Transcriptome Analysis of CYP450 Family Members in Fritillaria cirrhosa D. Don and Profiling of Key CYP450s Related to Isosteroidal Alkaloid Biosynthesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials, RNA Extraction, and Sequencing
2.2. Bioinformatics Analysis
2.3. Identification, Sequence Analyses and Phylogenetic of FcCYPs
2.4. RNA Extraction and qRT-PCR Analysis
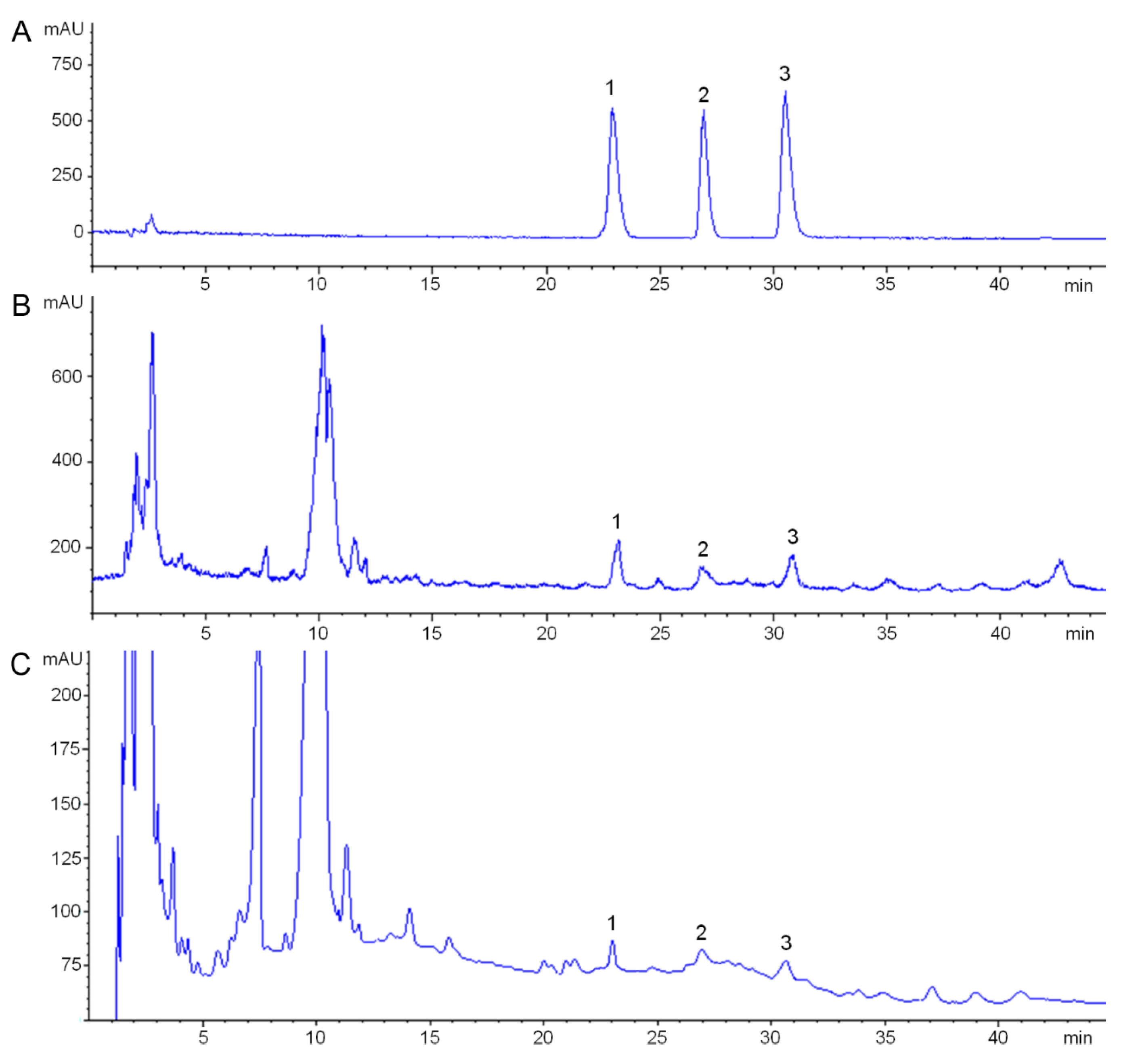
2.5. Determination of Alkaloids by HPLC-ELSD
2.6. Subcellular Localization Analysis
3. Results
3.1. SMRT Sequencing, Similarity Analysis, and Functional Annotation
3.2. Identification and Analyses of FcCYPs Sequences
3.3. Identification of Candidates CYPs Involved in ISA Biosynthesis
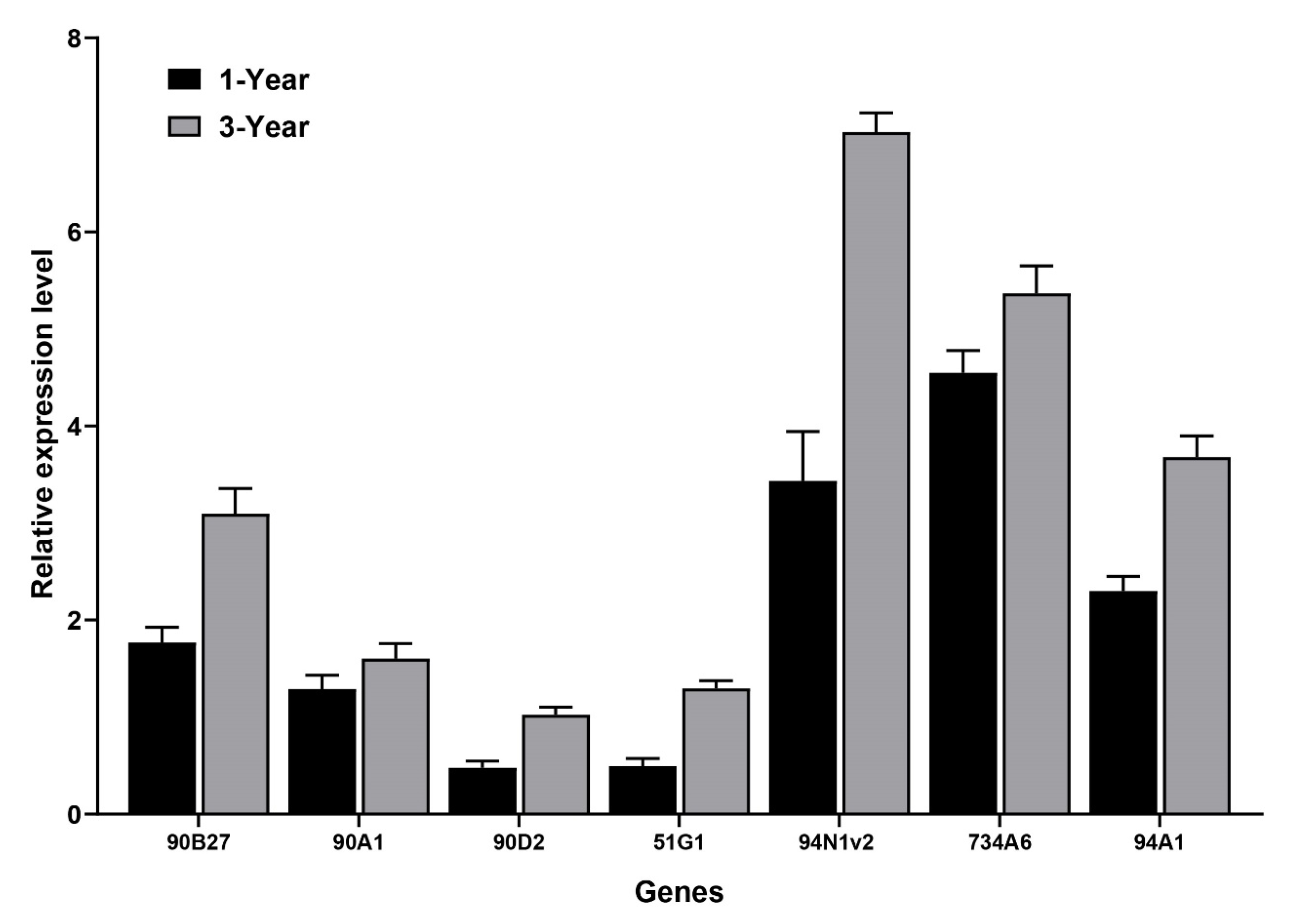
3.4. Analysis of Candidate CYPs Gene Expression and Key FISA Accumulation in Bulbs of Different Ages
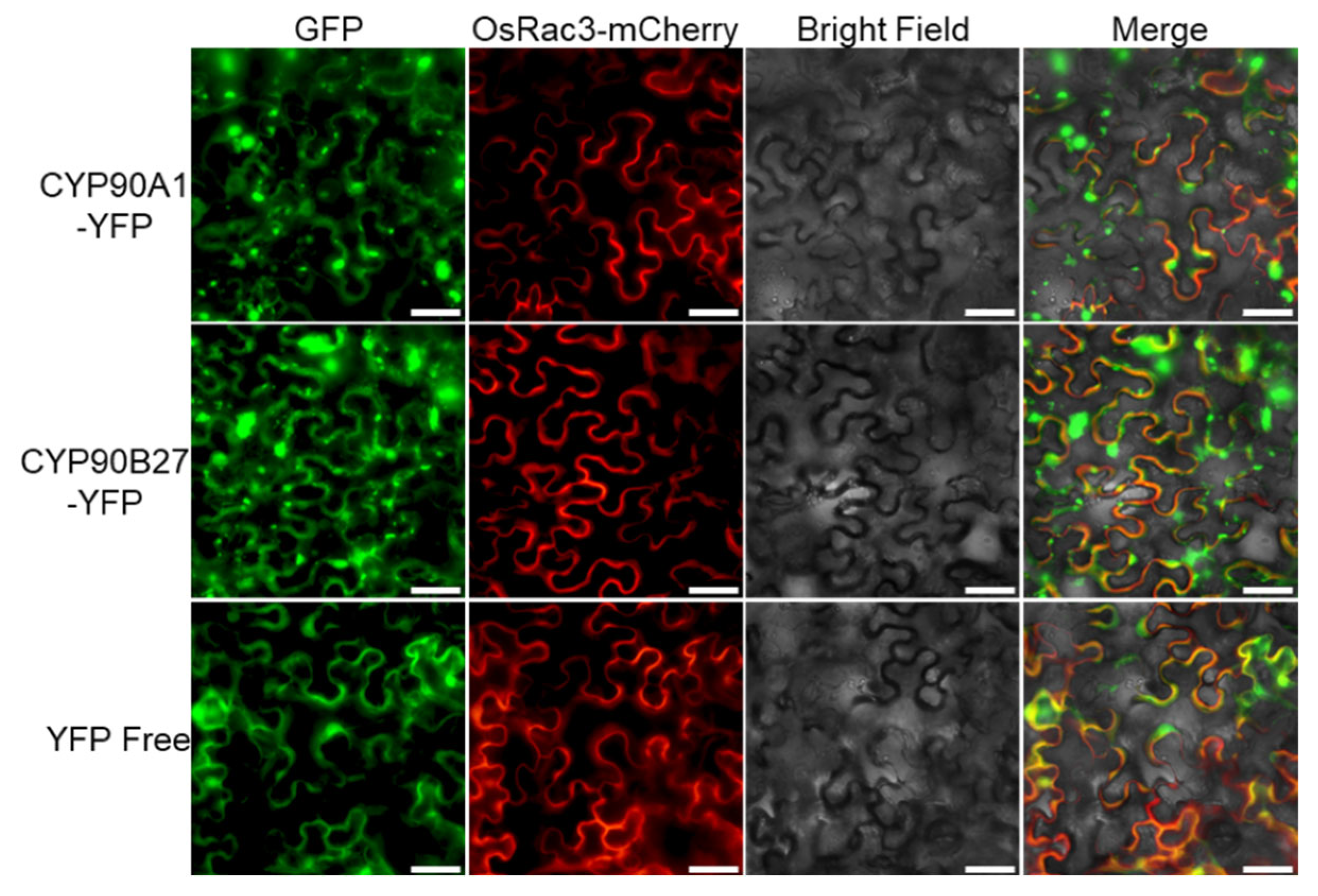
3.5. Assessment of the Subcellular Localization of Two CYP450 Fusion Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Da-Cheng, H.; Xiao-Jie, G.; Pei-Gen, X.; Yong, P. Phytochemical and biological research of Fritillaria medicine resources. Chin. J. Nat. Med. 2013, 11, 330–344. [Google Scholar]
- Cunningham, A.; Brinckmann, J.; Pei, S.-J.; Luo, P.; Schippmann, U.; Long, X.; Bi, Y.-F. High altitude species, high profits: Can the trade in wild harvested Fritillaria cirrhosa (Liliaceae) be sustained? Chin. J. Nat. Med. 2018, 223, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Quan, Y.; Li, L.; Yin, Z.; Chen, S.; Yi, J.; Lang, J.; Zhang, L.; Yue, Q.; Zhao, J. Bulbus Fritillariae cirrhosae as a Respiratory Medicine: Is There a Potential Drug in the Treatment of COVID-19? Front Pharmacol. 2021, 12, 784335. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhu, J.; Wang, S.; Wang, X.; Ou, Y.; Wei, D.; Li, X. Antitussive, expectorant and anti-inflammatory alkaloids from Bulbus Fritillariae cirrhosae. Chin. J. Nat. Med. 2011, 82, 1290–1294. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Yang, J.; Du, Q.; Li, H.; Wang, S. The total alkaloid fraction of bulbs of Fritillaria cirrhosa displays anti-inflammatory activity and attenuates acute lung injury. Front. Pharmacol. 2016, 193, 150–158. [Google Scholar] [CrossRef]
- Wang, D.; Wang, S.; Feng, Y.; Zhang, L.; Li, Z.; Ma, J.; Luo, Y.; Xiao, W. Antitumor effects of Bulbus Fritillariae cirrhosae on Lewis lung carcinoma cells in vitro and in vivo. Chin. J. Nat. Med. 2014, 54, 92–101. [Google Scholar] [CrossRef]
- Liu, S.; Yang, T.; Ming, T.W.; Gaun, T.K.W.; Zhou, T.; Wang, S.; Ye, B. Isosteroid alkaloids with different chemical structures from Fritillariae cirrhosae bulbus alleviate LPS-induced inflammatory response in RAW 264.7 cells by MAPK signaling pathway. Chin. J. Nat. Med. 2020, 78, 106047. [Google Scholar] [CrossRef]
- Wang, Y.; Hou, H.; Ren, Q.; Hu, H.; Yang, T.; Li, X. Natural drug sources for respiratory diseases from Fritillaria: Chemical and biological analyses. Chin. Med. 2021, 16, 40. [Google Scholar] [CrossRef]
- Wang, D.; Wang, S.; Du, Q.; Wang, N.; Liu, S.; Wang, X.; Jiang, J. Optimization of extraction and enrichment of steroidal alkaloids from bulbs of cultivated Fritillaria cirrhosa. Biomed Res. Int. 2014, 2014, 258402. [Google Scholar]
- Mathela, M.; Kumar, A.; Sharma, M.; Goraya, G.S. Hue and cry for Fritillaria cirrhosa D. Don, a threatened medicinal plant in the Western Himalaya. Discovery 2021, 2, 38. [Google Scholar]
- Chao, S.; Yongqiao, S.; Jingyuan, S.; Chenji, L.; Xiwen, L.; Xiaowei, Z.; Ying, L.; Songnian, H.; Hongmei, L.; Yingjie, Z. Discovery of genes related to steroidal alkaloid biosynthesis in Fritillaria cirrhosa by generating and mining a dataset of expressed sequence tags (ESTs). Plant Physiol. 2011, 5, 5307–5314. [Google Scholar]
- Zhao, Q.; Li, R.; Zhang, Y.; Huang, K.; Wang, W.; Li, J. Transcriptome analysis reveals in vitro-cultured regeneration bulbs as a promising source for targeted Fritillaria cirrhosa steroidal alkaloid biosynthesis. 3 Biotech 2018, 8, 191. [Google Scholar] [CrossRef]
- Kumar, P.; Acharya, V.; Warghat, A.R.J.P. Comparative transcriptome analysis infers bulb derived in vitro cultures as a promising source for sipeimine biosynthesis in Fritillaria cirrhosa D. Don (Liliaceae, syn. Fritillaria roylei Hook.)-High value Himalayan medicinal herb. Chin. J. Nat. Med. 2021, 183, 112631. [Google Scholar] [CrossRef]
- Lu, Q.; Li, R.; Liao, J.; Hu, Y.; Gao, Y.; Wang, M.; Li, J.; Zhao, Q. Integrative analysis of the steroidal alkaloids distribution and biosynthesis of bulbs Fritillariae cirrhosae through metabolome and transcriptome analyses. BMC Genomics 2022, 23, 511. [Google Scholar] [CrossRef]
- Qu, A.; Wu, Q.; Su, J.; Li, C.; Yang, L.; Wang, Z.; Wang, Z.; Li, Z.; Ruan, X.; Zhao, Y. A Review on the Composition and Biosynthesis of Alkaloids and on the Taxonomy, Domestication, and Cultivation of Medicinal Fritillaria Species. Agron. J. 2022, 12, 1844. [Google Scholar] [CrossRef]
- Li, H.-J.; Jiang, Y.; Li, P. Chemistry, bioactivity and geographical diversity of steroidal alkaloids from the Liliaceae family. Nat. Prod. Rep. 2006, 23, 735–752. [Google Scholar] [CrossRef]
- Nelson, D.R. The cytochrome p450 homepage. Hum. Genomics 2009, 4, 59. [Google Scholar] [CrossRef]
- Ma, B.; Ma, J.; Li, B.; Tao, Q.; Gan, J.; Yan, Z. Nutrition. Effects of different harvesting times and processing methods on the quality of cultivated Fritillaria cirrhosa D. Don. Food Sci. Nutr. 2021, 9, 2853–2861. [Google Scholar] [CrossRef]
- Tao, Y.; Zou, T.; Zhang, X.; Liu, R.; Chen, H.; Yuan, G.; Zhou, D.; Xiong, P.; He, Z.; Li, G. Secretory lipid transfer protein OsLTPL94 acts as a target of EAT1 and is required for rice pollen wall development. Plant J. 2021, 108, 358–377. [Google Scholar] [CrossRef]
- Malhotra, K.; Franke, J. Cytochrome P450 monooxygenase-mediated tailoring of triterpenoids and steroids in plants. Beilstein J. Org. Chem. 2022, 18, 1289–1310. [Google Scholar] [CrossRef]
- Christ, B.; Xu, C.; Xu, M.; Li, F.-S.; Wada, N.; Mitchell, A.J.; Han, X.-L.; Wen, M.-L.; Fujita, M.; Weng, J.-K. Repeated evolution of cytochrome P450-mediated spiroketal steroid biosynthesis in plants. Nat. Commun. 2019, 10, 3206. [Google Scholar] [CrossRef] [PubMed]
- Hansen, C.C.; Nelson, D.R.; Møller, B.L.; Werck-Reichhart, D. Plant cytochrome P450 plasticity and evolution. Mol. Plant 2021, 14, 1244–1265. [Google Scholar] [CrossRef] [PubMed]
- Szeliga, M.; Ciura, J.; Tyrka, M. Representational difference analysis of transcripts involved in jervine biosynthesis. Life 2020, 10, 88. [Google Scholar] [CrossRef] [PubMed]
- Hua, X.; Song, W.; Wang, K.; Yin, X.; Hao, C.; Duan, B.; Xu, Z.; Su, T.; Xue, Z. Effective prediction of biosynthetic pathway genes involved in bioactive polyphyllins in Paris polyphylla. Commun. Biol. 2022, 5, 50. [Google Scholar] [CrossRef]
- Sonawane, P.D.; Pollier, J.; Panda, S.; Szymanski, J.; Massalha, H.; Yona, M.; Unger, T.; Malitsky, S.; Arendt, P.; Pauwels, L. Plant cholesterol biosynthetic pathway overlaps with phytosterol metabolism. Nat. Plants 2016, 3, 16205. [Google Scholar] [CrossRef]
- Augustin, M.M.; Ruzicka, D.R.; Shukla, A.K.; Augustin, J.M.; Starks, C.M.; O′Neil-Johnson, M.; McKain, M.R.; Evans, B.S.; Barrett, M.D.; Smithson, A. Elucidating steroid alkaloid biosynthesis in Veratrum californicum: Production of verazine in Sf9 cells. Plant J. 2015, 82, 991–1003. [Google Scholar] [CrossRef]
- Sakamoto, T.; Ohnishi, T.; Fujioka, S.; Watanabe, B.; Mizutani, M. Rice CYP90D2 and CYP90D3 catalyze C-23 hydroxylation of brassinosteroids in vitro. Plant Physiol. Biochem. 2012, 58, 220–226. [Google Scholar] [CrossRef]
- Itkin, M.; Heinig, U.; Tzfadia, O.; Bhide, A.; Shinde, B.; Cardenas, P.; Bocobza, S.; Unger, T.; Malitsky, S.; Finkers, R. Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 2013, 341, 175–179. [Google Scholar] [CrossRef]
- Schuler, M.A.; Duan, H.; Bilgin, M.; Ali, S. Arabidopsis cytochrome P450s through the looking glass: A window on plant biochemistry. Phytochem. Rev. 2006, 5, 205–237. [Google Scholar] [CrossRef]
- Magwanga, R.O.; Lu, P.; Kirungu, J.N.; Dong, Q.; Cai, X.; Zhou, Z.; Wang, X.; Hou, Y.; Xu, Y.; Peng, R. Knockdown of cytochrome P450 genes Gh_D07G1197 and Gh_A13G2057 on chromosomes D07 and A13 reveals their putative role in enhancing drought and salt stress tolerance in Gossypium hirsutum. Genes 2019, 10, 226. [Google Scholar] [CrossRef]
- Chen, T.; Zhong, F.; Yao, C.; Chen, J.; Xiang, Y.; Dong, J.; Yan, Z.; Ma, Y.J.; Medicine, A. A systematic review on traditional uses, sources, phytochemistry, pharmacology, pharmacokinetics, and toxicity of Fritillariae cirrhosae bulbus. Evid. Based Complement Alternat. Med. 2020, 2020, 1536534. [Google Scholar] [CrossRef]
- Kelly, L.J.; Renny-Byfield, S.; Pellicer, J.; Macas, J.; Novák, P.; Neumann, P.; Lysak, M.A.; Day, P.D.; Berger, M.; Fay, M.F. Analysis of the giant genomes of Fritillaria (Liliaceae) indicates that a lack of DNA removal characterizes extreme expansions in genome size. New Phytol. 2015, 208, 596–607. [Google Scholar] [CrossRef]
- Peng, R.; Ma, P.; Mo, R.; Sun, N.J. Analysis of the bioactive components from different growth stages of Fritillaria taipaiensis PY Li. Acta Pharm. Sin. B 2013, 3, 167–173. [Google Scholar] [CrossRef]
- Geng, Z.; Liu, Y.; Gou, Y.; Zhou, Q.; He, C.; Guo, L.; Zhou, J.; Xiong, L.J. Metabolomics study of cultivated Bulbus Fritillariae cirrhosae at different growth stages using UHPLC-QTOF-MS coupled with multivariate data analysis. Phytochem. Anal. 2018, 29, 290–299. [Google Scholar] [CrossRef]
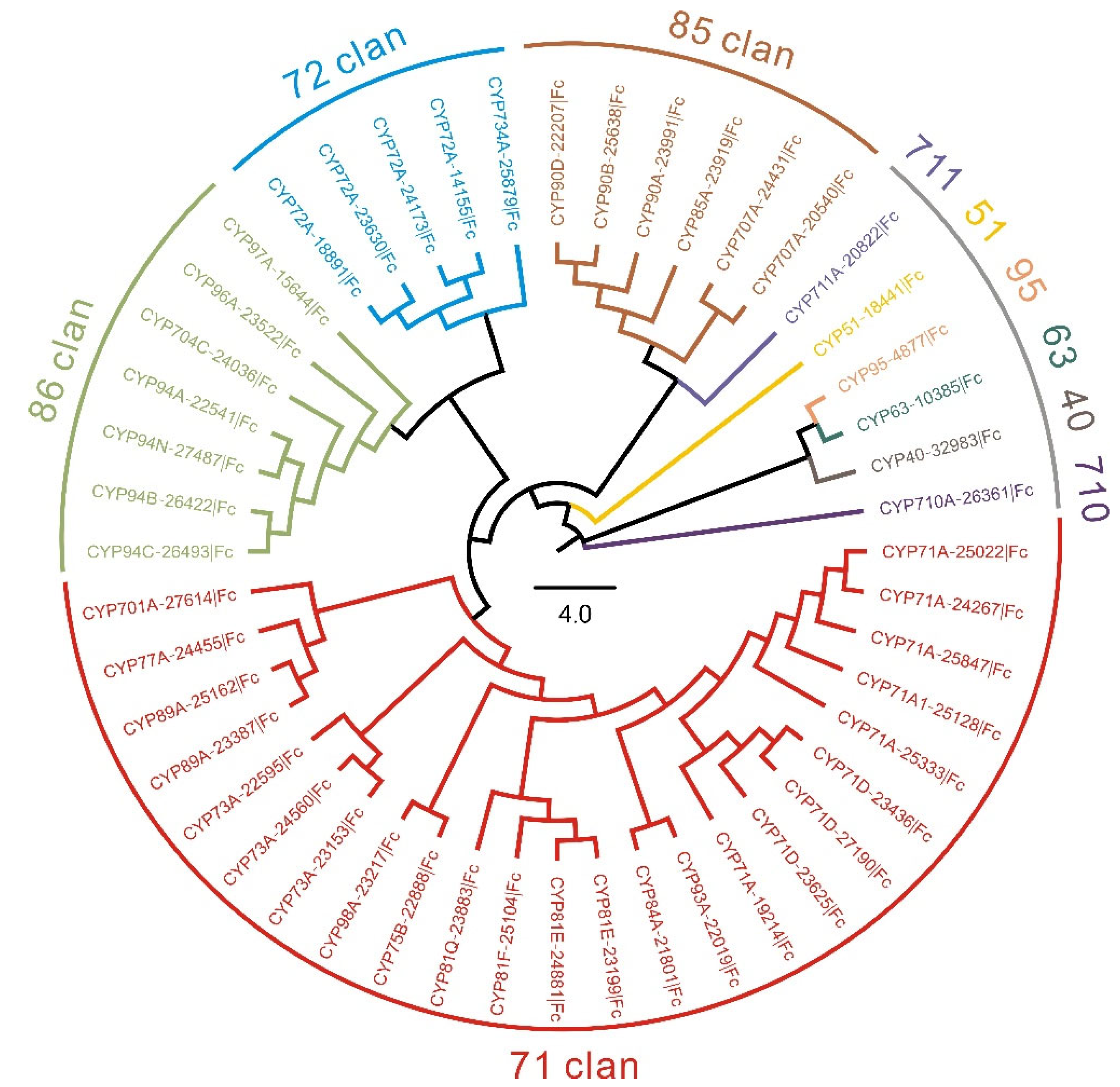
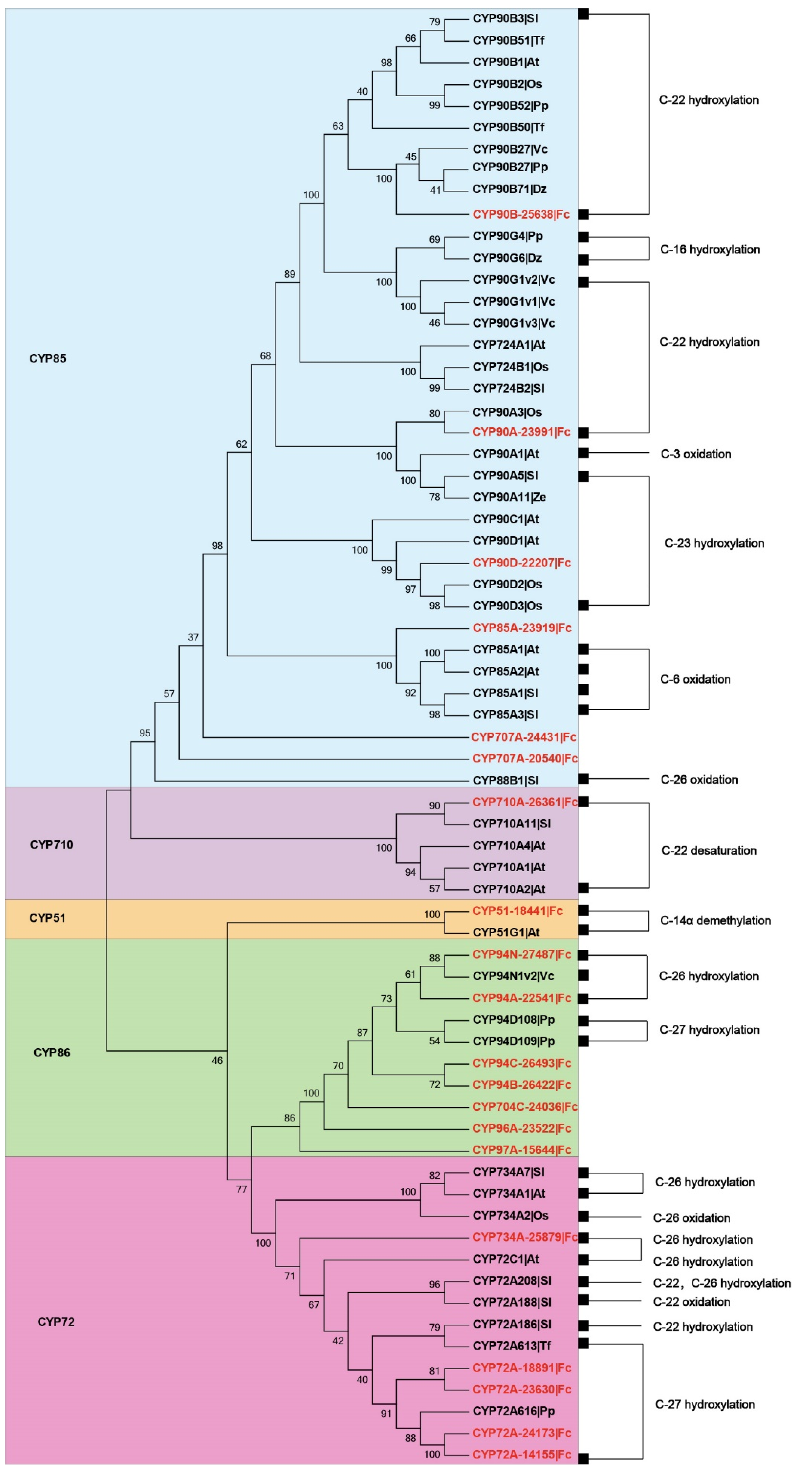
| No | Gene ID | Clan | Family | Subfamily | Amino Acid Residues | MW (kDa) | pI | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|
| 1 | 27614 | 71 | 701 | CYP701A6 | 429 | 49,317.84 | 5.73 | cytoplasmic |
| 2 | 25847 | 71 | 71 | CYP71A1 | 495 | 56,263.57 | 6.78 | chloroplast |
| 3 | 25128 | 71 | 71 | CYP71A1 | 496 | 56,013.99 | 8.22 | chloroplast |
| 4 | 25333 | 71 | 71 | CYP71A2 | 511 | 56,277.12 | 6.89 | chloroplast |
| 5 | 24267 | 71 | 71 | CYP71A25 | 503 | 57,395.89 | 7.69 | chloroplast |
| 6 | 25022 | 71 | 71 | CYP71A4 | 498 | 56,964.15 | 7.72 | vacular membrane |
| 7 | 19214 | 71 | 71 | CYP71A9 | 490 | 55,311.46 | 8.42 | chloroplast |
| 8 | 23625 | 71 | 71 | CYP71D381 | 495 | 55,669.95 | 9.04 | chloroplast |
| 9 | 23436 | 71 | 71 | CYP71D55 | 510 | 58,297.56 | 9.08 | chloroplast |
| 10 | 27190 | 71 | 71 | CYP71D8 | 498 | 56,365.75 | 6.48 | chloroplast |
| 11 | 24560 | 71 | 73 | CYP73A1 | 505 | 58,138.45 | 8.52 | plasma membrane |
| 12 | 22595 | 71 | 73 | CYP73A100 | 518 | 58,650.4 | 8.6 | plasma membrane |
| 13 | 23153 | 71 | 73 | CYP73A16 | 504 | 57,796.28 | 9.24 | plasma membrane |
| 14 | 22888 | 71 | 75 | CYP75B137 | 503 | 55,336.92 | 6.88 | chloroplast |
| 15 | 23199 | 71 | 81 | CYP81E8 | 507 | 56,385.25 | 7.15 | chloroplast |
| 16 | 24881 | 71 | 81 | CYP81E8 | 509 | 57,543.15 | 9.06 | chloroplast |
| 17 | 25104 | 71 | 81 | CYP81F3 | 494 | 55,327.71 | 6.19 | chloroplast |
| 18 | 23883 | 71 | 81 | CYP81Q32 | 513 | 57,720.7 | 7.26 | chloroplast |
| 19 | 21801 | 71 | 84 | CYP84A1 | 515 | 57,470.57 | 6.44 | chloroplast |
| 20 | 22019 | 71 | 93 | CYP93A3 | 502 | 56,432.31 | 5.96 | chloroplast |
| 21 | 25162 | 71 | 89 | CYP89A2 | 507 | 57,250.83 | 8.82 | chloroplast |
| 22 | 23387 | 71 | 89 | CYP89A9 | 498 | 56,047.58 | 8.41 | chloroplast |
| 23 | 23217 | 71 | 98 | CYP98A2 | 510 | 57,981.92 | 8.61 | chloroplast |
| 24 | 14155 | 72 | 72 | CYP72A14 | 522 | 59,146.97 | 9.19 | chloroplast |
| 25 | 18891 | 72 | 72 | CYP72A15 | 516 | 59,380.97 | 8.72 | cytoplasmic |
| 26 | 23630 | 72 | 72 | CYP72A219 | 518 | 59,496.39 | 9.47 | chloroplast |
| 27 | 24173 | 72 | 72 | CYP72A616 | 522 | 59,004.81 | 9.37 | chloroplast |
| 28 | 25879 | 72 | 734 | CYP734A6 | 515 | 58,157.25 | 9.44 | chloroplast |
| 29 | 20540 | 85 | 707 | CYP707A1 | 468 | 52,762.44 | 9.15 | chloroplast |
| 30 | 24431 | 85 | 707 | CYP707A2 | 470 | 53,332.54 | 8.96 | chloroplast |
| 31 | 23919 | 85 | 85 | CYP85A1 | 468 | 53,353.24 | 8.65 | chloroplast |
| 32 | 23991 | 85 | 90 | CYP90A1 | 510 | 57,539.32 | 8.7 | cytoplasmic |
| 33 | 25638 | 85 | 90 | CYP90B27 | 483 | 54,036.79 | 8.93 | chloroplast |
| 34 | 22207 | 85 | 90 | CYP90D2 | 482 | 54,659.17 | 8.11 | chloroplast |
| 35 | 24036 | 86 | 704 | CYP704C1 | 507 | 58,078.96 | 8.42 | chloroplast |
| 36 | 24455 | 71 | 77 | CYP77A4 | 509 | 57,610.25 | 8.77 | chloroplast |
| 37 | 22541 | 86 | 94 | CYP94A1 | 508 | 57,163.09 | 9.09 | chloroplast |
| 38 | 27487 | 86 | 94 | CYP94N1 | 499 | 55,985.22 | 8.47 | chloroplast |
| 39 | 26422 | 86 | 94 | CYP94B1 | 489 | 54,906.75 | 8.32 | chloroplast |
| 40 | 26493 | 86 | 94 | CYP94C1 | 495 | 55,621.93 | 6.43 | chloroplast |
| 41 | 23522 | 86 | 96 | CYP96A15 | 499 | 56,957.64 | 9.08 | chloroplast |
| 42 | 15644 | 86 | 97 | CYP97A3 | 614 | 68,383.46 | 5.86 | chloroplast |
| 43 | 4877 | 95 | 95 | CYP95 | 853 | 94,883.01 | 11.48 | nucleus |
| 44 | 26361 | 710 | 710 | CYP710A11 | 495 | 56,010.29 | 7.19 | plasma membrane |
| 45 | 20822 | 711 | 711 | CYP711A1 | 526 | 58,970.43 | 9.31 | plasma membrane |
| 46 | 32983 | 40 | 40 | CYP40 | 369 | 40,848.45 | 5.49 | cytoskeleton |
| 47 | 18441 | 51 | 51 | CYP51 | 488 | 55,580.08 | 8.13 | chloroplast |
| 48 | 10385 | 63 | 63 | CYP63 | 689 | 76,552.99 | 10.67 | nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, R.; Xiao, M.; Li, J.; Zhao, Q.; Wang, M.; Zhu, Z. Transcriptome Analysis of CYP450 Family Members in Fritillaria cirrhosa D. Don and Profiling of Key CYP450s Related to Isosteroidal Alkaloid Biosynthesis. Genes 2023, 14, 219. https://doi.org/10.3390/genes14010219
Li R, Xiao M, Li J, Zhao Q, Wang M, Zhu Z. Transcriptome Analysis of CYP450 Family Members in Fritillaria cirrhosa D. Don and Profiling of Key CYP450s Related to Isosteroidal Alkaloid Biosynthesis. Genes. 2023; 14(1):219. https://doi.org/10.3390/genes14010219
Chicago/Turabian StyleLi, Rui, Maotao Xiao, Jian Li, Qi Zhao, Mingcheng Wang, and Ziwei Zhu. 2023. "Transcriptome Analysis of CYP450 Family Members in Fritillaria cirrhosa D. Don and Profiling of Key CYP450s Related to Isosteroidal Alkaloid Biosynthesis" Genes 14, no. 1: 219. https://doi.org/10.3390/genes14010219
APA StyleLi, R., Xiao, M., Li, J., Zhao, Q., Wang, M., & Zhu, Z. (2023). Transcriptome Analysis of CYP450 Family Members in Fritillaria cirrhosa D. Don and Profiling of Key CYP450s Related to Isosteroidal Alkaloid Biosynthesis. Genes, 14(1), 219. https://doi.org/10.3390/genes14010219








