Skin Phototype and Disease: A Comprehensive Genetic Approach to Pigmentary Traits Pleiotropy Using PRS in the GCAT Cohort
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Phenotype Data Collection
2.3. Genetic Variants
2.4. Electronic Health Records Disease Prevalence and Phototype
2.5. Genome-Wide Association Analysis of Phototype
2.6. Trait Heritability and Pairwise Genetic Correlations
2.7. EHR Phenotype-Wide Association
2.8. Polygenic Risk Scores Association
3. Results
3.1. Systematic Search on Clinical Data (EHR) Identifies Diseases Associated with Phototype in the GCAT Cohort
3.2. GWAS Analysis Identifies Thousand Variants Associated with 10 Pigmentary Traits in the GCAT Cohort
3.3. Pigmentary Traits Correlation Are Partly Explained by Common Genetics
3.4. Genes Containing SNPs Associated with Pigmentary Traits, Show an Overrepresentation of Pigmentary Metabolic Processes, Skin, and Eye-Related Diseases
3.5. Analysis of Structural Variants Identified Small Intronic Deletions as the Most Common Structural Variation Associated with Eight Pigmentary Traits
3.6. PheWAS Analysis Confirmed the Pleiotropy among Pigmentary Traits Genes and Related Diseases
3.7. Polygenic Risk Scores Confirmed the Association of Pigmentary Traits and Eye Diseases
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, Y.; Hong, L.; Wakamatsu, K.; Ito, S.; Adhyaru, B.; Cheng, C.Y.; Bowers, C.R.; Simon, J.D. Comparison of structural and chemical properties of black and red human hair melanosomes. Photochem. Photobiol. 2005, 81, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Matts, P.J.; Dykes, P.J.; Marks, R. The distribution of melanin in skin determined in vivo. Br. J. Dermatol. 2007, 156, 620–628. [Google Scholar] [CrossRef] [PubMed]
- Sturm, R.A.; Frudakis, T.N. Eye color: Portals into pigmentation genes and ancestry. Trends Genet. 2004, 20, 327–332. [Google Scholar] [CrossRef]
- Thong, H.Y.; Jee, S.H.; Sun, C.C.; Boissy, R.E. The patterns of melanosome distribution in keratinocytes of human skin as one determining factor of skin color. Br. J. Dermatol. 2003, 149, 498–505. [Google Scholar] [CrossRef] [PubMed]
- Wielgus, A.R.; Sarna, T. Melanin in human irides of different color and age of donors. Pigment Cell Res. 2005, 18, 454–464. [Google Scholar] [CrossRef]
- Graf, J.; Hodgson, R.; van Daal, A. Single nucleotide polymorphisms in the MATP gene are associated with normal human pigmentation variation. Hum. Mutat. 2005, 25, 278–284. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Brenner, M.; Hearing, V.J. The regulation of skin pigmentation. J. Biol. Chem. 2007, 282, 27557–27561. [Google Scholar] [CrossRef]
- Wilson, P.D.; Kligman, A.M. Do freckles protect the skin from actinic damage? Br. J. Dermatol. 1982, 106, 27–32. [Google Scholar] [CrossRef]
- Cordero, R.J.B.; Casadevall, A. Melanin. Curr. Biol. 2020, 30, R142–R143. [Google Scholar] [CrossRef]
- Sakai, D.; Takagi, S.; Totani, K.; Yamamoto, M.; Matsuzaki, M.; Yamanari, M.; Sugiyama, S.; Yokota, S.; Maeda, A.; Hirami, Y.; et al. Retinal pigment epithelium melanin imaging using polarization-sensitive optical coherence tomography for patients with retinitis pigmentosa. Sci. Rep. 2022, 12, 7115. [Google Scholar] [CrossRef]
- Scherer, D.; Kumar, R. Genetics of pigmentation in skin cancer—A review. Mutat. Res. 2010, 705, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Hysi, P.G.; Valdes, A.M.; Liu, F.; Furlotte, N.A.; Evans, D.M.; Bataille, V.; Visconti, A.; Hemani, G.; McMahon, G.; Ring, S.M.; et al. Genome-wide association meta-analysis of individuals of European ancestry identifies new loci explaining a substantial fraction of hair color variation and heritability. Nat. Genet. 2018, 50, 652–656. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Xu, S. Adaptation of human skin color in various populations. Hereditas 2018, 155, 1. [Google Scholar] [CrossRef]
- Lao, O.; de Gruijter, J.M.; van Duijn, K.; Navarro, A.; Kayser, M. Signatures of positive selection in genes associated with human skin pigmentation as revealed from analyses of single nucleotide polymorphisms. Ann. Hum. Genet. 2007, 71, 354–369. [Google Scholar] [CrossRef] [PubMed]
- Makova, K.; Norton, H. Worldwide polymorphism at the MC1R locus and normal pigmentation variation in humans. Peptides 2005, 26, 1901–1908. [Google Scholar] [CrossRef] [PubMed]
- Valverde, P.; Healy, E.; Jackson, I.; Rees, J.L.; Thody, A.J. Variants of the melanocyte-stimulating hormone receptor gene are associated with red hair and fair skin in humans. Nat. Genet. 1995, 11, 328–330. [Google Scholar] [CrossRef]
- Morgan, M.D.; Pairo-Castineira, E.; Rawlik, K.; Canela-Xandri, O.; Rees, J.; Sims, D.; Tenesa, A.; Jackson, I.J. Genome-wide study of hair color in UK Biobank explains most of the SNP heritability. Nat. Commun. 2018, 9, 5271. [Google Scholar] [CrossRef]
- White, D.; Rabago-Smith, M. Genotype-phenotype associations and human eye color. J. Hum. Genet. 2011, 56, 5–7. [Google Scholar] [CrossRef]
- Lamason, R.L.; Mohideen, M.A.; Mest, J.R.; Wong, A.C.; Norton, H.L.; Aros, M.C.; Jurynec, M.J.; Mao, X.; Humphreville, V.R.; Humbert, J.E.; et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 2005, 310, 1782–1786. [Google Scholar] [CrossRef]
- Adhikari, K.; Fontanil, T.; Cal, S.; Mendoza-Revilla, J.; Fuentes-Guajardo, M.; Chacon-Duque, J.C.; Al-Saadi, F.; Johansson, J.A.; Quinto-Sanchez, M.; Acuna-Alonzo, V.; et al. A genome-wide association scan in admixed Latin Americans identifies loci influencing facial and scalp hair features. Nat. Commun. 2016, 7, 10815. [Google Scholar] [CrossRef]
- Liu, F.; Wen, B.; Kayser, M. Colorful DNA polymorphisms in humans. Semin. Cell Dev. Biol. 2013, 24, 562–575. [Google Scholar] [CrossRef] [PubMed]
- Meyer, O.S.; Lunn, M.M.B.; Garcia, S.L.; Kjaerbye, A.B.; Morling, N.; Borsting, C.; Andersen, J.D. Association between brown eye color in rs12913832:GG individuals and SNPs in TYR, TYRP1, and SLC24A4. PLoS ONE 2020, 15, e0239131. [Google Scholar] [CrossRef] [PubMed]
- Nan, H.; Kraft, P.; Hunter, D.J.; Han, J. Genetic variants in pigmentation genes, pigmentary phenotypes, and risk of skin cancer in Caucasians. Int. J. Cancer 2009, 125, 909–917. [Google Scholar] [CrossRef] [PubMed]
- MacArthur, J.; Bowler, E.; Cerezo, M.; Gil, L.; Hall, P.; Hastings, E.; Junkins, H.; McMahon, A.; Milano, A.; Morales, J.; et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017, 45, D896–D901. [Google Scholar] [CrossRef]
- Gomez-Acebo, I.; Dierssen-Sotos, T.; Palazuelos, C.; Fernandez-Navarro, P.; Castano-Vinyals, G.; Alonso-Molero, J.; Urtiaga, C.; Fernandez-Villa, T.; Ardanaz, E.; Rivas-Del-Fresno, M.; et al. Pigmentation phototype and prostate and breast cancer in a select Spanish population-A Mendelian randomization analysis in the MCC-Spain study. PLoS ONE 2018, 13, e0201750. [Google Scholar] [CrossRef] [PubMed]
- Hernando, B.; Sanz-Page, E.; Pitarch, G.; Mahiques, L.; Valcuende-Cavero, F.; Martinez-Cadenas, C. Genetic variants associated with skin photosensitivity in a southern European population from Spain. Photodermatol. Photoimmunol. Photomed. 2018, 34, 415–422. [Google Scholar] [CrossRef]
- Rocha, J. The Evolutionary History of Human Skin Pigmentation. J. Mol. Evol. 2020, 88, 77–87. [Google Scholar] [CrossRef]
- Beleza, S.; Santos, A.M.; McEvoy, B.; Alves, I.; Martinho, C.; Cameron, E.; Shriver, M.D.; Parra, E.J.; Rocha, J. The timing of pigmentation lightening in Europeans. Mol. Biol. Evol. 2013, 30, 24–35. [Google Scholar] [CrossRef]
- Jablonski, N.G.; Chaplin, G. Human skin pigmentation, migration and disease susceptibility. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012, 367, 785–792. [Google Scholar] [CrossRef]
- Amos, C.I.; Wang, L.E.; Lee, J.E.; Gershenwald, J.E.; Chen, W.V.; Fang, S.; Kosoy, R.; Zhang, M.; Qureshi, A.A.; Vattathil, S.; et al. Genome-wide association study identifies novel loci predisposing to cutaneous melanoma. Hum. Mol. Genet. 2011, 20, 5012–5023. [Google Scholar] [CrossRef]
- Duffy, D.L.; Zhao, Z.Z.; Sturm, R.A.; Hayward, N.K.; Martin, N.G.; Montgomery, G.W. Multiple pigmentation gene polymorphisms account for a substantial proportion of risk of cutaneous malignant melanoma. J. Investig. Dermatol. 2010, 130, 520–528. [Google Scholar] [CrossRef] [PubMed]
- Ibarrola-Villava, M.; Hu, H.H.; Guedj, M.; Fernandez, L.P.; Descamps, V.; Basset-Seguin, N.; Bagot, M.; Benssussan, A.; Saiag, P.; Fargnoli, M.C.; et al. MC1R, SLC45A2 and TYR genetic variants involved in melanoma susceptibility in southern European populations: Results from a meta-analysis. Eur. J. Cancer 2012, 48, 2183–2191. [Google Scholar] [CrossRef]
- Jorgenson, E.; Choquet, H.; Yin, J.; Hoffmann, T.J.; Banda, Y.; Kvale, M.N.; Risch, N.; Schaefer, C.; Asgari, M.M. Genetic ancestry, skin pigmentation, and the risk of cutaneous squamous cell carcinoma in Hispanic/Latino and non-Hispanic white populations. Commun. Biol. 2020, 3, 765. [Google Scholar] [CrossRef] [PubMed]
- Spritz, R.A.; Andersen, G.H. Genetics of Vitiligo. Dermatol. Clin. 2017, 35, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Cabaco, L.C.; Tomas, A.; Pojo, M.; Barral, D.C. The Dark Side of Melanin Secretion in Cutaneous Melanoma Aggressiveness. Front. Oncol. 2022, 12, 887366. [Google Scholar] [CrossRef]
- Kuan, V.; Martineau, A.R.; Griffiths, C.J.; Hypponen, E.; Walton, R. DHCR7 mutations linked to higher vitamin D status allowed early human migration to northern latitudes. BMC Evol. Biol. 2013, 13, 144. [Google Scholar] [CrossRef]
- Obon-Santacana, M.; Vilardell, M.; Carreras, A.; Duran, X.; Velasco, J.; Galvan-Femenia, I.; Alonso, T.; Puig, L.; Sumoy, L.; Duell, E.J.; et al. GCAT|Genomes for life: A prospective cohort study of the genomes of Catalonia. BMJ Open 2018, 8, e018324. [Google Scholar] [CrossRef]
- Fitzpatrick, T.B. The Validity and Practicality od Sun-Reactive Skin Types I Through VI. Arch. Dermatol. 1988, 124, 869–871. [Google Scholar] [CrossRef]
- Galvan-Femenia, I.; Obon-Santacana, M.; Pineyro, D.; Guindo-Martinez, M.; Duran, X.; Carreras, A.; Pluvinet, R.; Velasco, J.; Ramos, L.; Ausso, S.; et al. Multitrait genome association analysis identifies new susceptibility genes for human anthropometric variation in the GCAT cohort. J. Med. Genet. 2018, 55, 765–778. [Google Scholar] [CrossRef]
- Valls-Margarit, J.; Galvan-Femenia, I.; Matias-Sanchez, D.; Blay, N.; Puiggros, M.; Carreras, A.; Salvoro, C.; Cortes, B.; Amela, R.; Farre, X.; et al. GCAT|Panel, a comprehensive structural variant haplotype map of the Iberian population from high-coverage whole-genome sequencing. Nucleic Acids Res. 2022, 50, 2464–2479. [Google Scholar] [CrossRef]
- Agència de Qualitat i Avaluació Sanitàries de Catalunya (AQuAS). Available online: https://Aquas.Gencat.Cat (accessed on 1 September 2021).
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- de Leeuw, C.A.; Mooij, J.M.; Heskes, T.; Posthuma, D. MAGMA: Generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015, 11, e1004219. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Taskesen, E.; van Bochoven, A.; Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017, 8, 1826. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef] [PubMed]
- Bulik-Sullivan, B.K.; Loh, P.R.; Finucane, H.K.; Ripke, S.; Yang, J.; Schizophrenia Working Group of the Psychiatric Genomics Consortium; Patterson, N.; Daly, M.J.; Price, A.L.; Neale, B.M. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015, 47, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Gagliano Taliun, S.A.; VandeHaar, P.; Boughton, A.P.; Welch, R.P.; Taliun, D.; Schmidt, E.M.; Zhou, W.; Nielsen, J.B.; Willer, C.J.; Lee, S.; et al. Exploring and visualizing large-scale genetic associations by using PheWeb. Nat. Genet. 2020, 52, 550–552. [Google Scholar] [CrossRef]
- UKBiobank TOPMed-imputed PheWeb. Available online: Https://Pheweb.Org/UKB-TOPMed/ (accessed on 1 March 2022).
- Lambert, S.A.; Gil, L.; Jupp, S.; Ritchie, S.C.; Xu, Y.; Buniello, A.; McMahon, A.; Abraham, G.; Chapman, M.; Parkinson, H.; et al. The Polygenic Score Catalog as an open database for reproducibility and systematic evaluation. Nat. Genet. 2021, 53, 420–425. [Google Scholar] [CrossRef]
- Rotimi, C.N.; Jorde, L.B. Ancestry and disease in the age of genomic medicine. N. Engl. J. Med. 2010, 363, 1551–1558. [Google Scholar] [CrossRef]
- Jin, Y.; Andersen, G.; Yorgov, D.; Ferrara, T.M.; Ben, S.; Brownson, K.M.; Holland, P.J.; Birlea, S.A.; Siebert, J.; Hartmann, A.; et al. Genome-wide association studies of autoimmune vitiligo identify 23 new risk loci and highlight key pathways and regulatory variants. Nat. Genet. 2016, 48, 1418–1424. [Google Scholar] [CrossRef]
- Liu, F.; Visser, M.; Duffy, D.L.; Hysi, P.G.; Jacobs, L.C.; Lao, O.; Zhong, K.; Walsh, S.; Chaitanya, L.; Wollstein, A.; et al. Genetics of skin color variation in Europeans: Genome-wide association studies with functional follow-up. Hum. Genet. 2015, 134, 823–835. [Google Scholar] [CrossRef] [PubMed]
- Visconti, A.; Duffy, D.L.; Liu, F.; Zhu, G.; Wu, W.; Chen, Y.; Hysi, P.G.; Zeng, C.; Sanna, M.; Iles, M.M.; et al. Genome-wide association study in 176,678 Europeans reveals genetic loci for tanning response to sun exposure. Nat. Commun. 2018, 9, 1684. [Google Scholar] [CrossRef] [PubMed]
- Sinha, R.P.; Hader, D.P. UV-induced DNA damage and repair: A review. Photochem. Photobiol. Sci. 2002, 1, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Yamashita, Y.; Umezawa, K.; Hirobe, T.; Ito, S.; Wakamatsu, K. The Pro-Oxidant Activity of Pheomelanin is Significantly Enhanced by UVA Irradiation: Benzothiazole Moieties Are More Reactive than Benzothiazine Moieties. Int. J. Mol. Sci. 2018, 19, 2889. [Google Scholar] [CrossRef] [PubMed]
- Bernard, J.J.; Gallo, R.L.; Krutmann, J. Photoimmunology: How ultraviolet radiation affects the immune system. Nat. Rev. Immunol. 2019, 19, 688–701. [Google Scholar] [CrossRef]
- Vranic, L.; Mikolasevic, I.; Milic, S. Vitamin D Deficiency: Consequence or Cause of Obesity? Medicina 2019, 55, 541. [Google Scholar] [CrossRef]
- Azevedo, M.; Bandeira, L.; Luza, C.; Lemos, A.; Bandeira, F. Vitamin D Deficiency, Skin Phototype, Sun Index, and Metabolic Risk Among Patients with High Rates of Sun Exposure Living in the Tropics. J. Clin. Aesthet. Dermatol. 2018, 11, 15–18. [Google Scholar]
- Sulem, P.; Gudbjartsson, D.F.; Stacey, S.N.; Helgason, A.; Rafnar, T.; Magnusson, K.P.; Manolescu, A.; Karason, A.; Palsson, A.; Thorleifsson, G.; et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat. Genet. 2007, 39, 1443–1452. [Google Scholar] [CrossRef]
- Zhang, M.; Song, F.; Liang, L.; Nan, H.; Zhang, J.; Liu, H.; Wang, L.E.; Wei, Q.; Lee, J.E.; Amos, C.I.; et al. Genome-wide association studies identify several new loci associated with pigmentation traits and skin cancer risk in European Americans. Hum. Mol. Genet. 2013, 22, 2948–2959. [Google Scholar] [CrossRef]
- Prakash, T.; Sharma, V.K.; Adati, N.; Ozawa, R.; Kumar, N.; Nishida, Y.; Fujikake, T.; Takeda, T.; Taylor, T.D. Expression of conjoined genes: Another mechanism for gene regulation in eukaryotes. PLoS ONE 2010, 5, e13284. [Google Scholar] [CrossRef]
- Lee, P.J.; Yang, S.; Sun, Y.; Guo, J.U. Regulation of nonsense-mediated mRNA decay in neural development and disease. J. Mol. Cell Biol. 2021, 13, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Wintzen, M.; Gilchrest, B.A. Proopiomelanocortin, its derived peptides, and the skin. J. Investig. Dermatol. 1996, 106, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Millington, G.W. Proopiomelanocortin (POMC): The cutaneous roles of its melanocortin products and receptors. Clin. Exp. Dermatol. 2006, 31, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Gerstenblith, M.R.; Shi, J.; Landi, M.T. Genome-wide association studies of pigmentation and skin cancer: A review and meta-analysis. Pigment Cell Melanoma Res. 2010, 23, 587–606. [Google Scholar] [CrossRef]
- Schuh, M. An actin-dependent mechanism for long-range vesicle transport. Nat. Cell Biol. 2011, 13, 1431–1436. [Google Scholar] [CrossRef]
- Visser, M.; Palstra, R.J.; Kayser, M. Human skin color is influenced by an intergenic DNA polymorphism regulating transcription of the nearby BNC2 pigmentation gene. Hum. Mol. Genet. 2014, 23, 5750–5762. [Google Scholar] [CrossRef]
- Pospiech, E.; Kukla-Bartoszek, M.; Karlowska-Pik, J.; Zielinski, P.; Wozniak, A.; Boron, M.; Dabrowski, M.; Zubanska, M.; Jarosz, A.; Grzybowski, T.; et al. Exploring the possibility of predicting human head hair greying from DNA using whole-exome and targeted NGS data. BMC Genom. 2020, 21, 538. [Google Scholar] [CrossRef]
- D’Mello, S.A.; Finlay, G.J.; Baguley, B.C.; Askarian-Amiri, M.E. Signaling Pathways in Melanogenesis. Int. J. Mol. Sci. 2016, 17, 1144. [Google Scholar] [CrossRef]
- Gautron, A.; Migault, M.; Bachelot, L.; Corre, S.; Galibert, M.D.; Gilot, D. Human TYRP1: Two functions for a single gene? Pigment Cell Melanoma Res. 2021, 34, 836–852. [Google Scholar] [CrossRef]
- Sprong, H.; Degroote, S.; Claessens, T.; van Drunen, J.; Oorschot, V.; Westerink, B.H.; Hirabayashi, Y.; Klumperman, J.; van der Sluijs, P.; van Meer, G. Glycosphingolipids are required for sorting melanosomal proteins in the Golgi complex. J. Cell Biol. 2001, 155, 369–380. [Google Scholar] [CrossRef]
- Truschel, S.T.; Simoes, S.; Setty, S.R.; Harper, D.C.; Tenza, D.; Thomas, P.C.; Herman, K.E.; Sackett, S.D.; Cowan, D.C.; Theos, A.C.; et al. ESCRT-I function is required for Tyrp1 transport from early endosomes to the melanosome limiting membrane. Traffic 2009, 10, 1318–1336. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cai, Y.; Shi, M.; Jiang, S.; Cui, S.; Wu, Y.; Gao, X.H.; Chen, H.D. The Prevalence of Vitiligo: A Meta-Analysis. PLoS ONE 2016, 11, e0163806. [Google Scholar] [CrossRef] [PubMed]
- Barrea, L.; Savanelli, M.C.; Di Somma, C.; Napolitano, M.; Megna, M.; Colao, A.; Savastano, S. Vitamin D and its role in psoriasis: An overview of the dermatologist and nutritionist. Rev. Endocr. Metab. Disord. 2017, 18, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Leske, M.C.; Chylack, L.T.; Jr Wu, S.Y. The Lens Opacities Case-Control Study. Risk factors for cataract. Arch. Ophthalmol. 1991, 109, 244–251. [Google Scholar] [CrossRef]
- Cumming, R.G.; Mitchell, P.; Lim, R. Iris color and cataract: The Blue Mountains Eye Study. Am. J. Ophthalmol. 2000, 130, 237–238. [Google Scholar] [CrossRef] [PubMed]
- Yosipovitch, G.; DeVore, A.; Dawn, A. Obesity and the skin: Skin physiology and skin manifestations of obesity. J. Am. Acad. Dermatol. 2007, 56, 901–916. [Google Scholar] [CrossRef] [PubMed]
- Sultan, F.; Basu, R.; Murthy, D.; Kochar, M.; Attri, K.S.; Aggarwal, A.; Kumari, P.; Dnyane, P.; Tanwar, J.; Motiani, R.K.; et al. Temporal analysis of melanogenesis identifies fatty acid metabolism as key skin pigment regulator. PLoS Biol. 2022, 20, e3001634. [Google Scholar] [CrossRef]
- Jablonski, N.G.; Chaplin, G. The evolution of human skin coloration. J. Hum. Evol. 2000, 39, 57–106. [Google Scholar] [CrossRef]
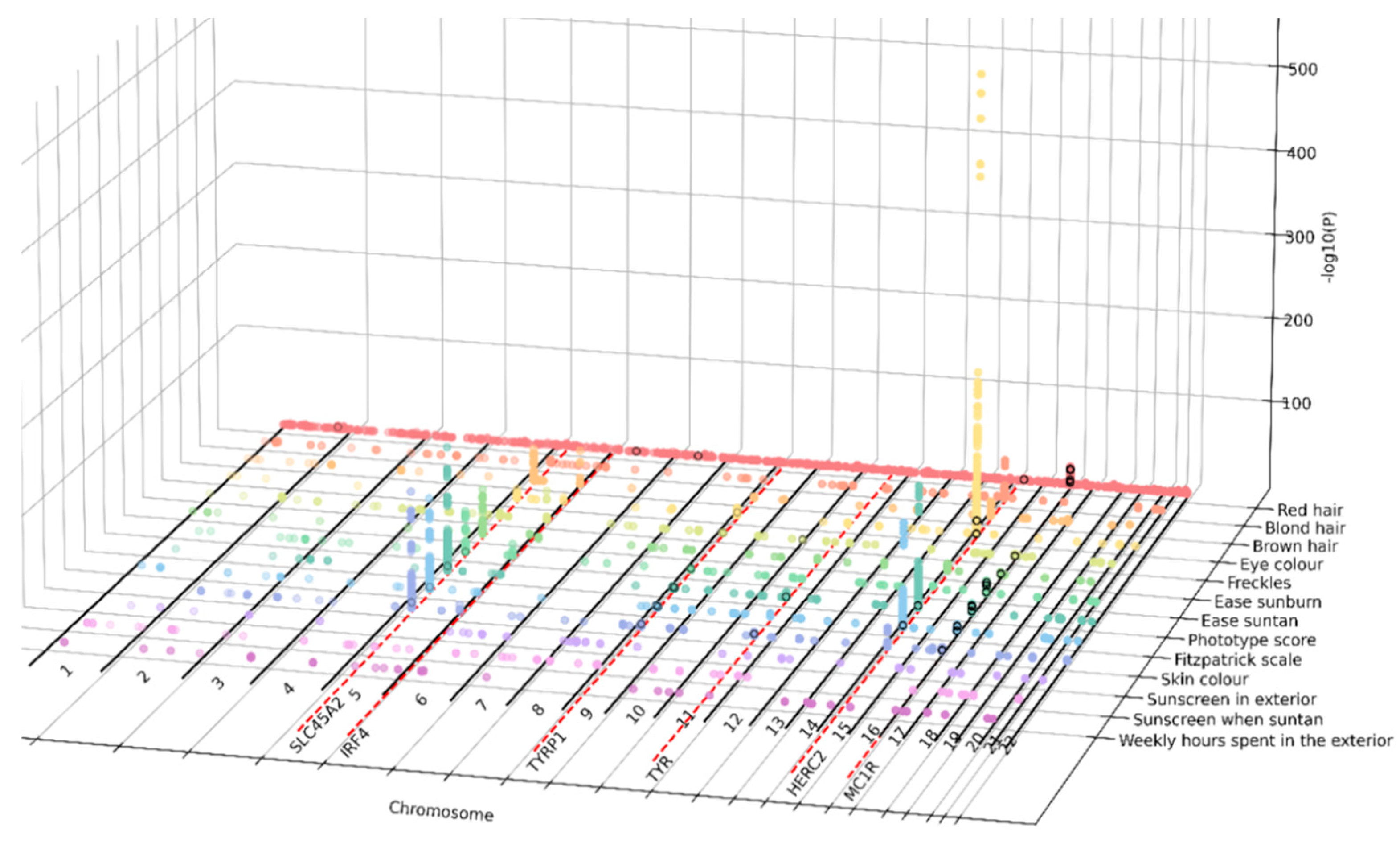
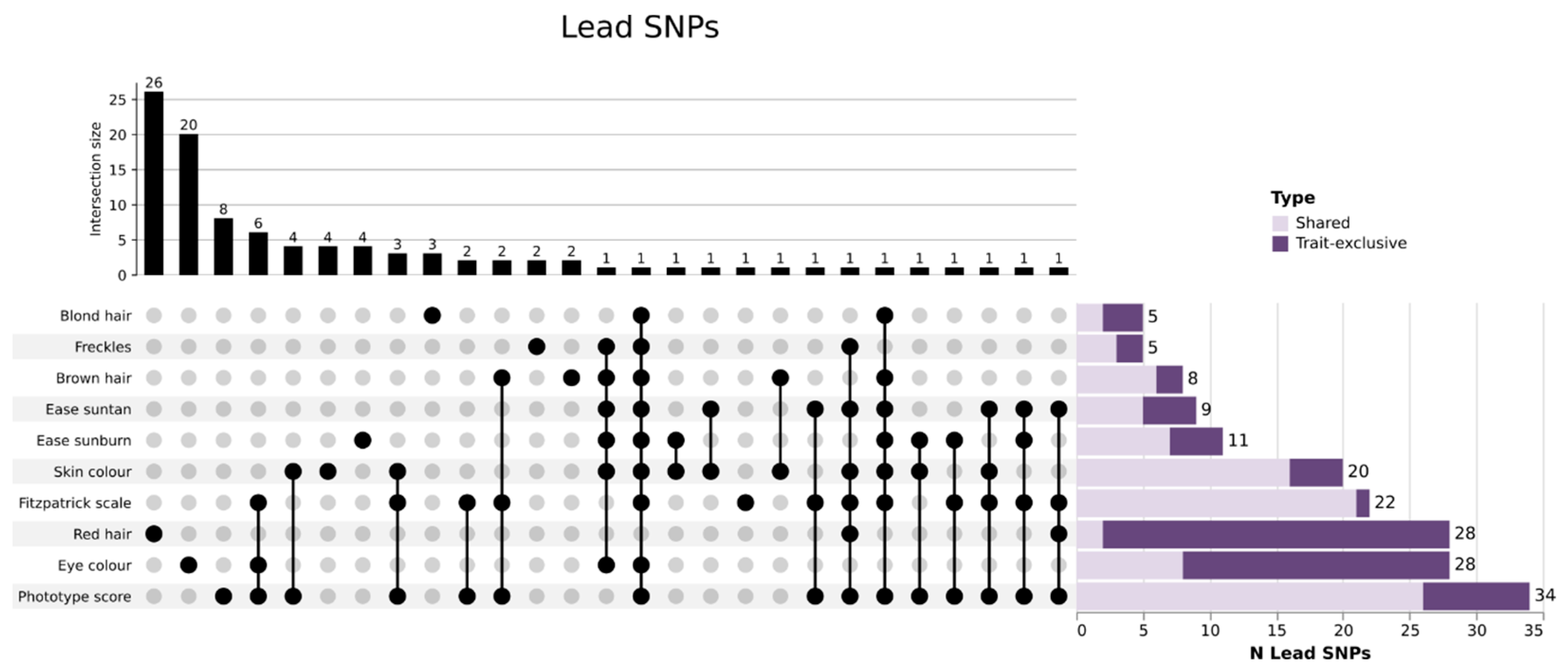

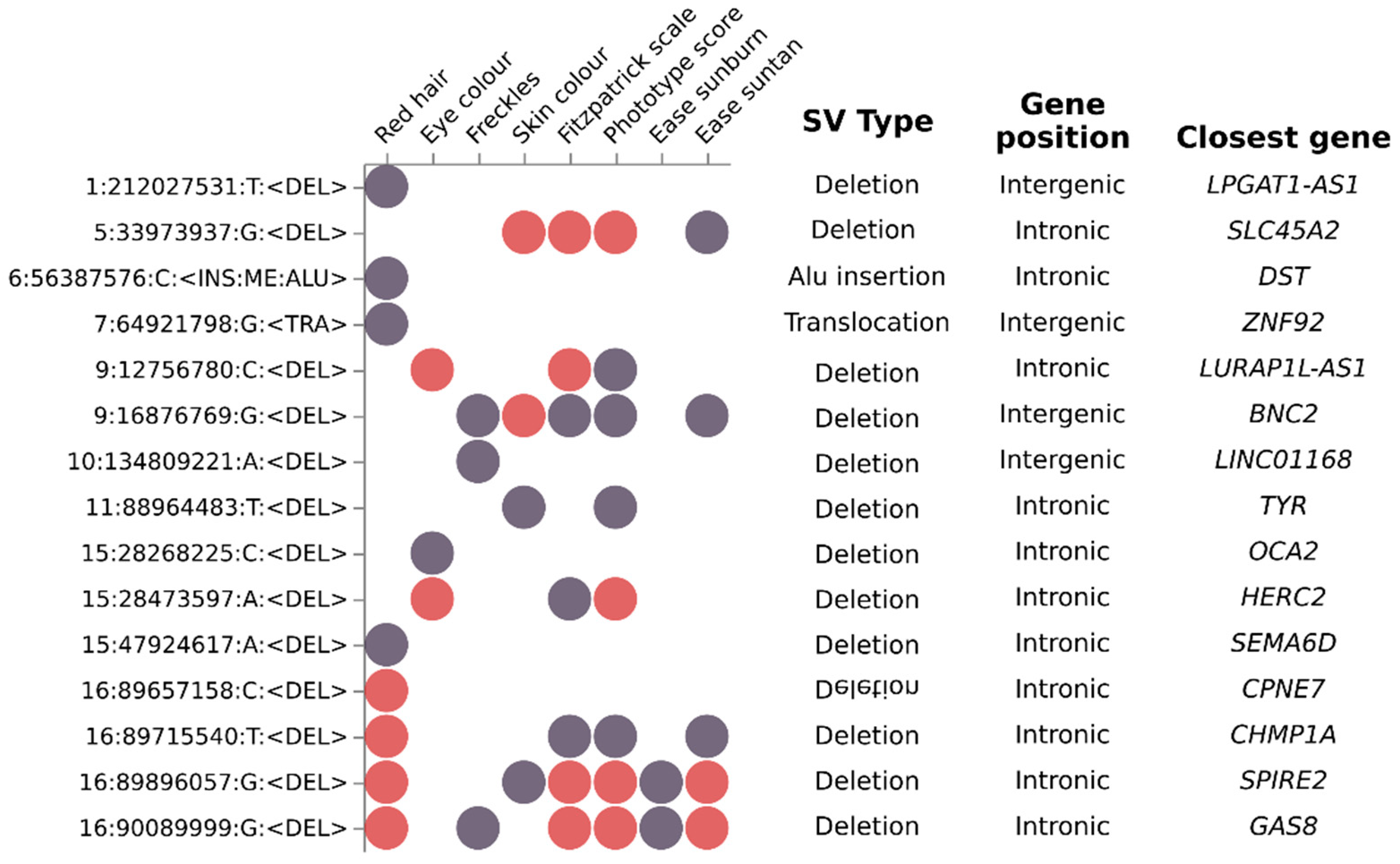

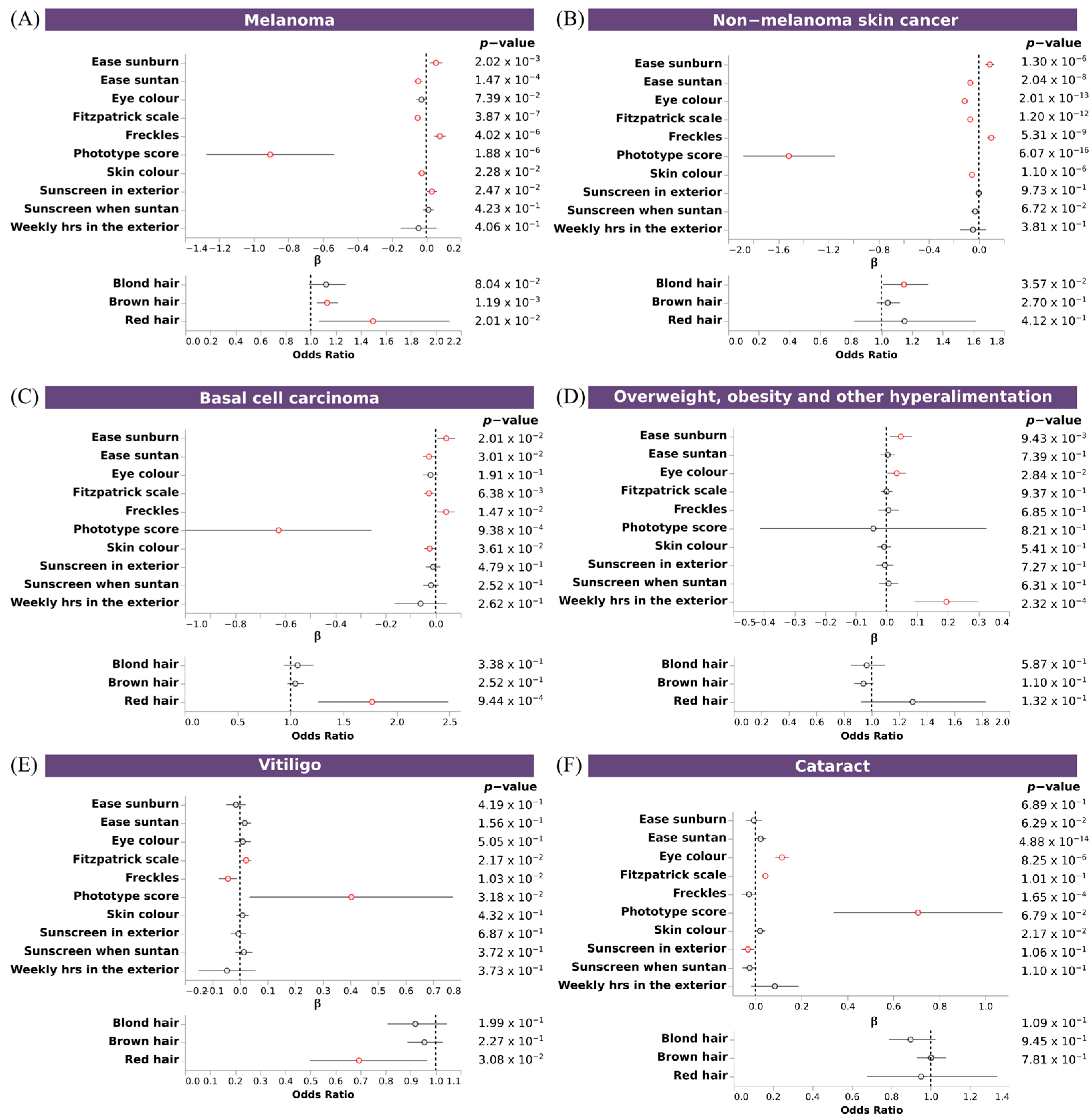
| Trait | Type | SNPs (n) | Independent SNPs (n) | Lead SNPs (n) | Genomic Risk Loci (n) | SVs (n) | λ |
|---|---|---|---|---|---|---|---|
| Red hair | Binary | 181 | 54 | 28 | 23 | 4 | 1.0946 |
| Blond hair | Binary | 209 | 17 | 5 | 3 | 0 | 1.0096 |
| Brown hair | Binary | 143 | 22 | 8 | 4 | 0 | 1.0171 |
| Eye color | Discrete | 829 | 97 | 28 | 6 | 2 | 1.0101 |
| Skin color | Discrete | 478 | 66 | 20 | 7 | 2 | 1.0180 |
| Freckles | Discrete | 30 | 7 | 5 | 4 | 0 | 0.9996 |
| Ease sunburn | Discrete | 170 | 28 | 11 | 6 | 0 | 1.0061 |
| Ease suntan | Discrete | 190 | 25 | 9 | 4 | 2 | 1.0138 |
| Sunscreen in exterior | Discrete | 0 | 0 | 0 | 0 | 0 | 1.0167 |
| Sunscreen when suntan | Discrete | 0 | 0 | 0 | 0 | 0 | 1.0007 |
| Weekly hours spent in the exterior | Continuous | 0 | 0 | 0 | 0 | 0 | 1.0061 |
| Phototype score | Continuous | 768 | 111 | 34 | 6 | 4 | 1.0011 |
| Fitzpatrick scale | Discrete | 564 | 82 | 22 | 5 | 4 | 1.0198 |
| Total | 1515 | 257 | 99 | 37 | 8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farré, X.; Blay, N.; Cortés, B.; Carreras, A.; Iraola-Guzmán, S.; de Cid, R. Skin Phototype and Disease: A Comprehensive Genetic Approach to Pigmentary Traits Pleiotropy Using PRS in the GCAT Cohort. Genes 2023, 14, 149. https://doi.org/10.3390/genes14010149
Farré X, Blay N, Cortés B, Carreras A, Iraola-Guzmán S, de Cid R. Skin Phototype and Disease: A Comprehensive Genetic Approach to Pigmentary Traits Pleiotropy Using PRS in the GCAT Cohort. Genes. 2023; 14(1):149. https://doi.org/10.3390/genes14010149
Chicago/Turabian StyleFarré, Xavier, Natalia Blay, Beatriz Cortés, Anna Carreras, Susana Iraola-Guzmán, and Rafael de Cid. 2023. "Skin Phototype and Disease: A Comprehensive Genetic Approach to Pigmentary Traits Pleiotropy Using PRS in the GCAT Cohort" Genes 14, no. 1: 149. https://doi.org/10.3390/genes14010149
APA StyleFarré, X., Blay, N., Cortés, B., Carreras, A., Iraola-Guzmán, S., & de Cid, R. (2023). Skin Phototype and Disease: A Comprehensive Genetic Approach to Pigmentary Traits Pleiotropy Using PRS in the GCAT Cohort. Genes, 14(1), 149. https://doi.org/10.3390/genes14010149








