Genome-Wide Identification of WD40 Proteins in Cucurbita maxima Reveals Its Potential Functions in Fruit Development
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatment
2.2. Acquisition and Identification of WD40 Proteins
2.3. Characterization of WD40 Protein in C. maxima
2.4. Chromosome Distribution and Replication of the WD40 Gene Family
2.5. Collinear Analysis
2.6. Phylogenetic Analysis
2.7. Gene Structure, Conserved Motifs, and Promoter Region Analysis
2.8. Expression Analysis
2.9. RNA Extraction and Quantitative Real-Time-PCR
3. Results
3.1. Identification of WD40 Genes in C. maxima
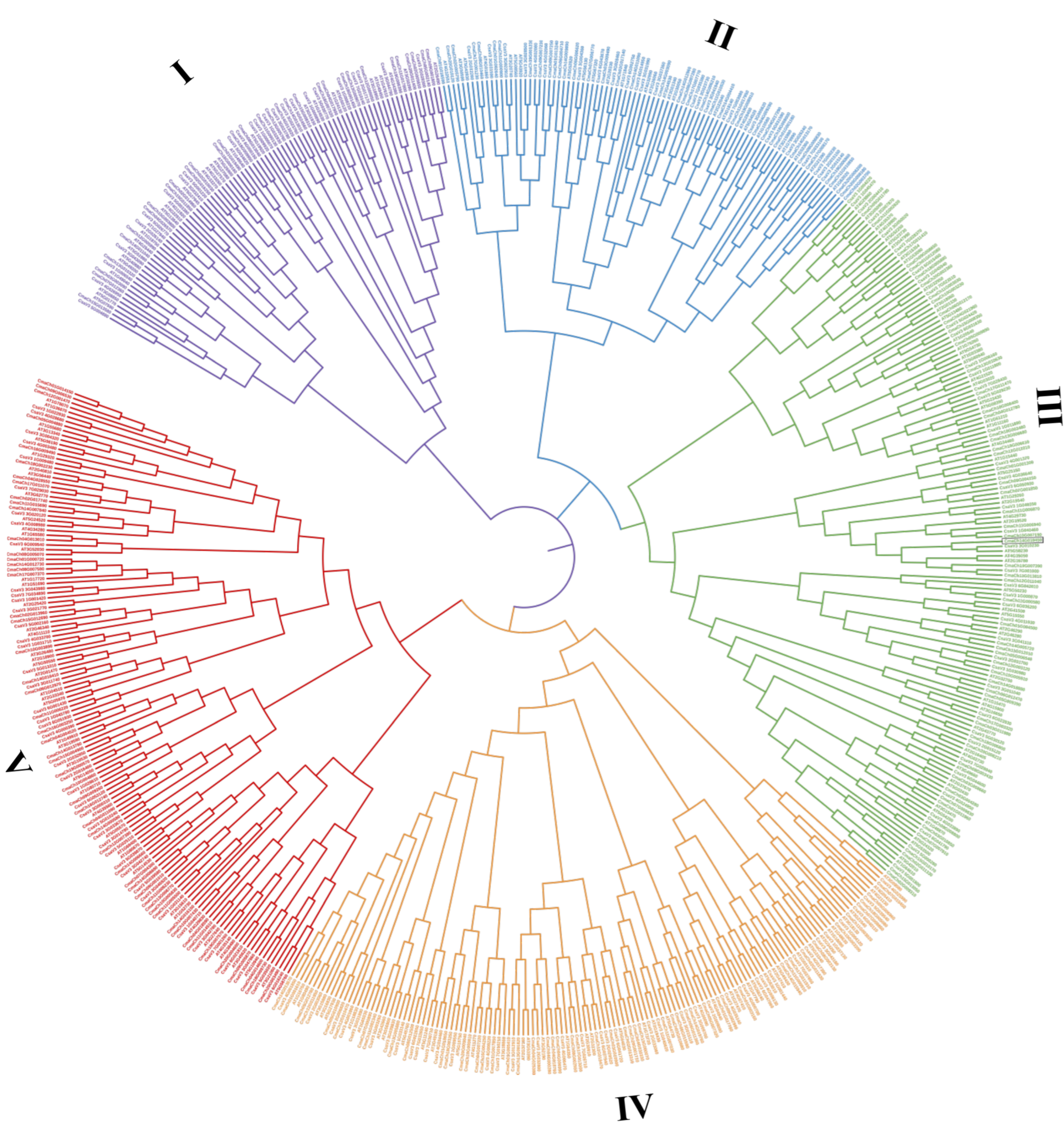
3.2. Protein Classification and Phylogenetic Analysis of the C. maxima WD40 Gene Family
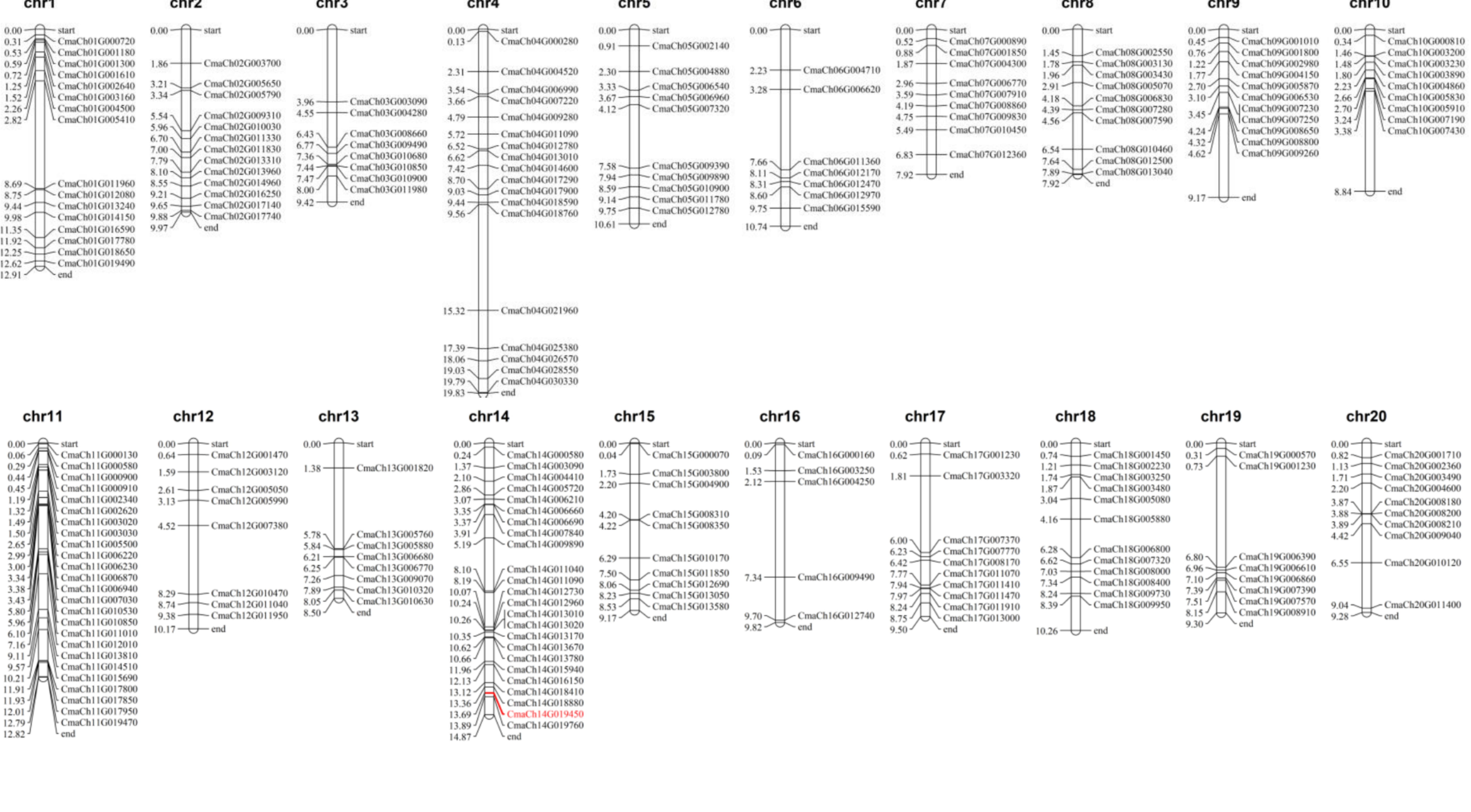
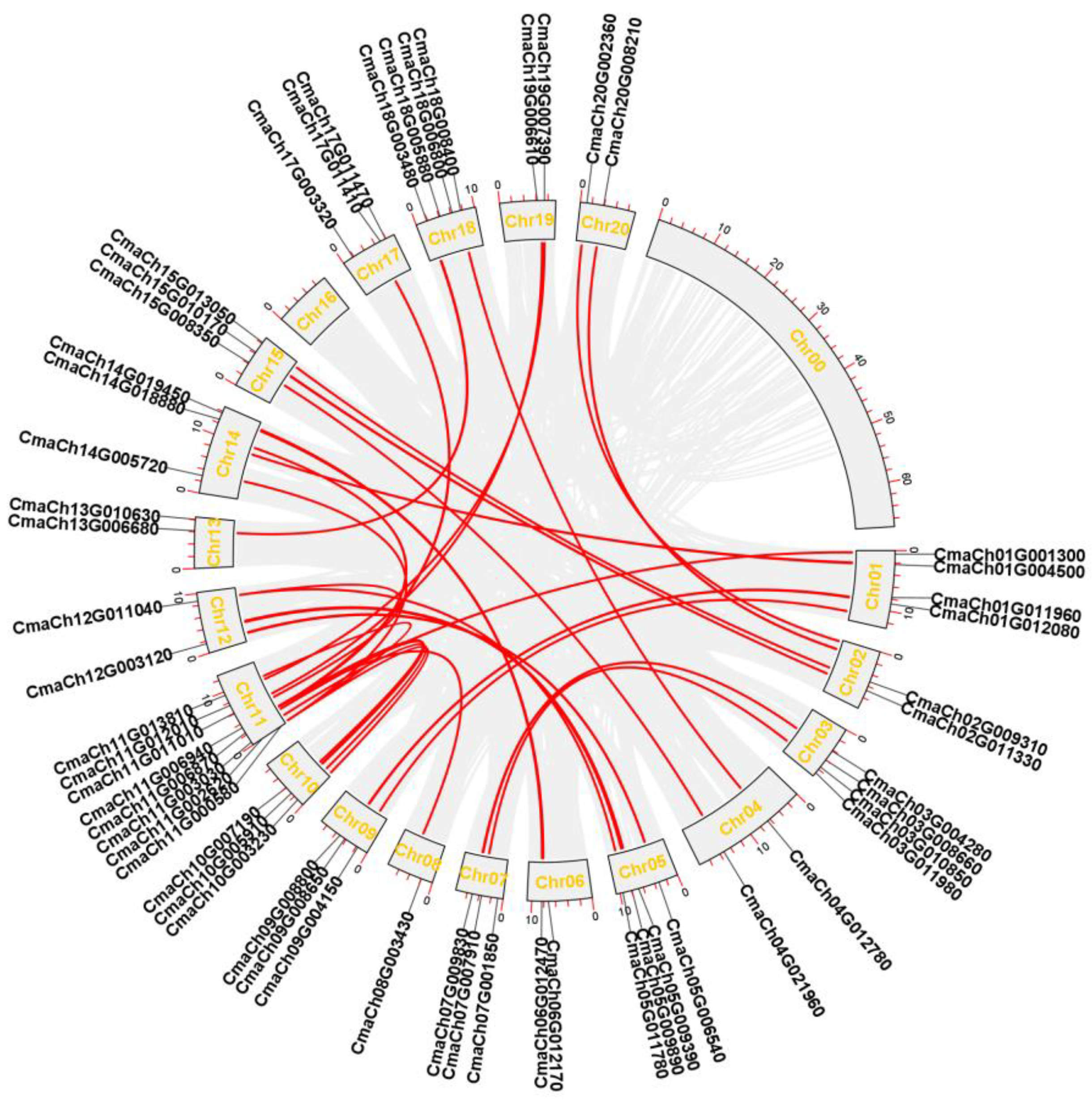
3.3. Chromosomal Distribution, Gene Replication Analysis of CmWD40s
3.4. Gene Structure and Promoter Region Analysis
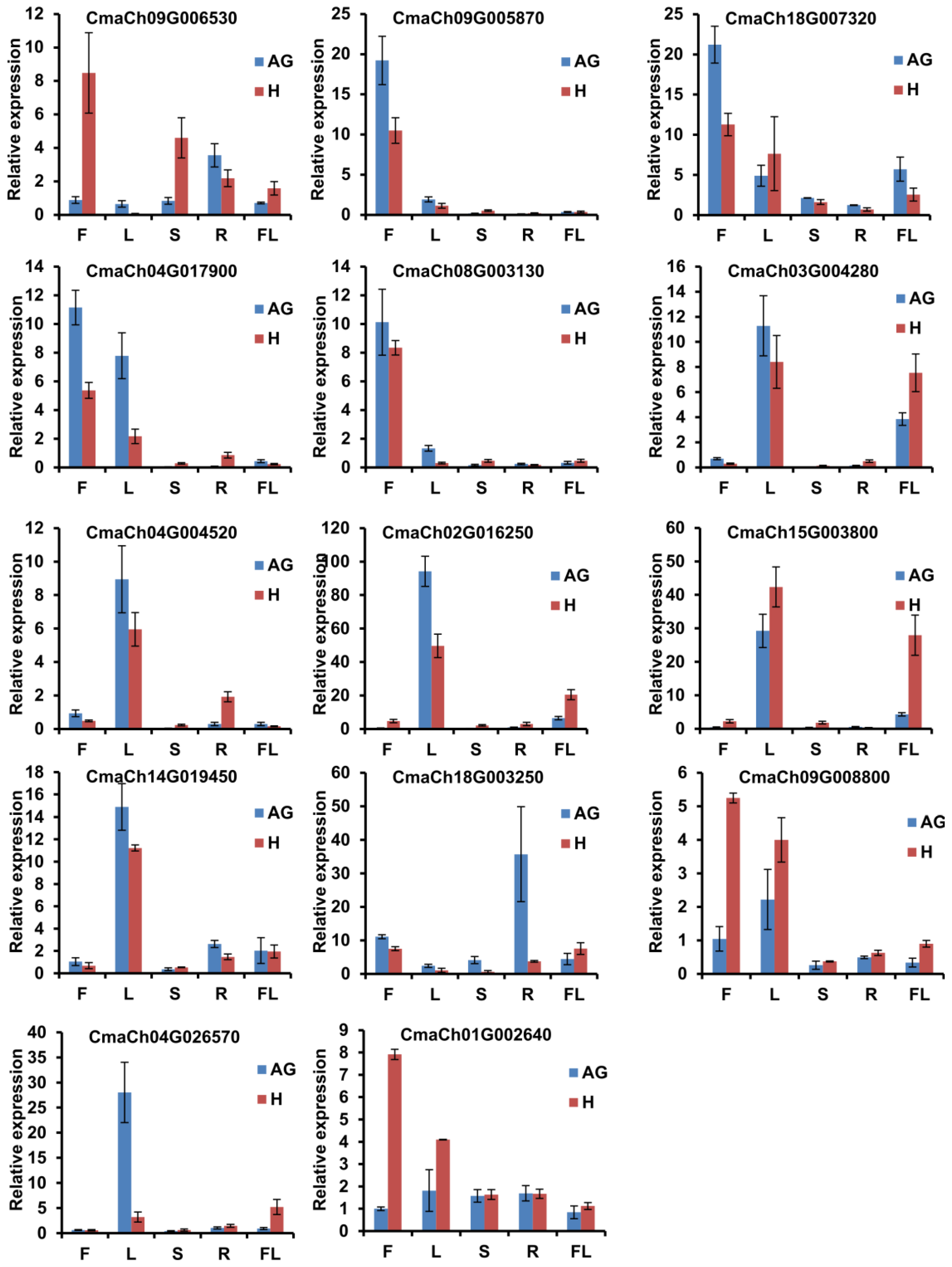
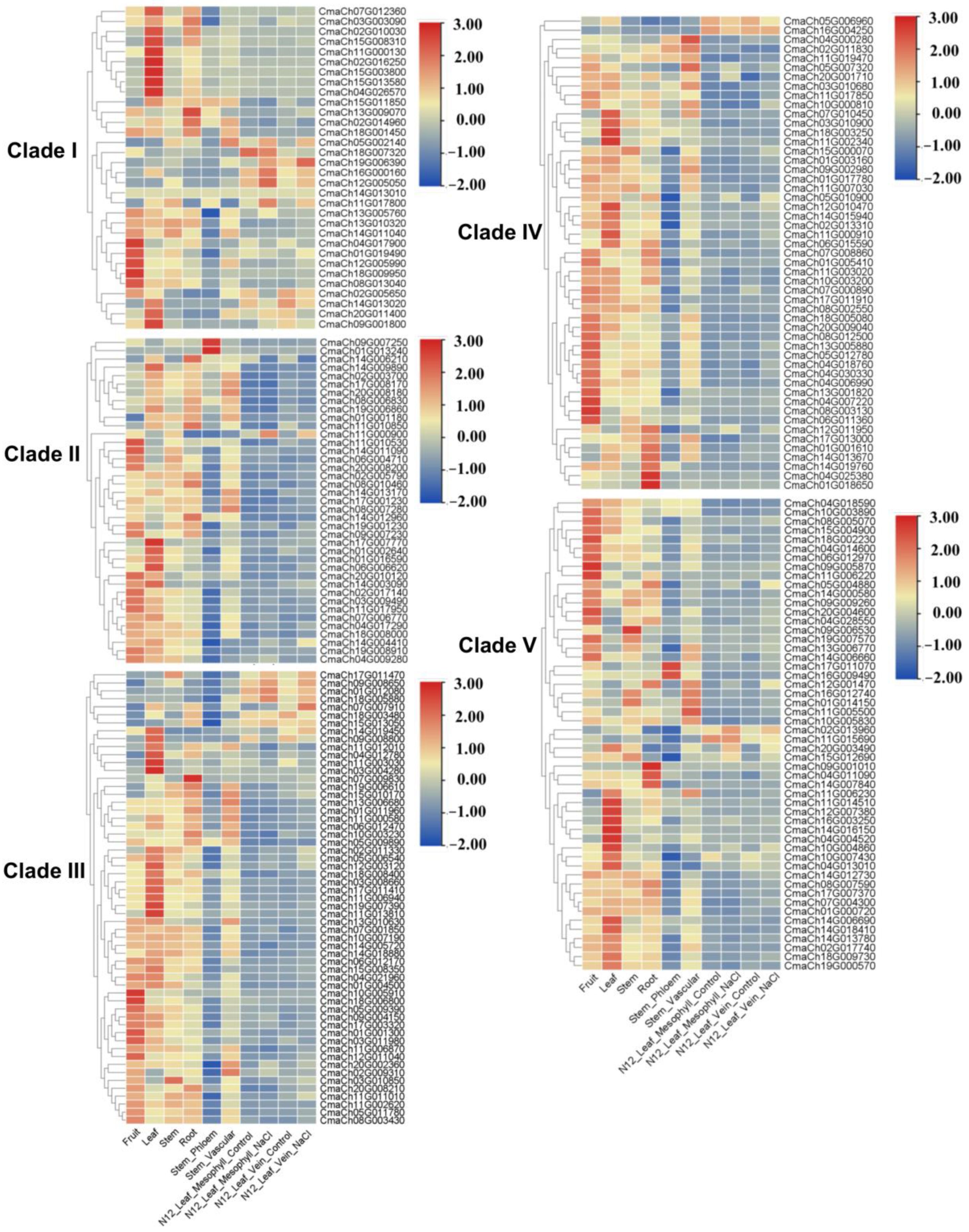
3.5. The Gene Expression Pattern Analysis of CmWD40s
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Neer, E.; Schmidt, C.; Nambudripad, R.; Smith, T. The ancient regulatory protein family of WD-repeat proteins. Nature 1994, 371, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.; Gaitatzes, C.; Saxena, K.; Neer, E.J. The WD repeat: A common architecture for diverse functions. Trends Biochem. Sci. 1999, 24, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Gibson, T. Cell regulation: Determined to signal discrete cooperation. Trends Biochem. Sci. 2009, 34, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Min, J. Structure and function of WD40 domain proteins. Protein Cell 2011, 2, 202–214. [Google Scholar] [CrossRef] [PubMed]
- Juhász, T.; Szeltner, Z.; Fülöp, V.; Polgár, L. Unclosed β-propellers display stable structures: Implications for substrate access to the active site of prolyloligopeptidase. J. Mol. Biol. 2005, 346, 907–917. [Google Scholar] [CrossRef]
- Fulop, V.; Bocskei, Z.; Polgar, L. Prolyl oligopeptidase: An unusual β-propeller domain regulates proteolysis. Cell 1998, 94, 161–170. [Google Scholar] [CrossRef]
- Van Nocker, S.; Ludwig, P. The WD-repeat protein superfamily in Arabidopsis: Conservation and divergence in structure and function. BMC Genom. 2003, 4, 50. [Google Scholar] [CrossRef]
- Jain, B.; Pandey, S. WD40 repeat proteins: Signalling scaffold with diverse functions. Protein J. 2018, 37, 391–406. [Google Scholar] [CrossRef]
- Dayebgadoh, G.; Sardiu, M.; Florens, L.; Washburn, M. Biochemical Reduction of the Topology of the Diverse WDR76 Protein Interactome. J. Proteome Res. 2019, 18, 3479–3491. [Google Scholar] [CrossRef]
- Hsia, K.; Stavropoulos, P.; Blobel, G.; Hoelz, A. Architecture of a coat for the nuclear pore membrane. Cell 2007, 131, 1313–1326. [Google Scholar] [CrossRef]
- Brohawn, S.; Leksa, N.; Spear, E.; Rajashankar, K.; Schwartz, T. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science 2008, 322, 1369–1373. [Google Scholar] [CrossRef] [PubMed]
- Saeki, M.; Irie, Y.; Ni, L.; Yoshida, M.; Itsuki, M.; Kamisaki, M. Monad, a WD40 repeat protein, promotes apoptosis induced by TNF-α. Biochem. Biophys. Res. Commun. 2006, 342, 568–572. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Puranik, S.; Bahadur, R.; Prasad, M. The DNA-binding activity of an AP2 protein is involved in transcriptional regulation of a stress-responsive gene, SiWD40, in foxtail millet. Genomics 2012, 100, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Xiao, J.; Gu, T.; Yu, X.; Zhang, Y.; Chang, J.; Yang, G.; He, G. Genome-wide identification and analysis of WD40 proteins in wheat (Triticum aestivum L.). BMC Genom. 2018, 19, 803. [Google Scholar]
- He, S.; Tong, X.; Han, M.; Hu, H.; Dai, F. Genome-wide identification and characterization of WD40 protein genes in the silkworm, bombyx mori. Int. J. Mol. Sci. 2018, 19, 527. [Google Scholar] [CrossRef]
- Zou, X.; Hu, X.; Ma, J.; Li, T.; Ye, Z. Genome-wide analysis of WD40 protein family in human. Sci. Rep. 2016, 6, 39262. [Google Scholar] [CrossRef]
- Feng, R.; Zhang, C.; Ma, R.; Cai, Z.; Lin, Y.; Yu, M. Identification and characterization of WD40 superfamily genes in peach. Gene 2019, 710, 291–306. [Google Scholar] [CrossRef]
- Salih, H.; Gong, W.; Mkulama, M.; Du, X. Genome-wide characterization, identification, and expression analysis of the WD40 protein family in cotton. Genome 2018, 61, 539–547. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, X.; Zhong, M.; Dong, X.; Yu, D.; Jiang, X.; Wang, D.; Cui, W.; Chen, J.; Hu, J. Genome-wide identification of WD40 genes reveals a functional diversification of COP1-like genes in Rosaceae. Plant Mol. Biol. 2020, 104, 81–95. [Google Scholar] [CrossRef]
- Li, Q.; Zhao, P.; Li, J.; Zhang, C.; Wang, L. Genome-wide analysis of the WD-repeat protein family in cucumber and Arabidopsis. Mol. Gen. Genom. 2014, 289, 103–124. [Google Scholar] [CrossRef]
- Ouyang, Y.; Huang, X.; Lu, Z.; Yao, J. Genomic survey, expression profile and co-expression network analysis of OsWD40 family in rice. BMC Genom. 2012, 13, 100. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Muthamilarasan, M.; Khan, Y.; Parida, S.; Prasad, M. Genome-wide investigation and expression analyses of WD40 protein family in the model plant foxtail millet (Setaria italica L.). PLoS ONE 2014, 9, e86852. [Google Scholar] [CrossRef]
- DeVetten, N.; Quattrocchio, F.; Mol, J.; Koes, R. The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev. 1997, 11, 1422–1434. [Google Scholar] [CrossRef] [PubMed]
- Simlat, M.; Nowak, M.; Brutkowski, K.; Hydzik, M.; Zielinski, A.; Mos, M. Expression of the aldehyde oxidase 3, ent-copalyl diphosphate synthase, and VIVIPAROUS 1 genes in wheat cultivars differing in their susceptibility to pre-harvest sprouting. Span. J. Agric. Res. 2017, 15, e07019. [Google Scholar] [CrossRef]
- Yi, C.; Wang, H.; Wei, N.; Deng, X. An initial biochemical and cell biological characterization of the mammalian homologue of a central plant developmental switch, COP1. BMC Cell Biol. 2002, 3, 30. [Google Scholar] [CrossRef] [PubMed]
- Marine, J. Spotlight on the role of COP1 in tumorigenesis. Nat. Rev. Cancer 2012, 12, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Gu, X.; He, Y. Establishment of the winter-annual growth habit via FRIGIDA-mediated histone methylation at FLOWERING LOCUS C in Arabidopsis. Plant Cell 2009, 21, 1733–1746. [Google Scholar] [CrossRef]
- Jiang, D.; Kong, N.; Gu, X.; Li, Z.; He, Y. Arabidopsis COMPASS-like complexes mediate histone H3 Lysine-4 trimethylation to control floral transition and plant development. PLoS Genet. 2011, 7, e1001330. [Google Scholar] [CrossRef]
- Sompornpailin, K.; Makita, Y.; Yamazaki, M.; Saito, K. A WD-repeat-containing putative regulatory protein in anthocyanin biosynthesis in Perilla frutescens. Plant Mol. Biol. 2002, 50, 485–495. [Google Scholar] [CrossRef]
- Carey, C.; Strahle, J.; Selinger, D.; Chandler, V. Mutations in the pale aleurone color1 regulatory gene of the Zea mays anthocyanin pathway have distinct phenotypes relative to the functionally similar TRANSPARENT TESTA GLABRA1 gene in Arabidopsis thaliana. Plant Cell 2004, 16, 450–464. [Google Scholar] [CrossRef]
- An, X.H.; Tian, Y.; Chen, K.; Wang, X.; Hao, Y. The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. J. Plant Physiol. 2012, 169, 710–717. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Barragan, A.; Ochoa-Alejo, N. Virus-induced silencing of MYB and WD40 transcription factor genes affects the accumulation of anthocyanins in chilli pepper fruit. Biol. Plant 2014, 58, 567–574. [Google Scholar] [CrossRef]
- Dong, W.; Niu, L.; Gu, J.; Gao, F. Isolation of a WD40-repeat gene regulating anthocyanin biosynthesis in storage roots of purple-fleshed sweet potato. Acta Physiol. Plant 2014, 36, 1123–1132. [Google Scholar] [CrossRef]
- Yao, P.; Zhao, H.; Luo, X.; Gao, F.; Li, C.; Yao, H.; Chen, H.; Un Park, S.; Wu, Q. Fagopyrum tataricum FtWD40 functions as a positive regulator of anthocyanin biosynthesis in transgenic tobacco. J. Plant Growth Regul. 2017, 36, 755–765. [Google Scholar] [CrossRef]
- Yang, X.; Wang, J.; Xia, X.; Zhang, Z.; He, J.; Nong, B.; Luo, T.; Feng, R.; Wu, Y.; Pan, Y.; et al. OsTTG1, a WD40 repeat gene, regulates anthocyanin biosynthesis in rice. Plant J. 2021, 107, 198–214. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Salih, H.; Htet, N.N.W.; Azeem, F.; Zhan, R. Genomic analysis of WD40 protein family in the mango reveals a TTG1 protein enhances root growth and abiotic tolerance in Arabidopsis. Sci. Rep. 2021, 11, 2266. [Google Scholar] [CrossRef]
- Fan, Z.; Zhai, Y.; Wang, Y.; Zhang, L.; Song, M.; Flaishman, M.; Ma, H. Genome-Wide Analysis of Anthocyanin Biosynthesis Regulatory WD40 Gene FcTTG1 and Related Family in Ficus carica L. Front Plant Sci. 2022, 13, 948084. [Google Scholar] [CrossRef]
- Quattrocchio, F.; Wing, J.; van der Woude, K.; Souer, E.; de Vetten, N.; Mol, J.; Koes, R. Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower colour. Plant Cell 1999, 11, 1433–1444. [Google Scholar] [CrossRef]
- Morita, Y.; Saitoh, M.; Hoshino, A.; Nitasaka, E.; Iida, S. Isolation of cDNAs for R2R3-MYB, bHLH and WDR transcriptional regulators and identification of c and ca mutations conferring white flowers in the Japanese morning glory. Plant Cell Physiol. 2006, 47, 457–470. [Google Scholar] [CrossRef]
- Kaya, H.; Shibahara, K.; Taoka, K.; Iwabuchi, M.; Stillman, B.; Araki, T. FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell 2001, 104, 131–142. [Google Scholar] [CrossRef]
- Franks, R.; Wang, C.; Levin, J.; Liu, Z. SEUSS, a member of a novel family of plant regulatory proteins, represses floral homeotic gene expression with LEUNIG. Development 2002, 129, 253–263. [Google Scholar] [CrossRef]
- Sorensen, M.; Chaudhury, A.; Robert, H.; Bancharel, E.; Berger, F. Polycomb group genes control pattern formation in plant seed. Curr. Biol. 2001, 11, 277–281. [Google Scholar] [CrossRef]
- Li, H.; He, Z.; Lu, G.; Lee, S.; Alonso, J.; Ecker, J.; Luan, S. AWD40 domain cyclophilin interacts with histone H3 and functions in gene repression and organogenesis in Arabidopsis. Plant Cell 2007, 19, 2403–2416. [Google Scholar] [CrossRef] [PubMed]
- Ullah, H.; Chen, J.; Temple, B.; Boyes, D.; Alonso, J.; Davis, K.; Ecker, J.; Jones, A. The β-subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes. Plant Cell 2003, 15, 393–409. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Chen, Z.; Zhang, J.; Li, X.; Chen, G.; Li, X.; Wu, C. OsLIS-L1 encoding a lissencephaly type-1-like protein with WD40 repeats is required for plant height and male gametophyte formation in rice. Planta 2011, 235, 713–727. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kim, M.; Hong, W.; Moon, S.; Kim, E.; Silva, J.; Lee, J.; Lee, S.; Kim, S.; Park, S.; et al. GORI, encoding the WD40 domain protein, is required for pollen tube germination and elongation in rice. Plant J. 2021, 105, 1645–1664. [Google Scholar] [CrossRef]
- Chen, W.; Chen, L.; Zhang, X.; Yang, N.; Guo, J.; Wang, M.; Ji, S.; Zhao, X.; Yin, P.; Cai, L.; et al. Convergent selection of a WD40 protein that enhances grain yield in maize and rice. Science 2022, 375, eabg7985. [Google Scholar] [CrossRef]
- Pan, L.; Wang, M.; Yang, Y.; Chen, C.; Dai, H.; Zhang, Z.; Hua, B.; Miao, M. Whole-genome resequencing identified QTLs, candidate genes and Kompetitive Allele-Specific PCR markers associated with the large fruit of Atlantic Giant (Cucurbita maxima). Front Plant Sci. 2022, 13, 942004. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.; Frank, M.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Godoy, F.; Kühn, N.; Muñoz, M.; Marchandon, G.; Gouthu, S.; Deluc, L.; Delrot, S.; Lauvergeat, V.; Arce-Johnson, P. The role of auxin during early berry development in grapevine as revealed by transcript profiling from pollination to fruit set. Hortic. Res. 2021, 8, 140. [Google Scholar] [CrossRef]
- de Jong, M.; Mariani, C.; Vriezen, W. The role of auxin and gibberellin in tomato fruit set. J. Exp. Bot. 2009, 60, 1523–1532. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Yamamuro, C. Interplays between auxin and GA signaling coordinate early fruit development. Hortic. Res. 2022, 9, uhab078. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Gu, K.; Wang, J.; Yu, J.; Wang, X.; Zhang, S.; You, C.; Hu, D.; Hao, Y. BTB-BACK-TAZ domain protein MdBT2-mediated MdMYB73 ubiquitination negatively regulates malate accumulation and vacuolar acidification in apple. Hortic. Res. 2020, 7, 151. [Google Scholar] [CrossRef] [PubMed]
- Rosado-Souza, L.; Yokoyama, R.; Sonnewald, U.; Fernie, A. Understanding source-sink interactions: Progress in model plants and translational research to crops in controlled and field conditions. Mol. Plant 2022, 28, 96–121. [Google Scholar]
- Wang, W.; Yang, D.; Feldmann, K. EFO1 and EFO2, encoding putative WD-domain proteins, have overlapping and distinct roles in the regulation of vegetative development and flowering of Arabidopsis. J. Exp. Bot. 2011, 62, 1077–1088. [Google Scholar] [CrossRef]
- Shi, D.; Liu, J.; Xiang, Y.; Ye, D.; Sundaresan, V.; Yang, W. SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18S ribosomal RNA biogenesis. Plant Cell 2005, 17, 2340–2354. [Google Scholar] [CrossRef]
- Velanis, C.; Perera, P.; Thomson, B.; de Leau, E.; Liang, S.; Hartwig, B.; Förderer, A.; Thornton, H.; Arede, P.; Chen, J.; et al. The domesticated transposase ALP2 mediates formation of a novel Polycomb protein complex by direct interaction with MSI1, a core subunit of Polycomb Repressive Complex 2 (PRC2). PLoS Genet. 2020, 16, e1008681. [Google Scholar] [CrossRef]
- Zhang, Y.; Feng, S.; Chen, F.; Chen, H.; Wang, J.; McCall, C.; Xiong, Y.; Deng, X. Arabidopsis DDB1-CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes. Plant Cell 2008, 20, 1437–1455. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.; Yang, Y.; Pan, L.; Xia, W.; Xu, L.; Hua, B.; Zhang, Z.; Miao, M. Genome-Wide Identification of WD40 Proteins in Cucurbita maxima Reveals Its Potential Functions in Fruit Development. Genes 2023, 14, 220. https://doi.org/10.3390/genes14010220
Chen C, Yang Y, Pan L, Xia W, Xu L, Hua B, Zhang Z, Miao M. Genome-Wide Identification of WD40 Proteins in Cucurbita maxima Reveals Its Potential Functions in Fruit Development. Genes. 2023; 14(1):220. https://doi.org/10.3390/genes14010220
Chicago/Turabian StyleChen, Chen, Yating Yang, Liu Pan, Wenhao Xia, Lanruoyan Xu, Bing Hua, Zhiping Zhang, and Minmin Miao. 2023. "Genome-Wide Identification of WD40 Proteins in Cucurbita maxima Reveals Its Potential Functions in Fruit Development" Genes 14, no. 1: 220. https://doi.org/10.3390/genes14010220
APA StyleChen, C., Yang, Y., Pan, L., Xia, W., Xu, L., Hua, B., Zhang, Z., & Miao, M. (2023). Genome-Wide Identification of WD40 Proteins in Cucurbita maxima Reveals Its Potential Functions in Fruit Development. Genes, 14(1), 220. https://doi.org/10.3390/genes14010220




