Abstract
The production and quality of apricots in China is currently limited by the availability of germplasm resource characterizations, including identification at the species and cultivar level. To help address this issue, the complete chloroplast genomes of Prunus armeniaca L., P. sibirica L. and kernel consumption apricot were sequenced, characterized, and phylogenetically analyzed. The three chloroplast (cp) genomes ranged from 157,951 to 158,224 bp, and 131 genes were identified, including 86 protein-coding genes, 37 rRNAs, and 8 tRNAs. The GC content ranged from 36.70% to 36.75%. Of the 170 repetitive sequences detected, 42 were shared by all three species, and 53–57 simple sequence repeats were detected with AT base preferences. Comparative genomic analysis revealed high similarity in overall structure and gene content as well as seven variation hotspot regions, including psbA-trnK-UUU, rpoC1-rpoB, rpl32-trnL-UAG, trnK-rps16, ndhG-ndhI, ccsA-ndhD, and ndhF-trnL. Phylogenetic analysis showed that the three apricot species clustered into one group, and the genetic relationship between P. armeniaca and kernel consumption apricot was the closest. The results of this study provide a theoretical basis for further research on the genetic diversity of apricots and the development and utilization of molecular markers for the genetic engineering and breeding of apricots.
1. Introduction
Apricot is a deciduous tree species, which belongs to section Armeniaca (Lam.) Koch genus Prunus of Rosaceae family (2n = 16) [1,2]. Ten species of apricot have been identified, and they are widely distributed worldwide, including P. armeniaca, P. sibirica, P. mandshurica, P. zhengheensis, P. dasycarpa, P. holosericea, P. zhidanensis, P. mume, P. limeixing, and P. byigantina. Four distinct species are most commonly recognized: P. armeniaca, P. mandshurica, P. sibirica, and P. mume [3]. Apricot is of Chinese origin and has been cultivated in China for more than 3000 years [4]. This study focused on P. armeniaca,P. sibirica, and kernel consumption apricots which are widely distributed in northern China [5]. The fruits of P. armeniaca are fresh with unique aroma, delicious taste, but also contains a variety of organic components, vitamins and inorganic salts, high nutritional value, wide range of uses, and can be processed into dried apricots, cultivated around the world, accounting for a high proportion of global fruit production [6,7]. P. sibirica has high ecological value as a pioneer tree species for vegetation restoration because it is cold, drought, and poor-soil tolerant. In addition, the bitter kernel of P. sibirica has an amygdalin content of 3.5–7.6%, and it is also rich in vitamins, selenium, calcium, phosphorus, iron, potassium and other nutrients. Thus, this kernel is a raw material in traditional Chinese medicine [8]. The kernel consumption apricots have typical characteristic of large and sweet kernels. And the kernels have a crude fat content of ~60% and thus can be used to produce kernel oil, and they have a protein content of ~30%, including eight kinds of essential amino acids for the human body. Thus, kernels are a high-quality plant protein raw material [9].
Chloroplasts are unique endosymbiotic organelles found in plants and photosynthetic algae [10], serving as the primary site of photosynthesis and supplying energy for plant growth and development and carbon intermediates for a number of critical metabolic reactions. In addition, chloroplasts play an important role in plant response to light, heat, drought, salt, and other stresses [11]. The chloroplast (cp) genome is maternally inherited in most angiosperms or paternally inherited in some gymnosperms. The sequence analysis of the double-stranded circular DNA cp genome is important in various areas of study, including the development of linked molecular markers, the reconstruction of phylogenetic relationships, and the genetic engineering and breeding of plants [12]. The cp genome exhibits a highly conserved organization composed of a pair of inverted repeats (IRs), a large single-copy (LSC) region, and a small single-copy (SSC) region [13,14]. The two IR regions, which are separated by the LSC and SSC regions, are equal in length and opposite in direction; however, variations have been observed in some plants, mainly presented as IR loss, contraction, expansion, and sequence direction changes [15]. Early research studies on the cp genome focused mainly on understanding the evolutionary history of chloroplasts and safeguarding uncommon and endangered plants [16]. With improvements in sequencing technology, the complete cp genome of Nicotiana tabacum was obtained for the first time in 1986 [17]. More recently, the cp genomes of Prunus cerasus (sour cherry) [18], P. phaeosticta (dark-spotted cherry) [19], P. kansuensis (Chinese bush peach) [20], and P. japonica (Japanese bush cherry) [21] were sequenced and analyzed, and their phylogenetic positions and genetic relationships were determined.
Morphological characteristic analyses can preliminarily reveal the morphological characteristics and genetic variations of plants [22]. However, the morphological characteristics of apricot are influenced by the environment and gene dominance, and the period required to obtain morphological characteristics is long [23]. With the rapid development of next-generation sequencing technology and phylogenetic genomics, cp genome sequencing has been widely used in molecular evolution and phylogenetic studies of many plant species [24]. More accurate classifications and phylogenetic relationships of apricot can be obtained through the combination of cp genome sequencing and phylogenetic genomics.
In this study, P. armeniaca, P. sibirica, and kernel consumption apricots were used as the research objects to obtain the cp genome sequences, and then, the sequences were spliced, annotated, and compared. A phylogenetic analysis was performed, and the evolutionary relationship between P. armeniaca, P. sibirica, and kernel consumption apricot was systematically studied at the cp genome sequence level. The results provide a reference for future taxonomic and phylogenetic analyses and molecular marker development of apricot and a molecular guide for genetic engineering and breeding.
2. Materials and Methods
2.1. Sample Material Collection, DNA Extraction, and Sequencing
Fresh tender leaves were collected from the cultivated variety P. armeniaca in Mentougou, Beijing (Sungold), wild resource of P. sibirica in Wanjiagou (F106), Inner Mongolia, and the cultivated variety of kernel consumption apricot growing in Wei County, Hebei (Youyi). The samples were stored at −80 °C.
Total genomic DNA was extracted using a Plant Genomic DNA Kit (Tiangen, Beijing, China). DNA quality and quantity were detected using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and 0.8% agarose gel electrophoresis, and the DNA was further fragmented for sequence library preparation following fragment purification and end repair. After testing the library preparation, DNA sequencing was performed using the Illumina HiSeq X high-throughput platform (Illumina, San Diego, CA, USA). Library preparation and sequencing were performed by BGI Genomics (Shenzhen, China).
2.2. Chloroplast Genome Assembly and Annotation
Using SOAPdenovo (http://soap.genomics.org.cn/soapdenovo.html, accessed on 5 September 2016), the reads were mapped to the cp genome of P. persica, which was downloaded from GenBank (NC_014697.1). Contigs obtained by de novo assembly mapping to the consensus sequence were obtained using the reference genome to check the errors or ambiguities resulting from either assembly method. Gapcloser (https://sourceforge.net/projects/soapdenovo2/files/GapCloser/, accessed on 7 September 2016) was used to modify the spaces between long contigs to obtain a complete cp genome. The three apricot cp genome sequences were preliminarily annotated using DOGMA and CpGAVAS (http://phylocluster, accessed on 11 September 2016), and the annotation was completed by manually modifying the start and stop codons of individual genes. Geneious was used for manual corrections. Finally, the annotated cp genomes of the three apricot species were submitted to GenomeVx (http://wolfe.gen.tcd.ie/GenomeVx/, accessed on 10 March 2023) to complete the physical mapping.
2.3. Repeat Sequences and Simple Sequence Repeat (SSR) Analysis
REPuter (http://bibiserv.techfak.uni-biele.org.de/reputer/, accessed on 15 March 2023) was used to predict the scattered repetitive sequences of the cp genomes, including forward (F), reverse (R), complementary (C), and palindromic (P) repeats. The conditions for repeat sequences included a sequence length ≥18 bp and similarity ≥ 90%. Simple sequence repeat loci in the apricot cp genome were detected using MISA (https://webblast.ipk-gatersleben.de/misa/index.php, accessed on 20 March 2023) with the following parameters: ≥6 mononucleotide repeats, ≥4 dinucleotide and trinucleotide repeats, and ≥3 tetranucleotide, pentanucleotide, and hexanucleotide repeats.
2.4. Chloroplast Genome Comparison and Analysis of Variations in IR/SC Boundaries
mVISTA (http://genome.lbl.gov/vista/index.shtml, accessed on 25 March 2023) was used to compare similarities and variations in the cp genomes among P. armeniaca, P. sibirica, kernel consumption apricot, P. mume, P. pyrifolia, and N. tabacum, with the P. persica cp genome sequence serving as the reference sequence. Based on the annotation information of the cp genome, the LSC, SSC, and IR boundary sequences in the apricot cp genomes were compared with those in the P. mume, P. persica, P. pyrifolia, and N. tabacum cp genomes. The IR-SC boundary of the cp genome was visualized using IRscope.
2.5. Phylogenetic Analysis
The cp genome sequences of 27 angiosperms and 2 gymnosperms were selected from the National Center for Biotechnology Information (NCBI) database for phylogenetic analysis (Supplemental Table S1). The phylogenetic tree was constructed by extracting 77 common protein-coding gene sequences from the 29 species and using Pinus thunbergia (NC_001631) and Ginkgo biloba (NC_016986) as outgroups. The 77 common protein coding genes were atpA, ndhA, atpB, atpE, ndhC, atpF, atpH, ndhD, atpI, ccsA, ndhE, cemA, clpP, ndhF, infA, matK, ndhK, petA, petB, ndhG, petD, petG, ndhH, petL, petN, ndhI, psaA, psaB, ndhJ, psaC, psaI, psbA, psaJ, rpl22, psbB, rpl23, rps16, psbC, rbcL, rpl14, psbD, rpl16, rpl2, psbE, rpl20, rpl32, psbF, rpl33, rpl36, psbH, rpoA, rpoB, psbI, rpoC1, rpoC2, psbJ, rps11, rps12, psbK, rps14, rps15, psbL, rps18, rps19, psbM, rps2, rps3, psbN, rps4, rps7, psbT, rps8, ycf3, psbZ, ycf2, and ycf4. MEGA was used for protein sequence alignment, and the maximum likelihood (ML) method of analysis was used to construct a phylogenetic tree. Visualization of the system tree was completed using Figtreev1.4.4.
3. Results
3.1. Organization and Features of the Chloroplast Genomes
The total cp genome lengths of P. armeniaca, P. sibirica, and kernel consumption apricot were 157,951, 158,224, and 157,994 bp, respectively. The cp genomes showed a typical tetrad structure, including two IRs (26,373 bp) that were separated by LSC (86,217–86,358 bp) and SSC (18,988–19,120 bp) regions. Of the total cp genome, the protein-coding region accounted for (49.63–49.72%); tRNA and rRNA accounted for (1.77–1.78%), and (5.72–5.73%), respectively; meanwhile, introns and intergenic spacers (IGSs) accounted for (11.40–11.41%) and (31.34–31.47%), respectively. The GC content of the cp genomes of P. armeniaca, P. sibirica, and kernel consumption apricot was 36.74%, 36.70%, and 36.75%, respectively. In addition, the protein-coding region accounted for 37.63–37.64% of the genome; tRNA and rRNA accounted for 53.25–53.35% and 55.50%, respectively; meanwhile, introns and IGSs accounted for 36.61–36.68%, and 30.65–30.83%, respectively. The GC content of the IR region (42.57–42.59%) was higher than that of the LSC region (34.52–34.56%) and SSC region (30.30–30.50%), suggesting the higher stability of the IR region than the LSC and SSC regions (Table 1).

Table 1.
Chloroplast genome characteristics of three apricot species.
The cp genomes of the three apricot species were assembled and annotated (Figure 1). Functional analyses classified them into four categories: self-replication-related, photosynthesis-related, other, and unknown functions (Table 2). A total of 131 genes were predicted and annotated, including 86 protein-coding genes, 37 tRNAs, and 8 rRNAs (Table 2). Among these, 18 genes (atpF, clpP, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps12, rps16, trnA-UGC, trnG-GCC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC, and ycf3) contained one intron, and two genes (clpP and ycf3) contained two introns (Supplemental Table S2).
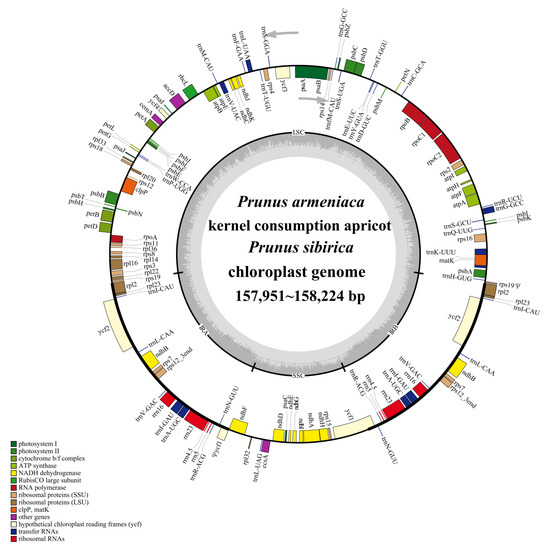
Figure 1.
Combined gene map of the chloroplast genome of the three apricot species.

Table 2.
Genes present in the chloroplast genomes of the three apricot species.
3.2. Characterization of Repeat Sequences and SSRs
There were 170 repeat sequences in the combined apricot cp genomes, including 57 forward repeats (F), 53 palindromic repeats (P), 56 reverse repeats (R), and 4 C complementary repeats (C). Among the apricots, P. sibirica had the most repeat sequences (63), followed by kernel consumption apricot (55) and P. armeniaca (52) (Figure 2a). Repeat sequence lengths of 18–20 bp (64.71%) and 21–25 bp (25.88%) dominated (Figure 2b). Of the 170 repeats identified, 42 were shared by the three species, while there were 13, 10, and 21 unique repeat sequences obtained for P. armeniaca, P. sibirica, and kernel consumption apricot, respectively (Figure 2c and Supplemental Table S3).
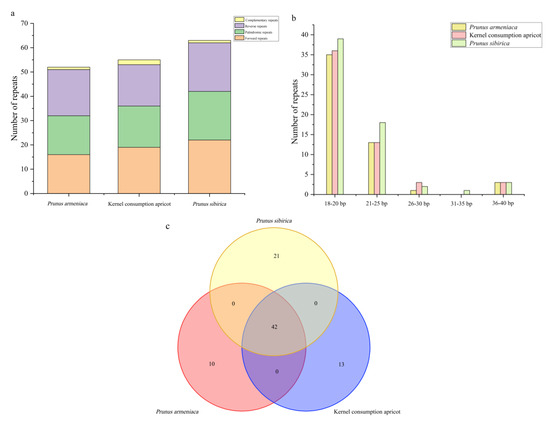
Figure 2.
Analysis of repeat sequences in the chloroplast genomes of the three apricot species. (a) Frequency of the repeat type. (b) Frequency of repeat sequences by length. (c) Number of common and unique chloroplast genome repeat sequences.
The three apricot cp genomes contained three forms of SSRs: mononucleotide, dinucleotide, and compound (Figure 3a). There were 53 SSRs in P. sibirica, 56 in P. armeniaca, and 57 in kernel consumption apricot. Mononucleotide repeats (ranging from 84.91% in P. sibirica to 94.64% in P. armeniaca) occurred most frequently, followed by dinucleotide (ranging from 3.57% in P. armeniaca to 10.53% in kernel consumption apricot) and compound SSRs (ranging from 1.76% in P. armeniaca to 3.77% in P. sibirica). The number of A/T mononucleotide repeats (ranging from 78.95% in kernel consumption apricot to 81.13% in P. sibirica) was greater than that of C/G repeats (ranging from 8.77% in kernel consumption apricot to 9.43% in P. sibirica). The quantity of dinucleotide repeats, including AT/TA repeats, ranged from 5.66% in P. sibirica to 8.93% in P. armeniaca (Supplemental Table S4). We further analyzed SSR distribution and found that most were distributed in the LSC region (83.93–85.96%); far fewer were in the SSC (10.53–12.50%) and IR (3.51–3.77%) regions (Figure 3b and Supplemental Table S4).
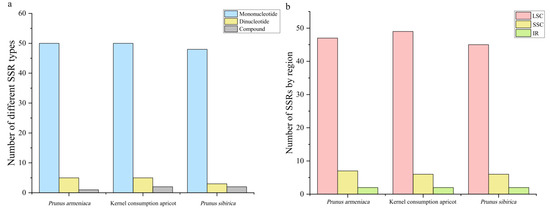
Figure 3.
Frequency of SSRs in three apricot species. (a) Number of SSRs by type. (b) Number of SSRs by genome region.
3.3. Comparative Analysis of Apricot Chloroplast Genomes
A comparison of the three apricot cp genomes indicated that the coding region is relatively conserved, with variations mainly occurring in the intergenic and intron regions. Intergenic spacer regions involving the psbA-trnK-UUU, rpoC1-rpoB, rpl32-trnL-UAG, trnK-rps16, ndhG-ndhI, ccsA-ndhD, and ndhF-trnL genes are hotspots for apricot cp genome variation (Figure 4). These hotspots can provide vital sequence information for the design of screening DNA barcodes and phylogenetic analyses of apricot species.
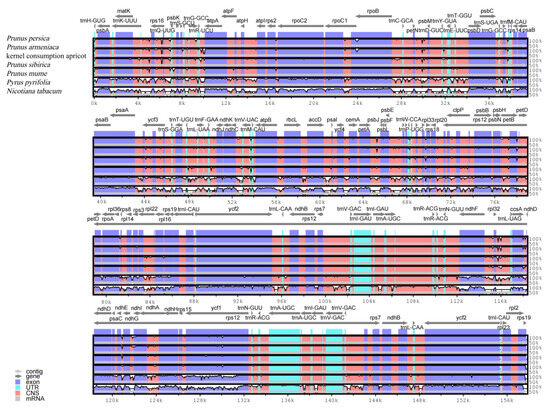
Figure 4.
Comparison of the chloroplast genome sequences of P. armeniaca, kernel consumption apricot, P. sibirica, P. mume, P. pyrifolia, and N. tabacum.
3.4. Analysis of Variations in the IR/SC Boundaries
We compared the cp genome IR regions of P. armeniaca, P. sibirica, and kernel consumption apricot with those of P. mume, P. persica, P. pyrifolia, and N. tabacum (Figure 5). The rps19 gene was detected at the IRb/LSC boundary in P. armeniaca, P. sibirica, kernel consumption apricot, P. mume, P. persica, and P. pyrifolia. The fragment size in the IRb region was 120–197 bp. In contrast, in N. tabacum, there was no pseudogene of rps19 in the IRb/LSC boundary region in N. tabacum. The ycf1 gene was found in the IRa/SSC boundary of P. armeniaca, P. sibirica, kernel consumption apricot, P. mume, P. persica, P. pyrifolia, and N. tabacum. The fragment size of Ψycf1 in the IRa region was 996–1073 bp. The Ψycf1 pseudogene in the IRb/SSC region of P. armeniaca, P. sibirica, kernel consumption apricot, P. mume, P. persica, and P. pyrifolia exhibited different lengths of overlap with that of ndhF, whereas ycf1 of N. tabacum did not overlap with that of ndhF.
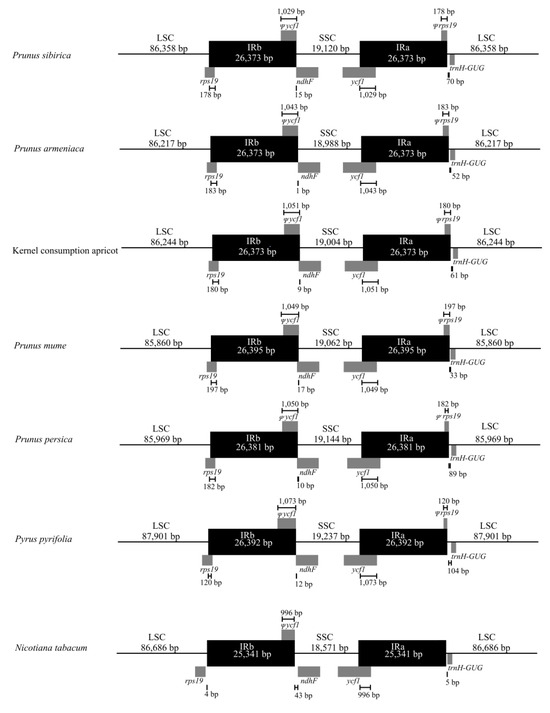
Figure 5.
Comparison of the IR-SC region of the chloroplast genome in P. sibirica, P. armeniaca, kernel consumption apricot, P. mume, P. persica, P. pyrifolia, and N. tabacum.
3.5. Phylogenetic Analysis
According to the constructed phylogenetic tree, the support rate for each branch was high (>80%), while the support rate for 21 of the 27 nodes was >90% (Figure 6). The 29 species analyzed were divided into seven groups: EUROSIDS I, EUROSIDS II, EUASTERIDS II, EUASTERIDS I, basal angiosperms, monocots, and gymnosperms. Rosales, Cucurbitales, Fabales, and Malpighiales were clustered together to form EUROSIDS I, and other plants were clustered together in turn. Gymnosperms and basal angiosperms were obviously clustered into one branch. In the phylogenetic tree, P. armeniaca, P. sibirica, and kernel consumption apricot clustered in the Rosales order within EUROSIDS I. The analysis shows that the relationship between P. armeniaca and kernel consumption apricot is the closest.
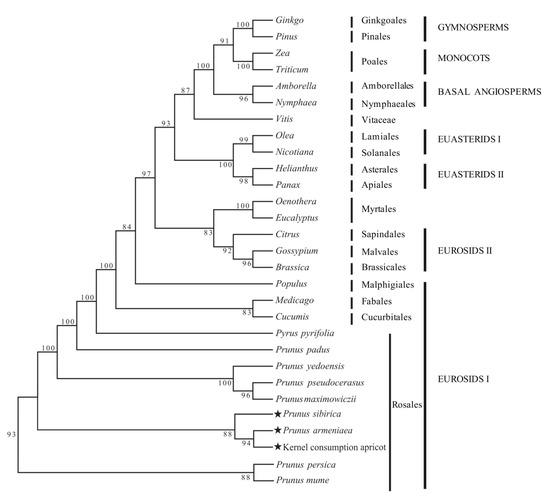
Figure 6.
Phylogenetic relationship of the three apricot species reconstructed using the maximum likelihood (ML) method. ★ The star is marked by the study of three apricots.
4. Discussion
In this study, the cp genomes of three apricot species were successfully sequenced, assembled, analyzed, and compared. The structural characteristics of the three cp genomes were similar, which exhibited typical tetrad structures [25,26]. The total length of the cp genomes ranged from 157,951 to 158,224 bp. The number of genes was consistent, with 131 genes each, including 86 protein-coding genes, 37 tRNAs, and 8 rRNAs, which is consistent with the previously reported cp genomes of P. mume [27], P. persica [26], and P. pyrifolia [28]. The GC content of the three apricot cp genomes was also similar (36.70–36.75%), and the GC content in the IR region was the highest (42.57–42.59%) owing to the GC-rich rRNA and tRNA in the IR region.
Repeat sequences play an important evolutionary role in the cp genome by promoting cp genome rearrangement, inducing genomic structural changes, and increasing population genetic diversity [29,30]. There were 170 repeats in the cp genome, among which, F, R, and C repeats were the main repetitive sequences (97.65%). Different abundances of palindromic repeats in the cp genomes may provide additional evolutionary information, as the presence and abundance of repeats in the cp genome may contain phylogenetic signals [31]. The total number and proportion of repeat types in the three apricot species showed a similar pattern, suggesting a similar evolutionary history and closer affinities among these species. SSRs related to genome rearrangement and recombination are widely distributed in the cp genome, and they are prone to dislocation during DNA replication, which leads to rich polymorphisms that provide information for marker development in population genetics and evolutionary research [24]. SSRs of plant chloroplast genes are mainly mononucleotide and dinucleotide repeats, and trinucleotide to hexanucleotide repeats are relatively less than mononucleotide and dinucleotide repeats [32]. The SSRs found in the three apricot species were dominated by mononucleotide repeats, especially poly (A/T), which is similar to that of other Rosaceae species [33]. The repeat sequences and SSRs detected in this study can provide useful information for future research on the evolution of apricot species.
Previous studies have shown that changes in genome size mainly occur in the SSC and LSC regions and are highly conserved in the IR regions [34,35]. Variation in the non-coding region was significantly higher than that in the coding region owing to the large selection pressure [36]. In particular, the intergenic regions, including psbA-trnK-UUU, rpoC1-rpoB, rpl32-trnL-UAG, trnK-rps16, ndhG-ndhI, ccsA-ndhD, and ndhF-trnL, are highly variable regions, which can be used as DNA barcodes for future phylogenetic analyses of apricots.
Contraction and expansion are the main causes of cp genome evolution, mainly occurring at the rps19, ycf1, trnH-GUG, and ndhF positions [37,38]. Although the gene distribution of the four main regional boundaries in the three apricot cp genomes showed the same pattern, there were differences in the microstructure, especially the location of rps19, ycf1, ndhF, and trnH-GUG. The rps19 crosses the boundary between the LSC region and the IR region, which is similar to previous observations in P. mume, P. armeniaca, and P. salicina [39]. The differences in the lengths of these four genes in the IR/SC boundary region can be used to identify P. armeniaca, P. sibirica, and kernel consumption apricot.
The cp genome of angiosperms is maternally inherited and an independent evolutionary system with a moderate rate and can be used for phylogenetic analyses of each classification level [40]. Especially in the study of the phylogenetic relationship between angiosperms and some controversial species, cp genome analysis is the preferred research method [41]. The results of the phylogenetic analysis showed that the 29 species included in the phylogenetic tree could be divided into 7 groups: EUROSIDS I, EUROSIDS II, EUASTERIDS II, EUASTERIDS I, basal angiosperms, monocots, and gymnosperms. The support rate for each branch was high (>80%), in which Rosales, Cucurbitales, Fabales, and Malpighiales were clustered together to form EUROSIDS I, which is consistent with the results of APG III [42]. Phylogenetic tree analysis in our study showed that P. armeniaca, P. sibirica, and kernel consumption apricot all clustered together, which was consistent with the results of traditional morphological analysis [43] and genetic diversity analysis [44]. This study has certain limitations, and the taxonomic status of apricot needs to be further analyzed to more clearly reveal the taxonomic status and origin of apricot.
5. Conclusions
In this study, we sequenced and analyzed the complete cp genome of three apricots. The results showed that the cp genomes showed a typical tetrad structure. Comparative analysis of the cp genomes revealed that the organization and gene order were highly conserved. The cp genome size, GC content, gene number, and gene arrangement order were similar in the three apricot species. We detected abundant long-repeat sequences and SSR loci in the three apricot species. The IR/SC boundary regions were similar but also exhibited some microstructural differences among the three species. Seven significant differences were identified in the non-coding regions of the three cp genomes, which can be exploited in the DNA barcoding of apricot. Finally, phylogenetic tree analysis supported a close relationship among the three apricot species. The results are of great significance for studies on the internal structure of the cp genome of apricots and the breeding, environmental adaptation, and hybrid breeding of apricots.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes14101959/s1, Supplemental Table S1: Species included in the phylogenetic analysis; Supplemental Table S2: Intron lengths in the chloroplast genomes of three apricots; Supplemental Table S3: Types of long repetitive sequences in three apricots; Supplemental Table S4: SSR information for three apricots.
Author Contributions
Conceptualization, T.-n.W., L.W. and W.B.; methodology, R.Y.; software, W.B.; validation, R.Y., W.B. and Y.-e.B.; formal analysis, D.A.; investigation, Y.-e.B.; resources, T.-n.W.; data curation, D.A.; writing—original draft preparation, R.Y. and W.B.; writing—review and editing, R.Y., W.B. and L.W.; visualization, Y.-e.B.; supervision, D.A.; project administration, W.B and T.-n.W.; funding acquisition, W.B. and T.-n.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Inner Mongolia Agricultural University Young Teachers Scientific Research Ability Promotion Project, grant number BR220114; the National Natural Science Foundation of CHINA, grant number 32360400; and the Program of Research and Demonstration of Key Technology of High-efficient Production in Kernel-aprico and Almond, grant number 2013BAD14B02.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The three chloroplast genome sequences are available in GenBank at the National Center for Biotechnology Information (NCBI) under accession numbers KY101151, KY101154, and KY101150 for P. armeniaca, P. sibirica, and kernel consumption apricot, respectively.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Basile, B.; Mataffo, A.; Forlani, M.; Corrado, G. Diversity in Morphometric, Pomological, and Fruit-Quality Traits of apricots (Prunus armeniaca) Traditional Varieties: Implications for Landrace Differentiation at Regional Scale. Diversity 2022, 14, 608. [Google Scholar] [CrossRef]
- Rehder, A. Manual of Cultivated Trees and Shrubs Hardy in North America, Exclusive of the Subtropical and Warmer Temperate Regions; Macmillan: New York, NY, USA, 1940. [Google Scholar]
- Mehlenbacher, S.A.; Cociu, V.; Hough, F.L. Apricots (Prunus). Acta Hortic. 1991, 290, 65–110. [Google Scholar] [CrossRef]
- Júlia, H.; Andrzej, P.; Attila, H. Origin and dissemination of the pollen-part mutated SC haplotype which confers self-compatibility in apricot (Prunus armeniaca). New Phytol. 2007, 176, 792–803. [Google Scholar]
- Hedia, S.I.; Christopher, S.; Zhebentyayeva, T.; Ledbetter, C.; Krška, B.; Remay, A.; D’Onofrio, C.; Iketani, H.; Christen, D.; Krichen, L.; et al. Genetic Structure of a Worldwide Germplasm Collection of Prunus armeniaca L. Reveals Three Major Diffusion Routes for Varieties Coming from the Species’ Center of Origin. Bourguiba. Front. Plant Sci. 2020, 11, 638. [Google Scholar]
- Karatas, N. Evaluation of Nutritional Content in Wild Apricot Fruits for Sustainable Apricot Production. Sustainability 2022, 14, 1063. [Google Scholar] [CrossRef]
- Cirillo, A.; De Luca, L.; Izzo, L.; Cepparulo, M.; Graziani, G.; Ritieni, A.; Romano, R.; Di Vaio, C. Biochemical and nutraceutical characterization of different accessions of the apricot (Prunus armeniaca L.). Horticulturae 2023, 9, 546. [Google Scholar] [CrossRef]
- Wang, X.; Wang, L.; Sun, Y.; Chen, J.; Liu, Q.; Dong, S. Genetic diversity and conservation of Siberian apricot (Prunus sibirica L.) based on microsatellite markers. Sci. Rep. 2023, 13, 11245. [Google Scholar] [CrossRef]
- Chen, L.; Sheng, Q.; Liu, J.; Jia, Y.; Zhao, W.; Li, P.; Zhang, A. A Review on the Functional Properties, Extraction and Microencapsulation of Almond Oil. Sci. Technol. Food Ind. 2023. [Google Scholar] [CrossRef]
- Daniell, H.; Jin, S.; Zhu, X.; Gitzendanner, M.A.; Soltis, D.E.; Soltis, P.S. Green giant—A tiny chloroplast genome with mighty power to produce high-value proteins: History and phylogeny. Plant Biotechnol. J. 2021, 19, 430–447. [Google Scholar] [CrossRef]
- Jiang, H.; Tian, J.; Yang, J.; Dong, X.; Zhong, Z.; Mwachala, G.; Zhang, C.; Hu, G.; Wang, Q. Comparative and phylogenetic analyses of six Kenya Polystachya (Orchidaceae) species based on the complete chloroplast genome sequences. BMC Plant Biol. 2022, 22, 177. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Fei, S.; Qiuping, Z.; Liu, N.; Zhang, Y.; Xu, M.; Liu, S.; Zhang, Y.; Ma, X.; Liu, W. Genetic diversity analysis of Chinese plum (Prunus salicina L.) based on whole-genome resequencing. Tree Genet. Genomes 2021, 17, 26. [Google Scholar]
- Oh, S.H.; Park, J. The complete chloroplast genome of Euscaphis japonica (Thunb.) Kanitz (Staphyleaceae) isolated in Korea. Mitochondr. DNA B Resour. 2020, 5, 3769–3771. [Google Scholar] [CrossRef] [PubMed]
- Zha, X.; Wang, X.; Li, J.; Gao, F.; Zhou, Y. Complete chloroplast genome of Sophora alopecuroides (Papilionoideae): Molecular structures, comparative genome analysis and phylogenetic analysis. J. Genet. 2020, 99, 1–14. [Google Scholar] [CrossRef]
- Du, X.; Zeng, T.; Feng, Q.; Hu, L.; Luo, X.; Weng, Q.; He, J.; Zhu, B. The complete chloroplast genome sequence of yellow mustard (Sinapis alba L.) and its phylogenetic relationship to other Brassicaceae species. Gene 2020, 731, 144340. [Google Scholar] [CrossRef] [PubMed]
- Bai, G.Q.; Zhou, T.; Zhao, J.X.; Li, W.M.; Han, G.J.; Li, S.F. The complete chloroplast genome of Kolkwitzia amabilis (Caprifoliaceae), an endangered horticultural plant in China. Mitochondr. DNA Part A 2017, 28, 296–297. [Google Scholar] [CrossRef]
- Shinozaki, K.; Ohme, M.; Tanaka, M.; Wakasugi, T.; Hayashida, N.; Matsubayashi, T.; Zaita, N.; Chunwongse, J.; Obokata, J.; Yamaguchi-Shinozaki, K.; et al. The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 1986, 5, 2043–2049. [Google Scholar] [CrossRef]
- Song, Y.F.; Yang, Q.H.; Yi, X.G.; Zhu, Z.Q.; Wang, X.R.; Li, M. Comparative analysis of codon usage patterns in chloroplast genomes of cherries. Forests 2022, 13, 1891. [Google Scholar] [CrossRef]
- Wu, J.Q.; Wang, Y.; Sun, P.; Sun, Z.S.; Shen, J.S. The complete chloroplast genome of Prunus phaeosticta (Hance) Maxim. (Rosaceae) and its phylogenetic implications. Mitochondr. DNA B 2023, 8, 136–140. [Google Scholar] [CrossRef]
- Zhongyu, D.; Ke, L.; Kai, Z.; He, Y.; Wang, H.; Chai, G.; Shi, J.; Duan, Y. The chloroplast genome of Amygdalus L. (Rosaceae) reveals the phylogenetic relationship and divergence time. BMC Genom. 2021, 22, 645. [Google Scholar]
- Mu, J.; Zhao, Y.; He, Y.; Sun, J.; Yuan, Q. The complete chloroplast genome of Prunus japonica thunb. (Rosaceae), an ornamental and medicinal plant. Mitochondr. DNA B 2021, 6, 112–114. [Google Scholar] [CrossRef] [PubMed]
- Hoang, T.K.; Wang, Y.; Hwang, Y.-J.; Lim, J.-H. Analysis of the morphological characteristics and karyomorphology of wild Chrysanthemum species in Korea. Hortic. Environ. Biotechnol. 2020, 61, 359–369. [Google Scholar] [CrossRef]
- Wani, A.A.; Zargar, S.A.; Malik, A.H.; Kashtwari, M.; Nazir, M.; Khuroo, A.A.; Ahmad, F.; Dar, T.A. Assessment of variability in morphological characters of apricot germplasm of Kashmir, India. Sci. Hortic. 2017, 225, 630–637. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhang, X.; Yan, S.; Feng, C.; Wang, D.; Yang, W.; Daud, M.K.; Xiang, J.; Mei, L. SSR identification and phylogenetic analysis in four plant species based on complete chloroplast genome sequences. Plasmid 2023, 125, 102670. [Google Scholar] [CrossRef]
- Alzahrani, D.; Albokhari, E.; Abba, A.; Yaradua, S. The first complete chloroplast genome sequences in Resedaceae: Genome structure and comparative analysis. Sci. Prog. 2021, 104, 368504211059973. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Zhu, J.; Ma, X.; Jia, J.; Luo, D.; Ding, Q.; Wang, X.; Huang, L. Comparative analysis of switchgrass chloroplast genomes provides insights into identification, phylogenetic relationships and evolution of different ecotypes. Ind. Crops Prod. 2023, 205, 117570. [Google Scholar] [CrossRef]
- Wang, S.; Gao, C.W.; Gao, L.Z. Plastid genome sequence of an ornamental and editable fruit tree of Rosaceae, Prunus mume. Mitochondr. DNA Part A 2016, 27, 4407–4408. [Google Scholar] [CrossRef]
- Terakami, S.; Matsumura, Y.; Kurita, K.; Kanamori, H.; Katayose, Y.; Yamamoto, T.; Katayama, H. Complete sequence of the chloroplast genome from pear (Pyrus pyrifolia): Genome structure and comparative analysis. Tree Genet. Genomes 2012, 8, 841–854. [Google Scholar] [CrossRef]
- Lu, R.S.; Li, P.; Qiu, Y.X. The complete chloroplast genomes of three Cardiocrinum (Liliaceae) species: Comparative genomic and phylogenetic analyses. Front. Plant Sci. 2016, 7, 2054. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef]
- Wang, W.C.; Chen, S.Y.; Zhang, X.Z. Chloroplast Genome Evolution in Actinidiaceae: clpP Loss, Heterogenous Divergence and Phylogenomic Practice. PLoS ONE 2016, 11, e0162324. [Google Scholar] [CrossRef]
- Asaf, S.; Khan, A.L.; Khan, M.A.; Waqas, M.; Kang, S.M.; Yun, B.W.; Lee, I.J. Chloroplast genomes of Arabidopsis halleri ssp. gemmifera and Arabidopsis lyrata ssp. petraea: Structures and comparative analysis. Sci. Rep. 2017, 7, 7556. [Google Scholar]
- Zhang, Y.; Du, L.; Liu, A.; Chen, J.; Wu, L.; Hu, W.; Zhang, W.; Kim, K.; Lee, S.-C.; Yang, T.-J.; et al. The complete chloroplast genome sequences of five Epimedium species: Lights into phylogenetic and taxonomic analyses. Front. Plant Sci. 2016, 7, 306. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Li, J.; Yu, L.; Bai, W.; Nie, D.; Xiong, Y.; Wu, S. Comparative chloroplast genomes of Prunus subgenus Cerasus (Rosaceae): Insights into sequence variations and phylogenetic relationships. Tree Genet. Genomes 2021, 17, 50. [Google Scholar] [CrossRef]
- Jansen, R.K.; Saski, C.; Lee, S.B.; Hansen, A.K.; Daniell, H. Complete plastid genome sequences of three Rosids (Castanea, Prunus, Theobroma): Evidence for at least two independent transfers of rpl22 to the nucleus. KRJ Mol. Biol. Evol. 2011, 28, 835–847. [Google Scholar] [CrossRef] [PubMed]
- Nan, W.; Ming, C.; Yong, C.; Liu, B.; Wang, X.; Li, G.; Zhou, Y.; Luo, P.; Xi, Z.; Yong, H.; et al. Natural variations in the non-coding region of ZmNAC080308 contributes maintaining grain yield under drought stress in maize. BMC Plant Biol. 2021, 21, 305. [Google Scholar]
- Lin, C.P.; Wu, C.S.; Huang, Y.Y.; Chaw, S.M. The complete chloroplast genome of Ginkgo biloba reveals the mechanism of inverted repeat contraction. Genome Biol. Evol. 2012, 4, 374–381. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Chen, X.; Geng, L.; Tang, C.; Wei, X. Phylogenetic Analysis of Sorbus ser Folgnerianae (Rosaceae). J. Nanjing For. Univ. 2023, pp. 1–15. Available online: https://kns.cnki.net/kcms/detail/32.1161.S.20230412.1754.006.html (accessed on 13 September 2023).
- Xue, S.; Shi, T.; Luo, W.; Ni, X.; Iqbal, S.; Ni, Z.; Huang, X.; Yao, D.; Shen, Z.; Gao, Z. Comparative analysis of the complete chloroplast genome among Prunus mume, P. Armeniaca, and P. salicina. Hortic. Res. 2019, 6, 89. [Google Scholar] [CrossRef]
- Jansen, R.K.; Cai, Z.; Raubeson, L.A.; Daniell, H.; Depamphilis, C.W.; Leebens-Mack, J.; Müller, K.F.; Guisinger-Bellian, M.; Haberle, R.C.; Hansen, A.K.; et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA 2007, 104, 19369–19374. [Google Scholar] [CrossRef] [PubMed]
- Raman, G.; Park, S. The complete chloroplast genome sequence of Ampelopsis: Gene organization, comparative analysis and phylogenetic relationships to other angiosperms. Front. Plant Sci. 2016, 7, 341. [Google Scholar] [CrossRef] [PubMed]
- Angiosperm Phylogeny Group. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot. J. Linn. Soc. 2009, 161, 105–121. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, W.; Liu, W.; Liu, N.; Zhang, Y.; Liu, S. Palynological Study on the Origin and Systematic Evolution of Kernel-using Apricots. Acta Hortic. Sin. 2010, 37, 1377–1387. [Google Scholar]
- Li, M.; Zheng, P.; Ni, B.; Hu, X.; Miao, X.; Zhao, Z. Genetic diversity analysis of apricot cultivars grown in China based on SSR markers. Eur. J. Hortic. Sci. 2018, 83, 18–27. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).