None | SPCS2 | 0.043 | 7.656182 | 2.64 × 10−9 |
| 15 | 28785486 | rs635852942 | XRRA1 | 0.107 | −6.364415 | 2.96 × 10−8 | |
| 24 | 18028701 | None | ENSCHIG00000022267, CELF4 | 0.350 | 3.811532 | 3.90 × 10−8 | |
| 15 | 28830886 | rs659007606 | NTM, TMEM45B | 0.123 | −6.175688 | 6.22 × 10−8 | |
| SNT | 26 | 10333285 | rs649605136 | BICRA | 0.090 | 1.08361 | 4.91 × 10−8 |
| 18 | 55634777 | None | CD83, ENSCHIG00000008871 | 0.150 | 0.7668384 | 6.02 × 10−8 | |
| 23 | 7992077 | rs662535387 | ATE1, FGFR2 | 0.070 | 1.119172 | 6.11 × 10−8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, X.; Gu, B.; Chen, M.; Sun, R.; Zhang, J.; Zhao, L.; Zhao, Y. Genome-Wide Association Study of the Reproductive Traits of the Dazu Black Goat (Capra hircus) Using Whole-Genome Resequencing. Genes 2023, 14, 1960. https://doi.org/10.3390/genes14101960
Fang X, Gu B, Chen M, Sun R, Zhang J, Zhao L, Zhao Y. Genome-Wide Association Study of the Reproductive Traits of the Dazu Black Goat (Capra hircus) Using Whole-Genome Resequencing. Genes. 2023; 14(10):1960. https://doi.org/10.3390/genes14101960
Chicago/Turabian StyleFang, Xingqiang, Bowen Gu, Meixi Chen, Ruifan Sun, Jipan Zhang, Le Zhao, and Yongju Zhao. 2023. "Genome-Wide Association Study of the Reproductive Traits of the Dazu Black Goat (Capra hircus) Using Whole-Genome Resequencing" Genes 14, no. 10: 1960. https://doi.org/10.3390/genes14101960
APA StyleFang, X., Gu, B., Chen, M., Sun, R., Zhang, J., Zhao, L., & Zhao, Y. (2023). Genome-Wide Association Study of the Reproductive Traits of the Dazu Black Goat (Capra hircus) Using Whole-Genome Resequencing. Genes, 14(10), 1960. https://doi.org/10.3390/genes14101960







