Systematic Pan-Cancer Analysis Reveals X-C Motif Chemokine Receptor 1 as a Prognostic and Immunological Biomarker
Abstract
:1. Introduction
2. Materials and Methods
2.1. Obtainment of Tissue Samples
2.2. RNA Extraction and Quantitative Real-Time PCR
2.3. Gene Expression Difference Analysis
2.4. Survival Prognosis Analysis and ROC Analysis
2.5. Genetic Alteration Analysis
2.6. Immune Infiltration Analysis
2.7. Gene Enrichment Analysis
2.8. Public Single-Cell Database Analysis
2.9. Statistical Analysis
3. Results
3.1. Pan-Cancer Screening of Chemokine Receptors Identifies Prognostic Value of XCR1
3.2. Higher XCR1 Expression Is Related with Better Patient Survival
3.3. XCR1 Was Decreased in Human Cancer
3.4. Low Expression of XCR1 in Advanced Cancer Stages
3.5. Mutation Features of XCR1 in Pan-Cancer
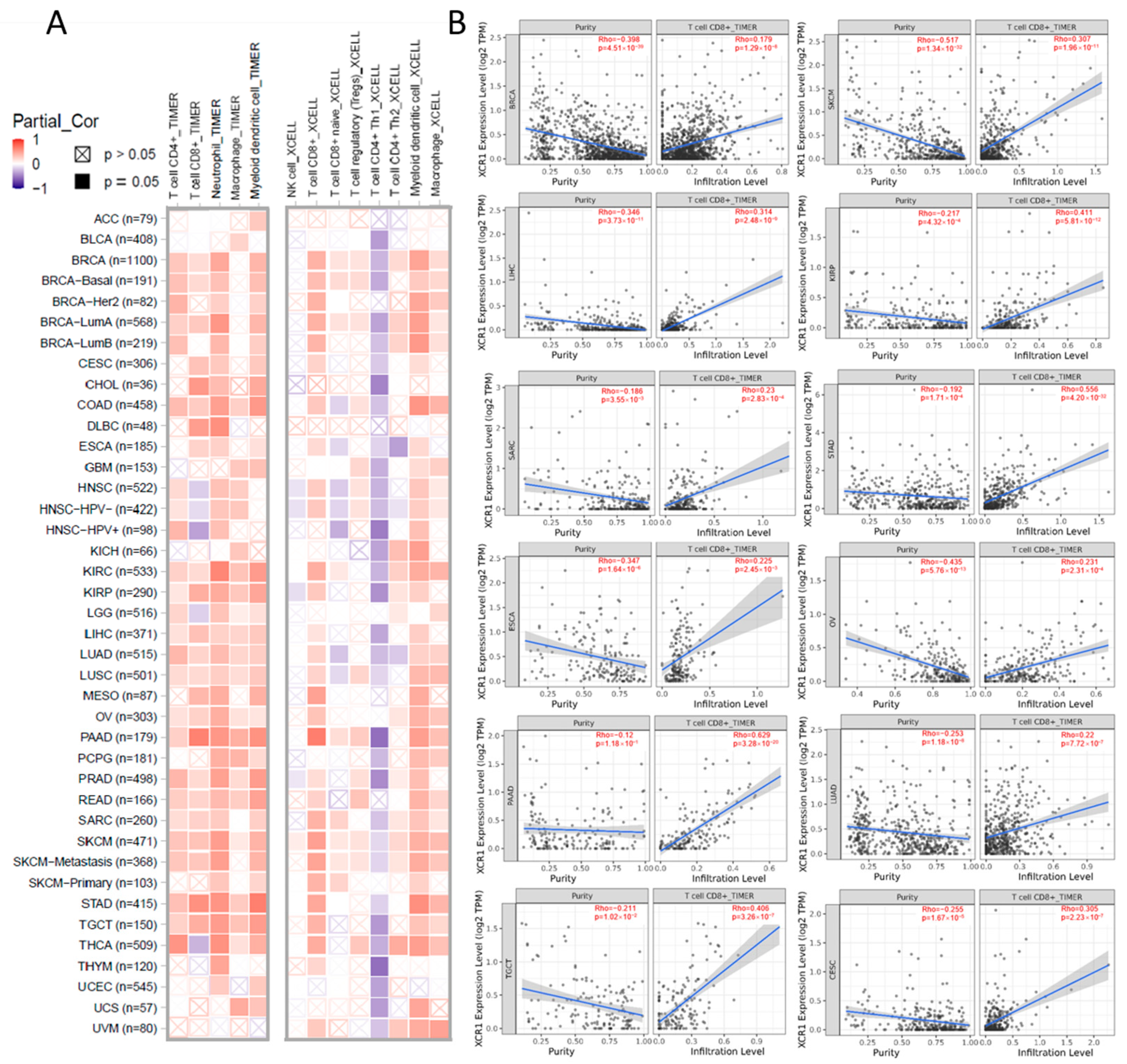
3.6. XCR1 Expression Correlation with Immune Infiltration of CD8+ T Cells
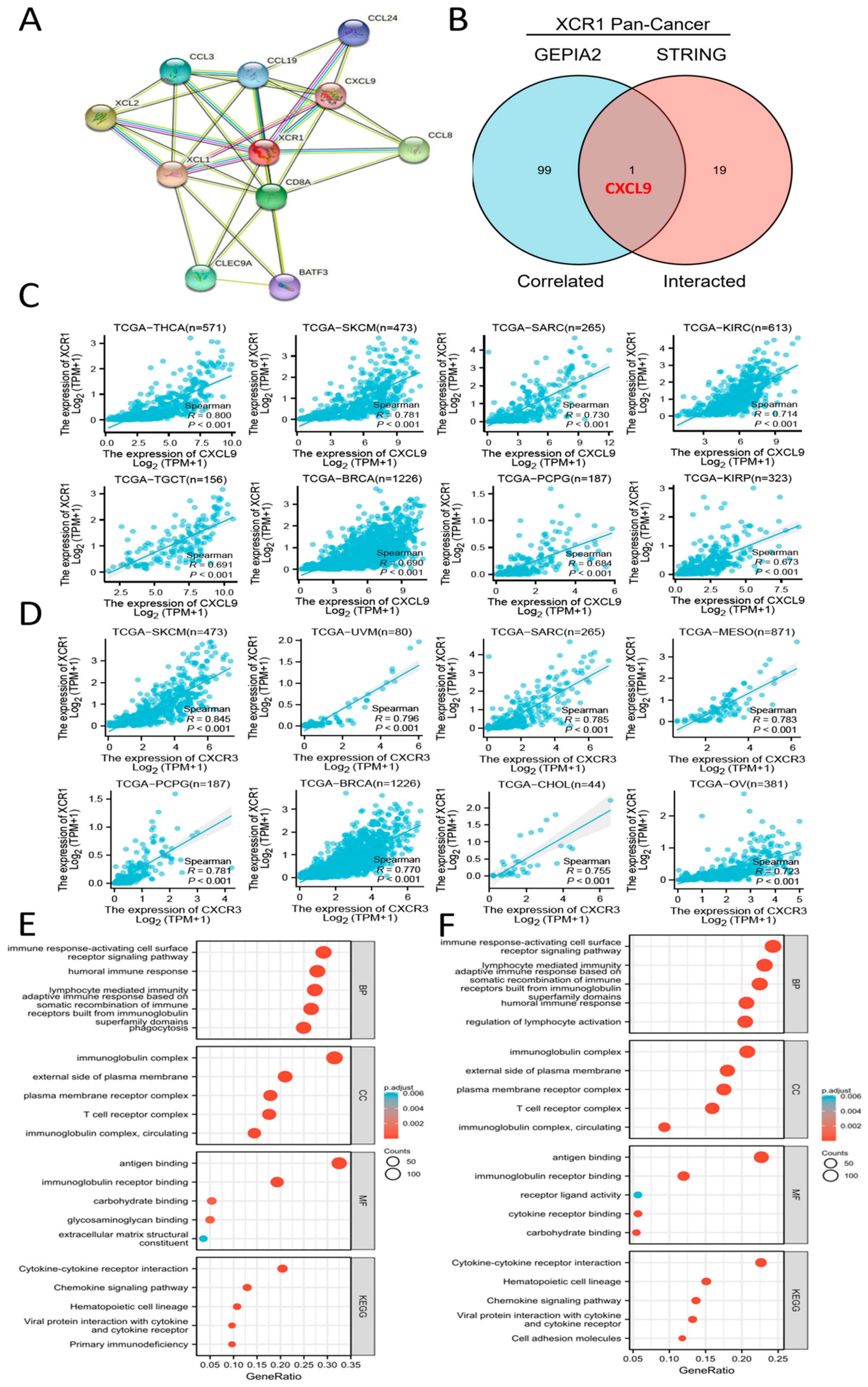
3.7. CXCL9-CXCR3 Axis Was Involved in Immune Infiltration by XCR1
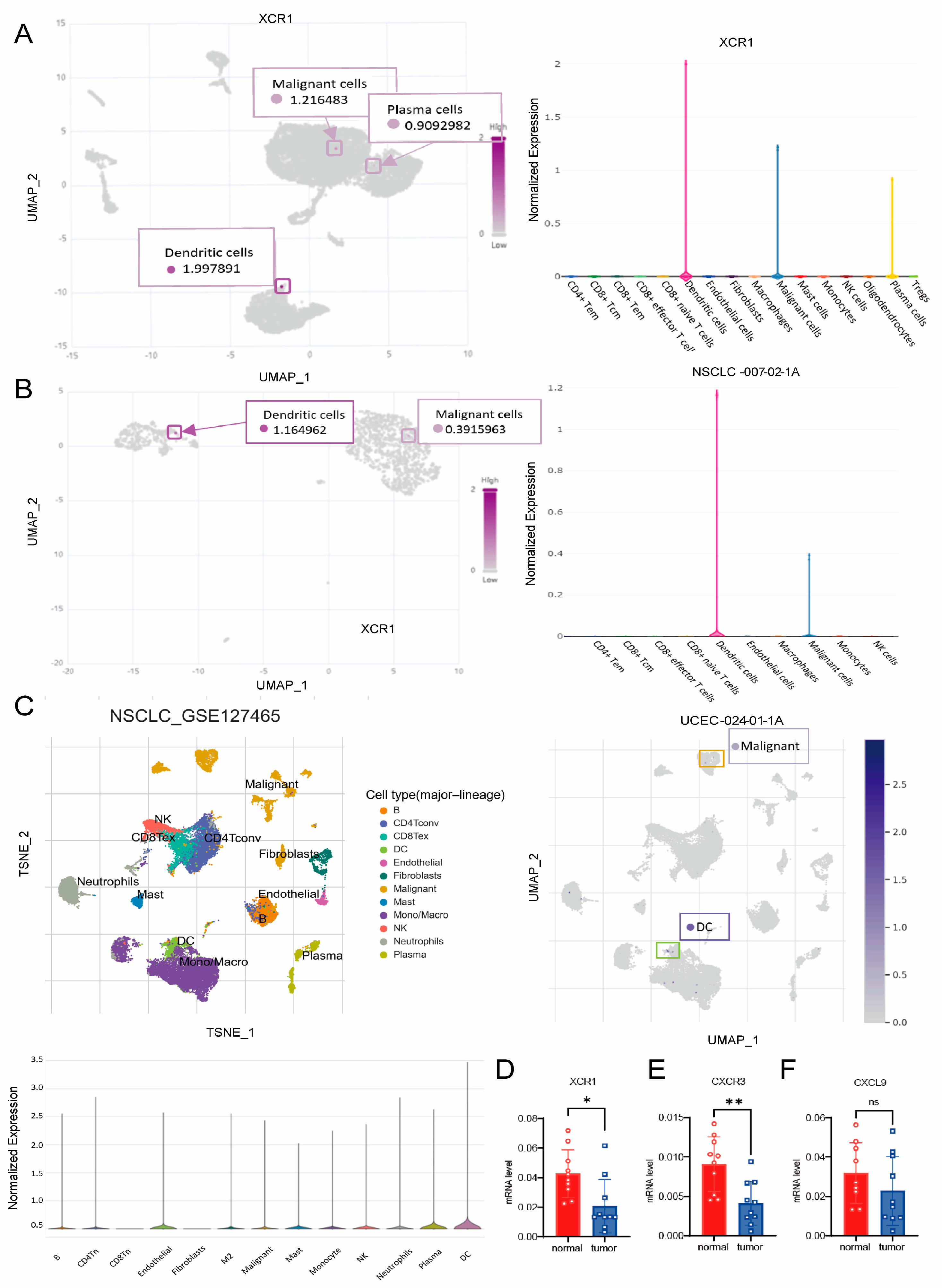
3.8. XCR1 Mainly Expressed in DCs and Malignant Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lei, Y.; Takahama, Y. XCL1 and XCR1 in the immune system. Microbes Infect. 2012, 14, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Kelner, G.S.; Kennedy, J.; Bacon, K.B.; Kleyensteuber, S.; Largaespada, D.A.; Jenkins, N.A.; Copeland, N.G.; Bazan, J.F.; Moore, K.W.; Schall, T.J.; et al. Lymphotactin: A cytokine that represents a new class of chemokine. Science 1994, 266, 1395–1399. [Google Scholar] [CrossRef] [PubMed]
- Hughes, C.E.; Nibbs, R.J.B. A guide to chemokines and their receptors. FEBS J. 2018, 285, 2944–2971. [Google Scholar] [CrossRef]
- Do, H.T.T.; Lee, C.H.; Cho, J. Chemokines and their Receptors: Multifaceted Roles in Cancer Progression and Potential Value as Cancer Prognostic Markers. Cancers 2020, 12, 287. [Google Scholar] [CrossRef] [PubMed]
- Nicolay, J.P.; Albrecht, J.D.; Alberti-Violetti, S.; Berti, E. CCR4 in cutaneous T-cell lymphoma: Therapeutic targeting of a pathogenic driver. Eur. J. Immunol. 2021, 51, 1660–1671. [Google Scholar] [CrossRef] [PubMed]
- Yoshie, O. CCR4 as a Therapeutic Target for Cancer Immunotherapy. Cancers 2021, 13, 5542. [Google Scholar] [CrossRef] [PubMed]
- Legler, D.F.; Uetz-von Allmen, E.; Hauser, M.A. CCR7: Roles in cancer cell dissemination, migration and metastasis formation. Int. J. Biochem. Cell Biol. 2014, 54, 78–82. [Google Scholar] [CrossRef]
- Nagarsheth, N.; Wicha, M.S.; Zou, W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol. 2017, 17, 559–572. [Google Scholar] [CrossRef]
- Liu, G.T.; Chen, H.T.; Tsou, H.K.; Tan, T.W.; Fong, Y.C.; Chen, P.C.; Yang, W.H.; Wang, S.W.; Chen, J.C.; Tang, C.H. CCL5 promotes VEGF-dependent angiogenesis by down-regulating miR-200b through PI3K/Akt signaling pathway in human chondrosarcoma cells. Oncotarget 2014, 5, 10718–10731. [Google Scholar] [CrossRef]
- Suffee, N.; Hlawaty, H.; Meddahi-Pelle, A.; Maillard, L.; Louedec, L.; Haddad, O.; Martin, L.; Laguillier, C.; Richard, B.; Oudar, O.; et al. RANTES/CCL5-induced pro-angiogenic effects depend on CCR1, CCR5 and glycosaminoglycans. Angiogenesis 2012, 15, 727–744. [Google Scholar] [CrossRef]
- Korbecki, J.; Grochans, S.; Gutowska, I.; Barczak, K.; Baranowska-Bosiacka, I. CC Chemokines in a Tumor: A Review of Pro-Cancer and Anti-Cancer Properties of Receptors CCR5, CCR6, CCR7, CCR8, CCR9, and CCR10 Ligands. Int. J. Mol. Sci. 2020, 21, 7619. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Xu, W.; Wei, C.; Huang, J.; Xu, J.; Zhang, Y.; Zhao, Y.; Chen, J.; Dong, S.; Liu, B.; et al. CCL14 serves as a novel prognostic factor and tumor suppressor of HCC by modulating cell cycle and promoting apoptosis. Cell Death Dis. 2019, 10, 796. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Li, X.; Bi, Y.; Zheng, Y.; Wang, J.; Li, X.; Huang, Z.; Chen, L.; Huang, Y.; Huang, Y. CCL14 is a prognostic biomarker and correlates with immune infiltrates in hepatocellular carcinoma. Aging 2020, 12, 784–807. [Google Scholar] [CrossRef] [PubMed]
- Ramazzotti, D.; Lal, A.; Wang, B.; Batzoglou, S.; Sidow, A. Multi-omic tumor data reveal diversity of molecular mechanisms that correlate with survival. Nat. Commun. 2018, 9, 4453. [Google Scholar] [CrossRef] [PubMed]
- Repana, D.; Nulsen, J.; Dressler, L.; Bortolomeazzi, M.; Venkata, S.K.; Tourna, A.; Yakovleva, A.; Palmieri, T.; Ciccarelli, F.D. The Network of Cancer Genes (NCG): A comprehensive catalogue of known and candidate cancer genes from cancer sequencing screens. Genome Biol. 2019, 20, 1. [Google Scholar] [CrossRef]
- Vilgelm, A.E.; Richmond, A. Chemokines Modulate Immune Surveillance in Tumorigenesis, Metastasis, and Response to Immunotherapy. Front. Immunol. 2019, 10, 333. [Google Scholar] [CrossRef]
- Kato, T.; Fujita, Y.; Nakane, K.; Mizutani, K.; Terazawa, R.; Ehara, H.; Kanimoto, Y.; Kojima, T.; Nozawa, Y.; Deguchi, T.; et al. CCR1/CCL5 interaction promotes invasion of taxane-resistant PC3 prostate cancer cells by increasing secretion of MMPs 2/9 and by activating ERK and Rac signaling. Cytokine 2013, 64, 251–257. [Google Scholar] [CrossRef]
- Anderson, M.W.; Zhao, S.; Ai, W.Z.; Tibshirani, R.; Levy, R.; Lossos, I.S.; Natkunam, Y. C-C chemokine receptor 1 expression in human hematolymphoid neoplasia. Am. J. Clin. Pathol. 2010, 133, 473–483. [Google Scholar] [CrossRef]
- Zi, J.; Yuan, S.; Qiao, J.; Zhao, K.; Xu, L.; Qi, K.; Xu, K.; Zeng, L. Treatment with the C-C chemokine receptor type 5 (CCR5)-inhibitor maraviroc suppresses growth and induces apoptosis of acute lymphoblastic leukemia cells. Am. J. Cancer Res. 2017, 7, 869–880. [Google Scholar]
- Hemmatazad, H.; Berger, M.D. CCR5 is a potential therapeutic target for cancer. Expert. Opin. Ther. Targets 2021, 25, 311–327. [Google Scholar] [CrossRef]
- Jiao, X.; Nawab, O.; Patel, T.; Kossenkov, A.V.; Halama, N.; Jaeger, D.; Pestell, R.G. Recent Advances Targeting CCR5 for Cancer and Its Role in Immuno-Oncology. Cancer Res. 2019, 79, 4801–4807. [Google Scholar] [CrossRef]
- Zou, W.; Machelon, V.; Coulomb-L’Hermin, A.; Borvak, J.; Nome, F.; Isaeva, T.; Wei, S.; Krzysiek, R.; Durand-Gasselin, I.; Gordon, A.; et al. Stromal-derived factor-1 in human tumors recruits and alters the function of plasmacytoid precursor dendritic cells. Nat. Med. 2001, 7, 1339–1346. [Google Scholar] [CrossRef]
- Zhou, W.; Guo, S.; Liu, M.; Burow, M.E.; Wang, G. Targeting CXCL12/CXCR4 Axis in Tumor Immunotherapy. Curr. Med. Chem. 2019, 26, 3026–3041. [Google Scholar] [CrossRef]
- Janssens, R.; Struyf, S.; Proost, P. The unique structural and functional features of CXCL12. Cell Mol. Immunol. 2018, 15, 299–311. [Google Scholar] [CrossRef]
- Daniel, S.K.; Seo, Y.D.; Pillarisetty, V.G. The CXCL12-CXCR4/CXCR7 axis as a mechanism of immune resistance in gastrointestinal malignancies. Semin. Cancer Biol. 2020, 65, 176–188. [Google Scholar] [CrossRef]
- Pollard, J.W. Tumour-educated macrophages promote tumour progression and metastasis. Nat. Rev. Cancer 2004, 4, 71–78. [Google Scholar] [CrossRef]
- Hao, Q.; Vadgama, J.V.; Wang, P. CCL2/CCR2 signaling in cancer pathogenesis. Cell Commun. Signal 2020, 18, 82. [Google Scholar] [CrossRef]
- Kadomoto, S.; Izumi, K.; Mizokami, A. Roles of CCL2-CCR2 Axis in the Tumor Microenvironment. Int. J. Mol. Sci. 2021, 22, 8530. [Google Scholar] [CrossRef]
- Kim, M.; Rooper, L.; Xie, J.; Rayahin, J.; Burdette, J.E.; Kajdacsy-Balla, A.A.; Barbolina, M.V. The lymphotactin receptor is expressed in epithelial ovarian carcinoma and contributes to cell migration and proliferation. Mol. Cancer Res. MCR 2012, 10, 1419–1429. [Google Scholar] [CrossRef]
- Wang, T.; Han, S.; Wu, Z.; Han, Z.; Yan, W.; Liu, T.; Wei, H.; Song, D.; Zhou, W.; Yang, X.; et al. XCR1 promotes cell growth and migration and is correlated with bone metastasis in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2015, 464, 635–641. [Google Scholar] [CrossRef]
- Yang, X.L.; Qi, L.G.; Lin, F.J.; Ou, Z.L. The role of the chemokine receptor XCR1 in breast cancer cells. Breast Cancer (Dove Med. Press) 2017, 9, 227–236. [Google Scholar] [CrossRef]
- Khurram, S.A.; Whawell, S.A.; Bingle, L.; Murdoch, C.; McCabe, B.M.; Farthing, P.M. Functional expression of the chemokine receptor XCR1 on oral epithelial cells. J. Pathol. 2010, 221, 153–163. [Google Scholar] [CrossRef]
- Vivian, J.; Rao, A.A.; Nothaft, F.A.; Ketchum, C.; Armstrong, J.; Novak, A.; Pfeil, J.; Narkizian, J.; Deran, A.D.; Musselman-Brown, A.; et al. Toil enables reproducible, open source, big biomedical data analyses. Nat. Biotechnol. 2017, 35, 314–316. [Google Scholar] [CrossRef]
- Ren, Z.; He, Y.; Yang, Q.; Guo, J.; Huang, H.; Li, B.; Wang, D.; Yang, Z.; Tian, X. A Comprehensive Analysis of the Glutathione Peroxidase 8 (GPX8) in Human Cancer. Front. Oncol. 2022, 12, 812811. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Deng, M.; Peng, L.; Li, J.; Liu, X.; Xia, X.; Li, G. PPP1R14B Is a Prognostic and Immunological Biomarker in Pan-Cancer. Front. Genet. 2021, 12, 763561. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- von Mering, C.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018, 173, 400–416.e11. [Google Scholar] [CrossRef]
- Gershenwald, J.E.; Scolyer, R.A.; Hess, K.R.; Sondak, V.K.; Long, G.V.; Ross, M.I.; Lazar, A.J.; Faries, M.B.; Kirkwood, J.M.; McArthur, G.A.; et al. Melanoma staging: Evidence-based changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J. Clin. 2017, 67, 472–492. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Masterman, K.A.; Haigh, O.L.; Tullett, K.M.; Leal-Rojas, I.M.; Walpole, C.; Pearson, F.E.; Cebon, J.; Schmidt, C.; O’Brien, L.; Rosendahl, N.; et al. Human CLEC9A antibodies deliver NY-ESO-1 antigen to CD141(+) dendritic cells to activate naïve and memory NY-ESO-1-specific CD8(+) T cells. J. Immunother. Cancer 2020, 8, e000691. [Google Scholar] [CrossRef] [PubMed]
- Giampazolias, E.; Schulz, O.; Lim, K.H.J.; Rogers, N.C.; Chakravarty, P.; Srinivasan, N.; Gordon, O.; Cardoso, A.; Buck, M.D.; Poirier, E.Z.; et al. Secreted gelsolin inhibits DNGR-1-dependent cross-presentation and cancer immunity. Cell 2021, 184, 4016–4031.e22. [Google Scholar] [CrossRef] [PubMed]
- Tsao, H.W.; Kaminski, J.; Kurachi, M.; Barnitz, R.A.; DiIorio, M.A.; LaFleur, M.W.; Ise, W.; Kurosaki, T.; Wherry, E.J.; Haining, W.N.; et al. Batf-mediated epigenetic control of effector CD8(+) T cell differentiation. Sci. Immunol. 2022, 7, eabi4919. [Google Scholar] [CrossRef]
- de Mingo Pulido, Á.; Hänggi, K.; Celias, D.P.; Gardner, A.; Li, J.; Batista-Bittencourt, B.; Mohamed, E.; Trillo-Tinoco, J.; Osunmakinde, O.; Peña, R.; et al. The inhibitory receptor TIM-3 limits activation of the cGAS-STING pathway in intra-tumoral dendritic cells by suppressing extracellular DNA uptake. Immunity 2021, 54, 1154–1167.e7. [Google Scholar] [CrossRef]
- Eisenbarth, S.C. Dendritic cell subsets in T cell programming: Location dictates function. Nat. Rev. Immunol. 2019, 19, 89–103. [Google Scholar] [CrossRef]
- Kvedaraite, E.; Ginhoux, F. Human dendritic cells in cancer. Sci. Immunol. 2022, 7, eabm9409. [Google Scholar] [CrossRef]
- Schlitzer, A.; Sivakamasundari, V.; Chen, J.; Sumatoh, H.R.; Schreuder, J.; Lum, J.; Malleret, B.; Zhang, S.; Larbi, A.; Zolezzi, F.; et al. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat. Immunol. 2015, 16, 718–728. [Google Scholar] [CrossRef]
- Collin, M.; Bigley, V. Human dendritic cell subsets: An update. Immunology 2018, 154, 3–20. [Google Scholar] [CrossRef]
- Edelson, B.T.; Kc, W.; Juang, R.; Kohyama, M.; Benoit, L.A.; Klekotka, P.A.; Moon, C.; Albring, J.C.; Ise, W.; Michael, D.G.; et al. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J. Exp. Med. 2010, 207, 823–836. [Google Scholar] [CrossRef]
- Gurka, S.; Hartung, E.; Becker, M.; Kroczek, R.A. Mouse Conventional Dendritic Cells Can be Universally Classified Based on the Mutually Exclusive Expression of XCR1 and SIRPα. Front. Immunol. 2015, 6, 35. [Google Scholar] [CrossRef] [PubMed]
- Hacker, C.; Kirsch, R.D.; Ju, X.S.; Hieronymus, T.; Gust, T.C.; Kuhl, C.; Jorgas, T.; Kurz, S.M.; Rose-John, S.; Yokota, Y.; et al. Transcriptional profiling identifies Id2 function in dendritic cell development. Nat. Immunol. 2003, 4, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Desch, A.N.; Randolph, G.J.; Murphy, K.; Gautier, E.L.; Kedl, R.M.; Lahoud, M.H.; Caminschi, I.; Shortman, K.; Henson, P.M.; Jakubzick, C.V. CD103+ pulmonary dendritic cells preferentially acquire and present apoptotic cell-associated antigen. J. Exp. Med. 2011, 208, 1789–1797. [Google Scholar] [CrossRef] [PubMed]
- Sancho, D.; Mourão-Sá, D.; Joffre, O.P.; Schulz, O.; Rogers, N.C.; Pennington, D.J.; Carlyle, J.R.; Reis e Sousa, C. Tumor therapy in mice via antigen targeting to a novel, DC-restricted C-type lectin. J. Clin. Investig. 2008, 118, 2098–2110. [Google Scholar] [CrossRef] [PubMed]
- Bachem, A.; Hartung, E.; Güttler, S.; Mora, A.; Zhou, X.; Hegemann, A.; Plantinga, M.; Mazzini, E.; Stoitzner, P.; Gurka, S.; et al. Expression of XCR1 Characterizes the Batf3-Dependent Lineage of Dendritic Cells Capable of Antigen Cross-Presentation. Front. Immunol. 2012, 3, 214. [Google Scholar] [CrossRef]
- Dorner, B.G.; Dorner, M.B.; Zhou, X.; Opitz, C.; Mora, A.; Güttler, S.; Hutloff, A.; Mages, H.W.; Ranke, K.; Schaefer, M.; et al. Selective expression of the chemokine receptor XCR1 on cross-presenting dendritic cells determines cooperation with CD8+ T cells. Immunity 2009, 31, 823–833. [Google Scholar] [CrossRef]
- Mikolajczyk, T.P.; Szczepaniak, P.; Vidler, F.; Maffia, P.; Graham, G.J.; Guzik, T.J. Role of inflammatory chemokines in hypertension. Pharmacol. Ther. 2021, 223, 107799. [Google Scholar] [CrossRef]
- Reschke, R.; Gajewski, T.F. CXCL9 and CXCL10 bring the heat to tumors. Sci. Immunol. 2022, 7, eabq6509. [Google Scholar] [CrossRef]
- Mikucki, M.E.; Fisher, D.T.; Matsuzaki, J.; Skitzki, J.J.; Gaulin, N.B.; Muhitch, J.B.; Ku, A.W.; Frelinger, J.G.; Odunsi, K.; Gajewski, T.F.; et al. Non-redundant requirement for CXCR3 signalling during tumoricidal T-cell trafficking across tumour vascular checkpoints. Nat. Commun. 2015, 6, 7458. [Google Scholar] [CrossRef]
- Spranger, S.; Dai, D.; Horton, B.; Gajewski, T.F. Tumor-Residing Batf3 Dendritic Cells Are Required for Effector T Cell Trafficking and Adoptive T Cell Therapy. Cancer Cell 2017, 31, 711–723.e4. [Google Scholar] [CrossRef]
- Humblin, E.; Kamphorst, A.O. CXCR3-CXCL9: It’s All in the Tumor. Immunity 2019, 50, 1347–1349. [Google Scholar] [CrossRef] [PubMed]
- Neo, S.Y.; Lundqvist, A. The Multifaceted Roles of CXCL9 Within the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1231, 45–51. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, L.; Zhu, L.; Chen, J.; Li, C.; Yu, Y.; Xu, S. Systematic Pan-Cancer Analysis Reveals X-C Motif Chemokine Receptor 1 as a Prognostic and Immunological Biomarker. Genes 2023, 14, 1961. https://doi.org/10.3390/genes14101961
Cui L, Zhu L, Chen J, Li C, Yu Y, Xu S. Systematic Pan-Cancer Analysis Reveals X-C Motif Chemokine Receptor 1 as a Prognostic and Immunological Biomarker. Genes. 2023; 14(10):1961. https://doi.org/10.3390/genes14101961
Chicago/Turabian StyleCui, Likun, Liye Zhu, Jie Chen, Chunzhen Li, Yizhi Yu, and Sheng Xu. 2023. "Systematic Pan-Cancer Analysis Reveals X-C Motif Chemokine Receptor 1 as a Prognostic and Immunological Biomarker" Genes 14, no. 10: 1961. https://doi.org/10.3390/genes14101961
APA StyleCui, L., Zhu, L., Chen, J., Li, C., Yu, Y., & Xu, S. (2023). Systematic Pan-Cancer Analysis Reveals X-C Motif Chemokine Receptor 1 as a Prognostic and Immunological Biomarker. Genes, 14(10), 1961. https://doi.org/10.3390/genes14101961