Integrated Metabolomics and Transcriptomics Analyses Reveal the Candidate Genes Regulating the Meat Quality Change by Castration in Yudong Black Goats (Capra hircus)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Diets
2.2. Measurements, Sample Collection and Chemical Analysis
2.3. Metabolite Extraction and Metabolomic Analysis
2.4. RNA Extraction and Illumina Sequencing
2.5. Sample Relationship Analysis
2.6. RNA Extraction and Real-Time Quantitative PCR (RT-qPCR)
2.7. Joint Analysis of Transcriptomic and Metabolomic Data
2.8. Statistical Analyses
3. Results
3.1. Slaughter Performance, Carcass Traits and Meat Quality
3.2. Metabolic Profiling Analysis
3.3. Transcriptomic Analysis
3.4. Validation of mRNAs by qPCR
3.5. Transcriptomics–Metabolomics Joint Analysis on Meat Quality
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Berry, D.; Conroy, S.; Pabiou, T.; Cromie, A. Animal breeding strategies can improve meat quality attributes within entire populations. Meat Sci. 2017, 132, 6–18. [Google Scholar] [CrossRef]
- Liu, Y.; Li, R.; Ying, Y.; Zhang, Y.; Huang, Y.; Wu, H.; Lin, K. Non-genetic factors affecting the meat quality and flavor of Inner Mongolian lambs: A review. Front. Vet. Sci. 2022, 9, 1067880. [Google Scholar] [CrossRef]
- Ivanović, S.; Pavlović, M.; Pavlović, I.; Tasić, A.; Janjić, J.; Baltić, M. Influence of breed on selected quality parameters of fresh goat meat. Arch. Anim. Breed. 2020, 63, 219–229. [Google Scholar] [CrossRef]
- Bai, J.; Dong, Z.; Yang, Y.; Li, Z.; Lu, X.; Gong, H. Comparative Analysis of Meat Quality in Different Parts of Small-Tailed Han Sheep. Pak. J. Zool. 2021, 53, 1193–1195. [Google Scholar] [CrossRef]
- Sales, J. Quantification of the effects of castration on carcass and meat quality of sheep by meta-analysis. Meat Sci. 2014, 98, 858–868. [Google Scholar] [CrossRef]
- Chen, Y.-A.; Chen, J.-Y.; Chen, W.-Q.; Wang, W.-Y.; Wu, H.-H. Effects of Castration Age on the Growth Performance of Nubian Crossbred Male Goats. Animals 2022, 12, 3516. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, Y.; Shan, H.; Zheng, Y.; Xuan, Z.; Hu, J.; Wei, M.; Wang, Z.; Liu, Q.; Li, Z. Novel Insights into the Differences in Nutrition Value, Gene Regulation and Network Organization between Muscles from Pasture-Fed and Barn-Fed Goats. Foods 2022, 11, 381. [Google Scholar] [CrossRef]
- Pan, P.; Qin, Z.; Xie, W.; Chen, B.; Guan, Z.; Xie, B. Identification of Differentially Expressed Genes in the Longissimus Dorsi Muscle of Luchuan and Duroc Pigs by Transcriptome Sequencing. Genes 2023, 14, 132. [Google Scholar] [CrossRef]
- Wu, X.; Zhou, X.; Chu, M.; Guo, X.; Pei, J.; Xiong, L.; Ma, X.; Bao, P.; Liang, C.; Yan, P. Whole transcriptome analyses and comparison reveal the metabolic differences between oxidative and glycolytic skeletal muscles of yak. Meat Sci. 2022, 194, 108948. [Google Scholar] [CrossRef]
- Zhang, X.; Han, L.; Hou, S.; Raza, S.H.A.; Gui, L.; Sun, S.; Wang, Z.; Yang, B.; Yuan, Z.; Simal-Gandara, J.; et al. Metabolomics approach reveals high energy diet improves the quality and enhances the flavor of black Tibetan sheep meat by altering the composition of rumen microbiota. Front. Nutr. 2022, 9, 915558. [Google Scholar] [CrossRef]
- Liu, H.; Wei, B.; Tang, Q.; Chen, C.; Li, Y.; Yang, Q.; Wang, J.; Li, J.; Qi, J.; Xi, Y.; et al. Non-target metabolomics reveals the changes of small molecular substances in duck breast meat under different preservation time. Food Res. Int. 2022, 161, 111859. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. Feature Counts: An efficient general-purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.Y.; Dillies, M.A. SAR Tools: A DESeq2- and Edger-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007, 36, D480–D484. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Li, T.; Yan, H.; Geng, Y.; Shi, H.; Li, H.; Wang, S.; Wang, Y.; Xu, J.; Zhao, G.; Lu, X. Target genes associated with lipid and glucose metabolism in non-alcoholic fatty liver disease. Lipids Health Dis. 2019, 18, 211. [Google Scholar] [CrossRef]
- Rajkumar, V.; Verma, A.K.; Ramachandran, N.; Pal, S. Carcass traits and meat quality characteristics and fatty acid profile of Barbari goats as influenced by castration and slaughter age. Indian J. Anim. Sci. 2017, 87, 1149–1154. [Google Scholar] [CrossRef]
- Koyuncu, M.; Duru, S.; Uzun, K.; Öziş, Ş.; Tuncel, E. Effect of castration on growth and carcass traits in hair goat kids under a semi-intensive system in the south-Marmara region of Turkey. Small Rumin. Res. 2007, 72, 38–44. [Google Scholar] [CrossRef]
- Abdullah, A.Y.; Musallam, H.S. Effect of different levels of energy on carcass composition and meat quality of male black goat kids. Livest. Sci. 2007, 107, 70–80. [Google Scholar] [CrossRef]
- Fang, Y.; Vilella-Bach, M.; Bachmann, R.; Flanigan, A.; Chen, J. Phosphatidic Acid-Mediated Mitogenic Activation of mTOR Signaling. Science 2001, 294, 1942–1945. [Google Scholar] [CrossRef]
- Shi, K.; Zhao, Q.; Shao, M.; Duan, Y.; Li, D.; Lu, Y.; Tang, Y.; Feng, C. Untargeted Metabolomics Reveals the Effect of Selective Breeding on the Quality of Chicken Meat. Metabolites 2022, 12, 367. [Google Scholar] [CrossRef]
- Zhan, H.; Xiong, Y.; Wang, Z.; Dong, W.; Zhou, Q.; Xie, S.; Li, X.; Zhao, S.; Ma, Y. Integrative analysis of transcriptomic and metabolomic profiles reveal the complex molecular regulatory network of meat quality in Enshi black pigs. Meat Sci. 2022, 183, 108642. [Google Scholar] [CrossRef]
- Li, F.-L.; Fu, V.; Liu, G.; Tang, T.; Konradi, A.W.; Peng, X.; Kemper, E.; Cravatt, B.F.; Franklin, J.M.; Wu, Z.; et al. Hippo pathway regulation by phosphatidylinositol transfer protein and phosphoinositide. Nat. Chem. Biol. 2022, 18, 1076–1086. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, Y.; Wu, J.; Qiao, M.; Xu, Z.; Peng, X.; Mei, S. Proteomic and lipidomic analyses reveal saturated fatty acids, phosphatidylinositol, phosphatidylserine, and associated proteins contributing to intramuscular fat deposition. J. Proteom. 2021, 241, 104235. [Google Scholar] [CrossRef]
- Takahashi, Y. The Role of Growth Hormone and Insulin-Like Growth Factor-I in the Liver. Int. J. Mol. Sci. 2017, 18, 1447. [Google Scholar] [CrossRef]
- Zhou, G.; Wang, X.; Yuan, C.; Kang, D.; Xu, X.; Zhou, J.; Geng, R.; Yang, Y.; Yang, Z.; Chen, Y. Integrating miRNA and mRNA Expression Profiling Uncovers MiRNAs Underlying Fat Deposition in Sheep. BioMed Res. Int. 2017, 2017, 6274854. [Google Scholar] [CrossRef]
- Indurain, G.; Beriain, M.; Sarries, M.; Insausti, K. Effect of weight at slaughter and breed on beef intramuscular lipid classes and fatty acid profile. Animal 2010, 4, 1771–1780. [Google Scholar] [CrossRef]
- van Diepen, J.A.; Jansen, P.A.; Ballak, D.B.; Hijmans, A.; Hooiveld, G.J.; Rommelaere, S.; Galland, F.; Naquet, P.; Rutjes, F.P.; Mensink, R.P.; et al. PPAR-alpha dependent regulation of vanin-1 mediates hepatic lipid metabolism. J. Hepatol. 2014, 61, 366–372. [Google Scholar] [CrossRef]
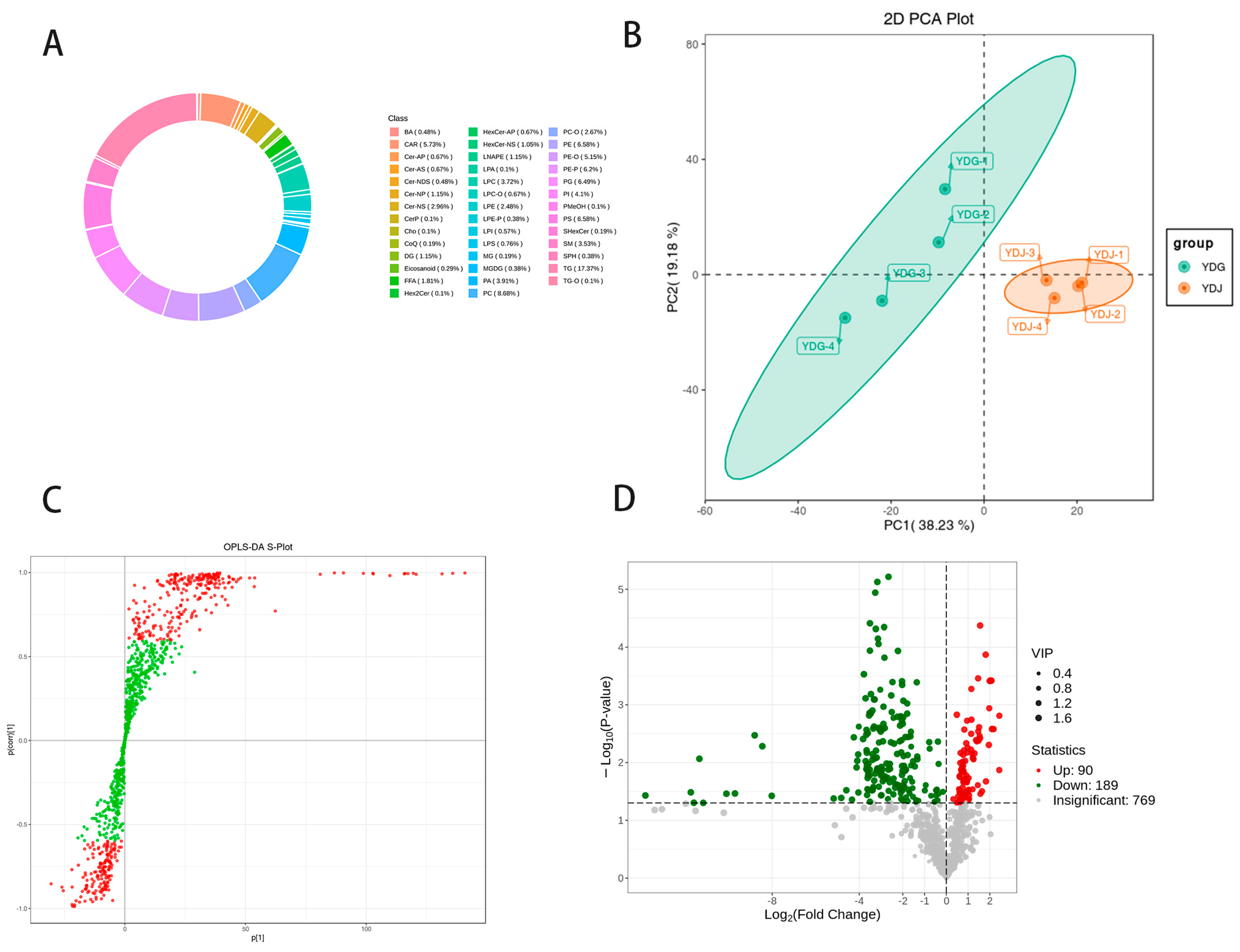

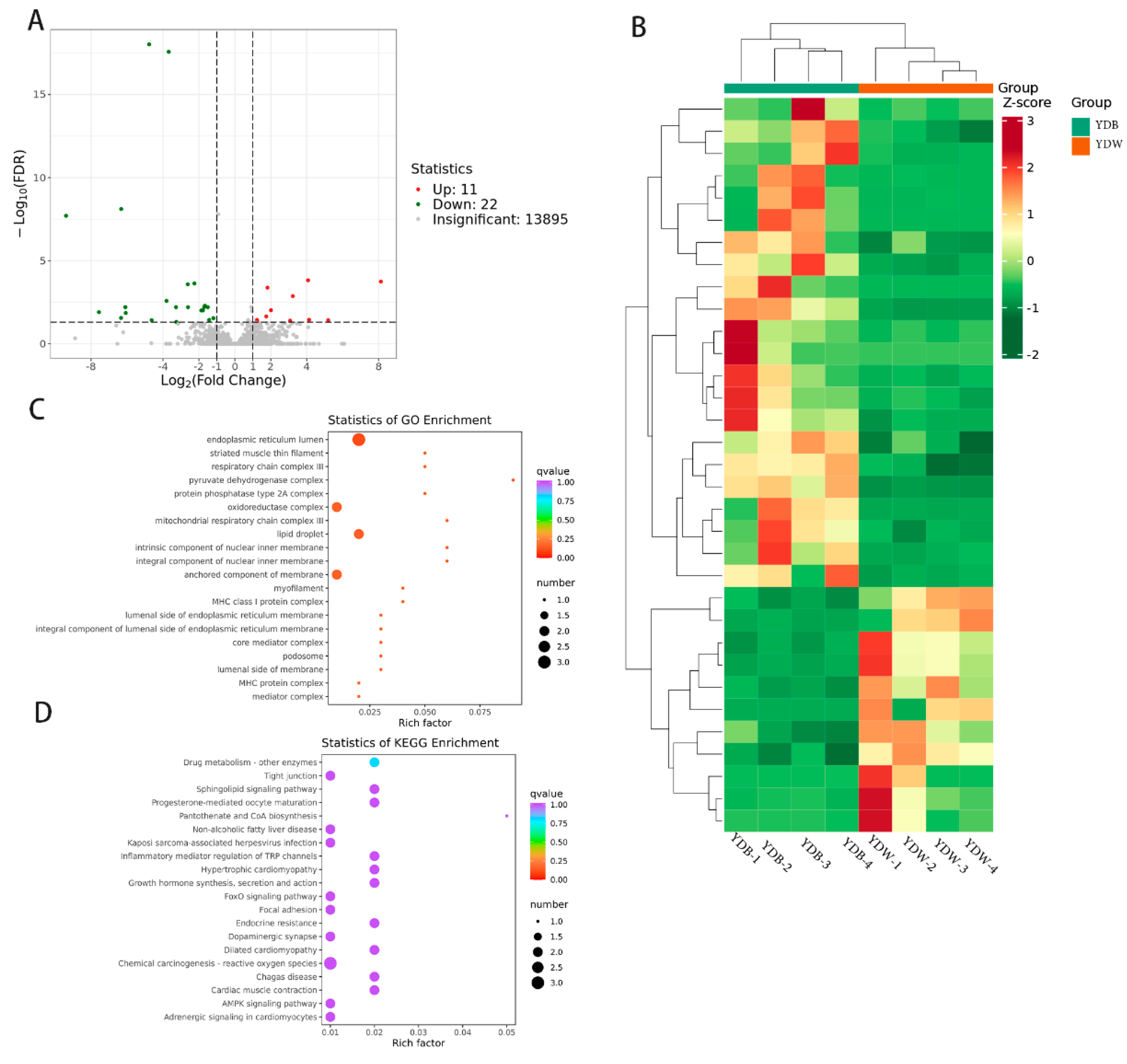
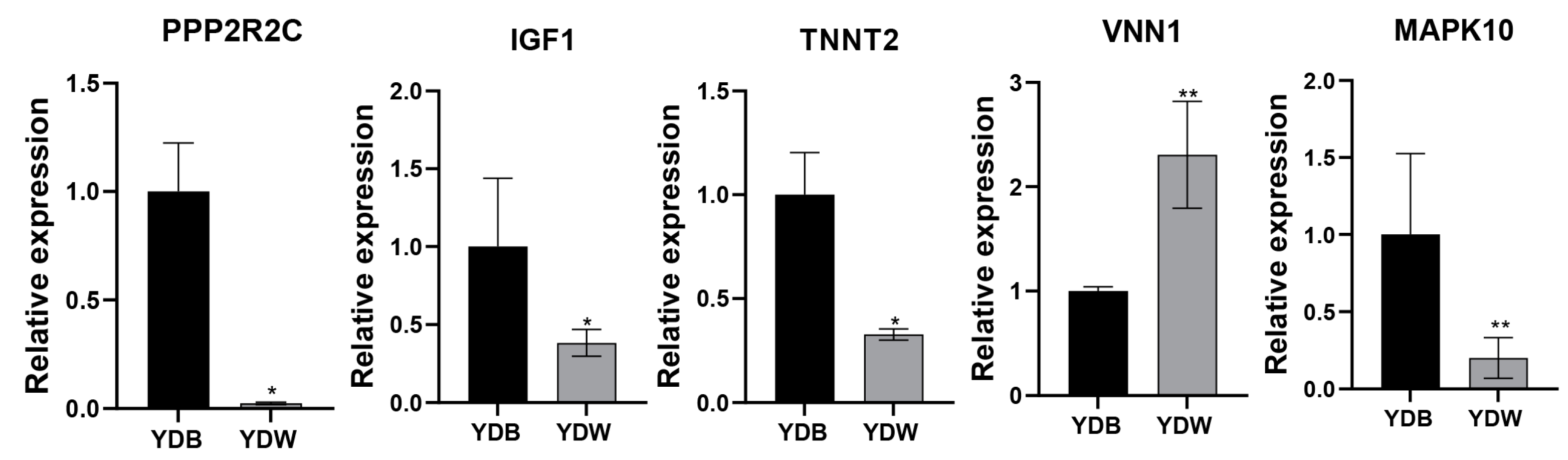
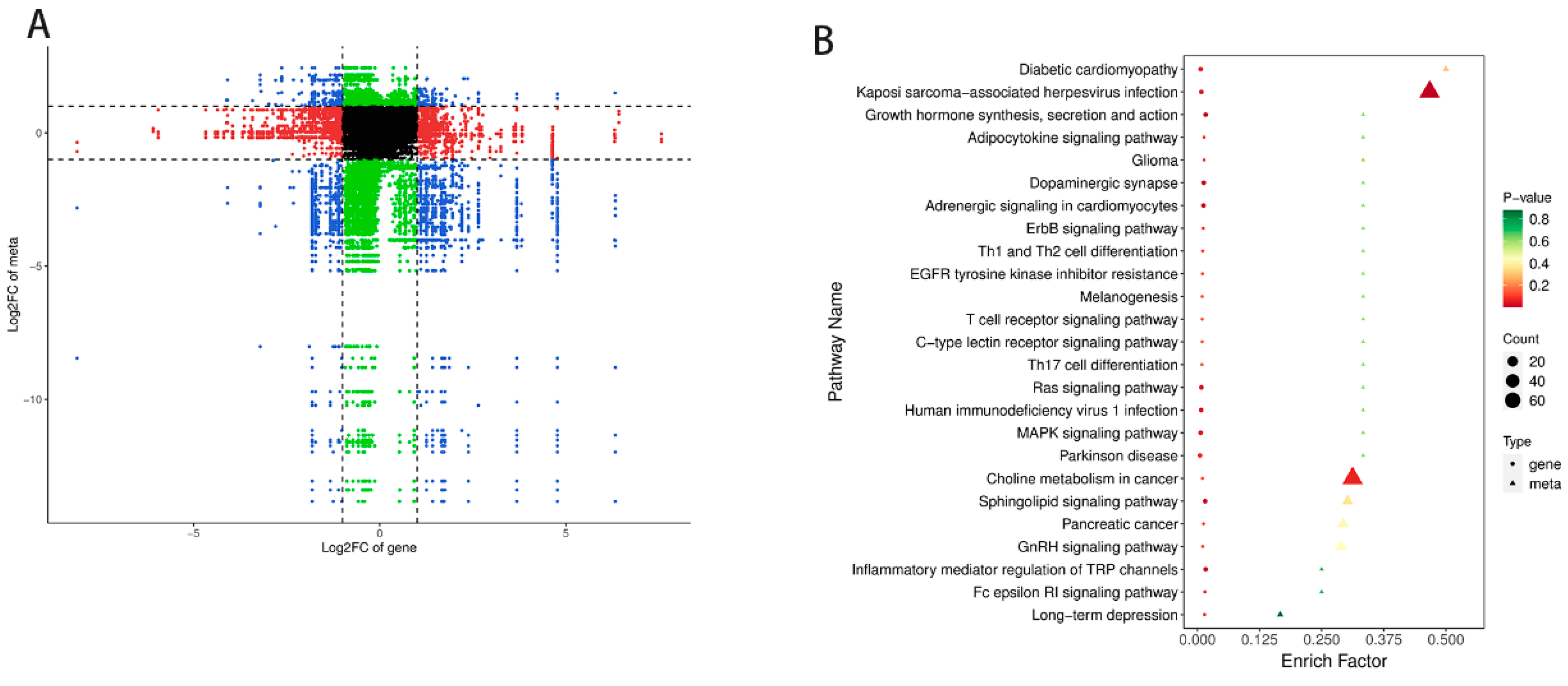
| Items | YDW | YDB |
|---|---|---|
| SLW (kg) | 38.45 ± 5.16 | 42.63 ± 10.61 |
| Carcass weight (kg) | 18.43 ± 2.01 | 22.00 ± 5.86 |
| Dressing percentage (%) | 48.10 ± 2.36 | 51.45 ± 2.29 |
| Meat weight (kg) | 14.15 ± 2.02 | 17.20 ± 6.08 |
| Meat yield (%) | 36.78 ± 1.14 | 39.58 ± 4.32 |
| LEA (cm2) | 13.95 ± 1.61 | 15.45 ± 1.76 |
| GR (mm) | 5.25 ± 0.81 | 6.23 ± 1.74 |
| Backfat thickness (mm) | 0.10 ± 0.03 | 0.10 ± 0.02 |
| Items | YDW | YDB |
|---|---|---|
| Asp (%) | 2.18 ± 0.14 | 2.29 ± 0.24 |
| Thr (%) | 0.85 ± 0.06 | 0.90 ± 0.06 |
| Ser (%) | 0.78 ± 0.06 | 0.82 ± 0.06 |
| Glu (%) | 3.37 ± 0.23 | 3.48 ± 0.28 |
| Gly (%) | 0.92 ± 0.09 | 0.93 ± 0.05 |
| Ala (%) | 1.29 ± 0.09 | 1.32 ± 0.07 |
| Val (%) | 1.09 ± 0.07 | 1.14 ± 0.09 |
| Met (%) | 0.62 ± 0.06 | 0.67 ± 0.09 |
| Lle (%) | 1.02 ± 0.07 | 1.10 ± 0.11 |
| Leu (%) | 1.89 ± 0.15 | 1.99 ± 0.24 |
| Tyr (%) | 0.76 ± 0.05 | 0.81 ± 0.08 |
| Phe (%) | 0.84 ± 0.03 | 0.89 ± 0.11 |
| His (%) | 0.76 ± 0.15 | 0.85 ± 0.04 |
| Lys (%) | 1.82 ± 0.14 | 1.93 ± 0.13 |
| Arg (%) | 1.33 ± 0.07 | 1.39 ± 0.14 |
| Pro (%) | 0.50 ± 0.04 | 0.47 ± 0.09 |
| Items | YDW | YDB |
|---|---|---|
| L* | 26.89 ± 5.00 | 29.51 ± 8.97 |
| a* | 17.28 ± 2.63 | 8.80 ± 2.92 * |
| b* | 9.67 ± 0.86 | 8.54 ± 2.59 |
| PH | 6.77 ± 0.35 | 6.23 ± 0.35 |
| Cooking loss (%) | 1.14 ± 0.15 | 2.09 ± 0.58 * |
| Shear force (N) | 91.33 ± 12.75 | 85.89 ± 13.99 |
| IMF (%) | 2.36 ± 0.82 | 1.47 ± 0.54 |
| Water (g/100 g) | 68.45 ± 2.26 | 71.95 ± 2.90 |
| Fatty Acids (%) | YDW | YDB |
|---|---|---|
| Myristic acid | 2.00 ± 0.09 | 1.98 ± 0.33 |
| Pentadecanoic acid | 0.66 ± 0.10 | 0.15 ± 0.30 * |
| Palmitic acid | 23.90 ± 0.78 | 24.53 ± 0.97 |
| Heptadecanoic acid | 1.67 ± 0.27 | 0.63 ± 0.74 |
| Stearic acid | 25.13 ± 3.62 | 23.78 ± 4.63 |
| Trans-9-octadecanecarboxylic acid | 2.38 ± 0.21 | 3.64 ± 0.26 * |
| Cis-9-octadecenoic acid | 39.48 ± 3.25 | 39.00 ± 3.90 |
| Linoleic acid | 2.07 ± 0.14 | 4.52 ± 1.24 * |
| Cis-11-eicosenoic acid | 0.95 ± 0.16 | ND |
| Saturated fatty acid | 53.35 ± 3.16 | 51.05 ± 4.55 |
| Monounsaturated fatty acid | 43.73 ± 3.30 | 43.20 ± 4.31 |
| Polyunsaturated fatty acid | 2.89 ± 0.45 | 5.54 ± 1.94 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, S.; Zhang, X.; Li, X.; Zheng, J.; Zhao, L.; Fan, C.; Zhao, Y. Integrated Metabolomics and Transcriptomics Analyses Reveal the Candidate Genes Regulating the Meat Quality Change by Castration in Yudong Black Goats (Capra hircus). Genes 2024, 15, 43. https://doi.org/10.3390/genes15010043
Yang S, Zhang X, Li X, Zheng J, Zhao L, Fan C, Zhao Y. Integrated Metabolomics and Transcriptomics Analyses Reveal the Candidate Genes Regulating the Meat Quality Change by Castration in Yudong Black Goats (Capra hircus). Genes. 2024; 15(1):43. https://doi.org/10.3390/genes15010043
Chicago/Turabian StyleYang, Songjian, Xinyue Zhang, Xingchun Li, Jikang Zheng, Le Zhao, Chengli Fan, and Yongju Zhao. 2024. "Integrated Metabolomics and Transcriptomics Analyses Reveal the Candidate Genes Regulating the Meat Quality Change by Castration in Yudong Black Goats (Capra hircus)" Genes 15, no. 1: 43. https://doi.org/10.3390/genes15010043
APA StyleYang, S., Zhang, X., Li, X., Zheng, J., Zhao, L., Fan, C., & Zhao, Y. (2024). Integrated Metabolomics and Transcriptomics Analyses Reveal the Candidate Genes Regulating the Meat Quality Change by Castration in Yudong Black Goats (Capra hircus). Genes, 15(1), 43. https://doi.org/10.3390/genes15010043





