ADGR: Admixture-Informed Differential Gene Regulation
Abstract
1. Introduction
2. Materials and Methods
3. Results
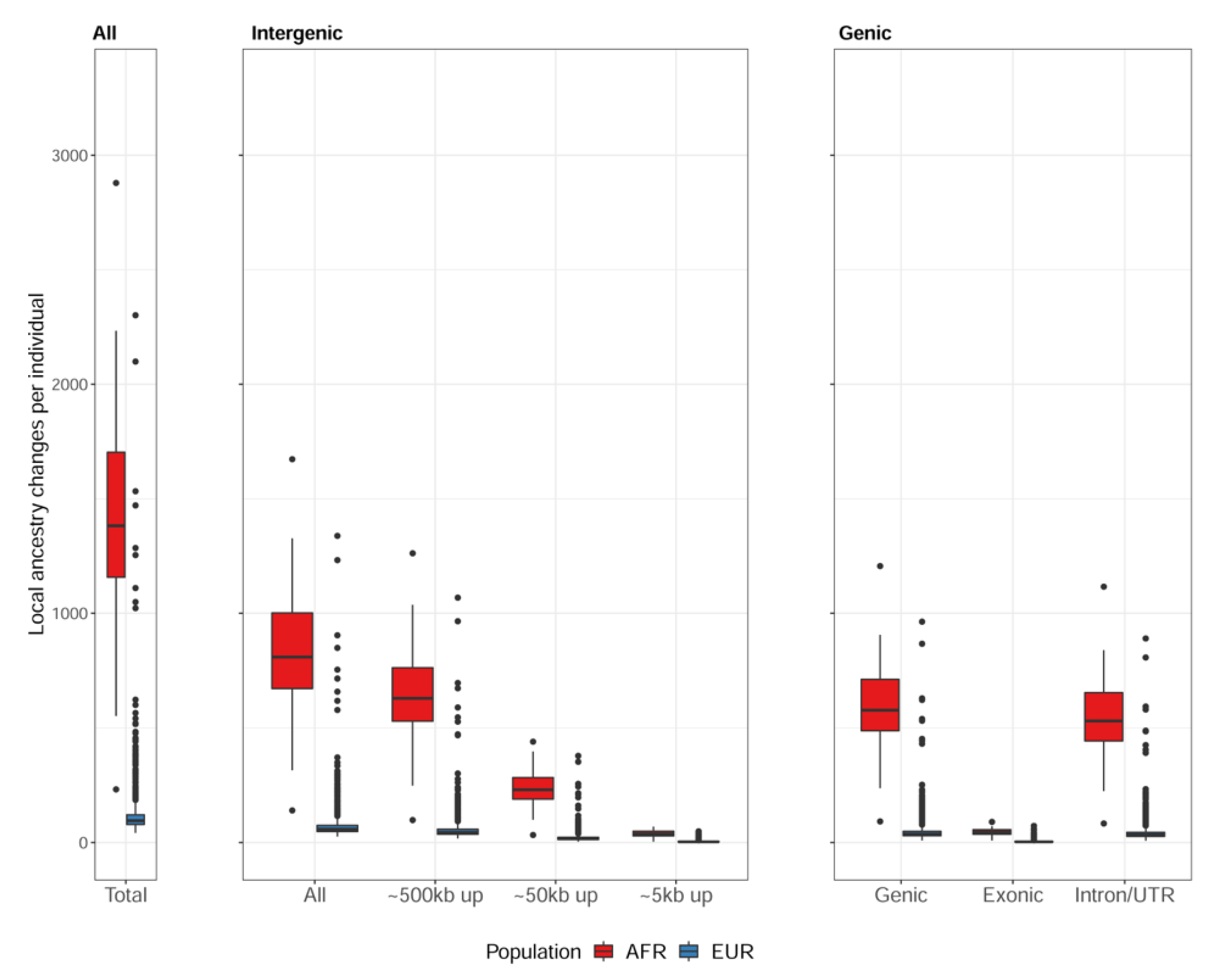
3.1. Local Ancestral Structure of 838 Individuals
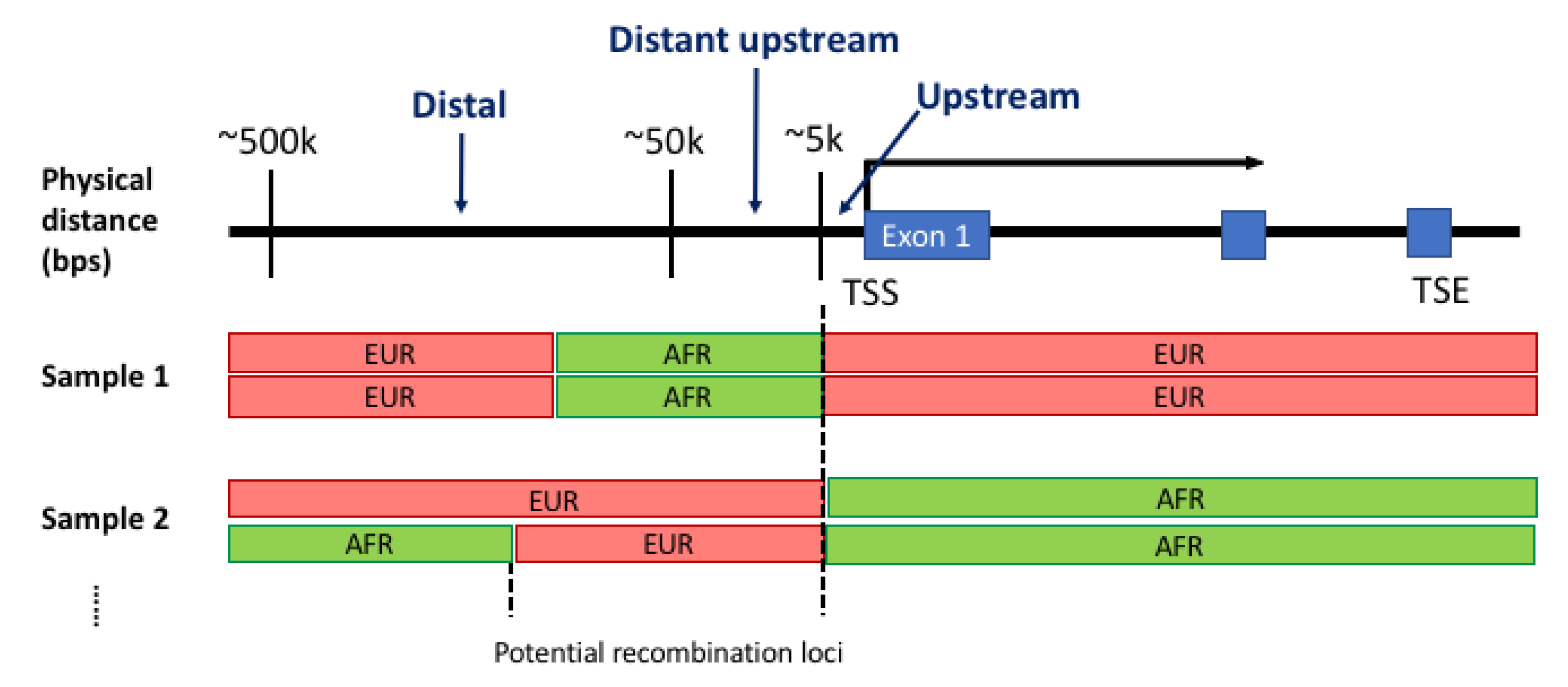
3.2. Transition of Local Ancestral Structure and Gene Model
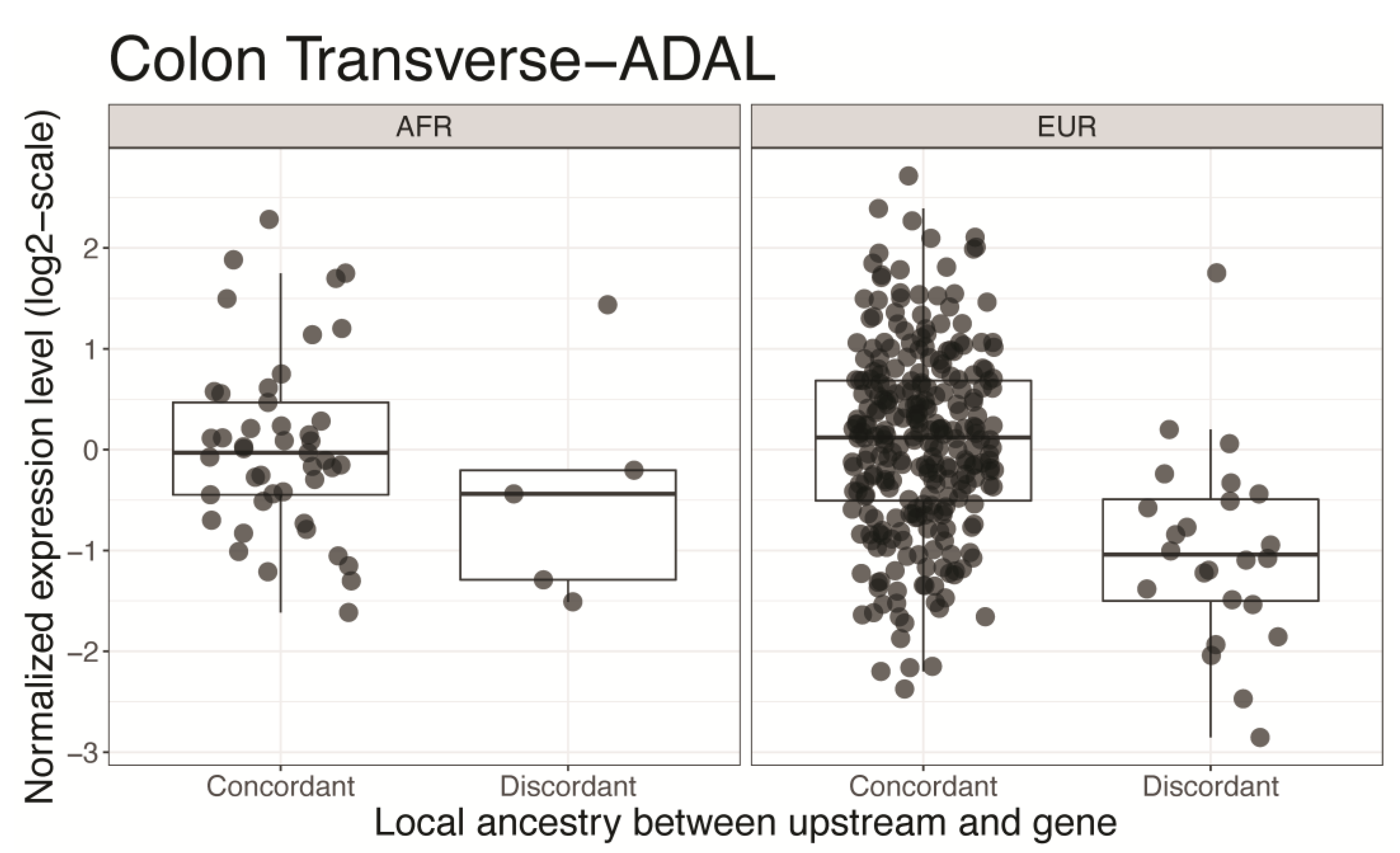
3.3. Gene Expression Levels Associated with Admixed Ancestral Structure in the Regulatory Region
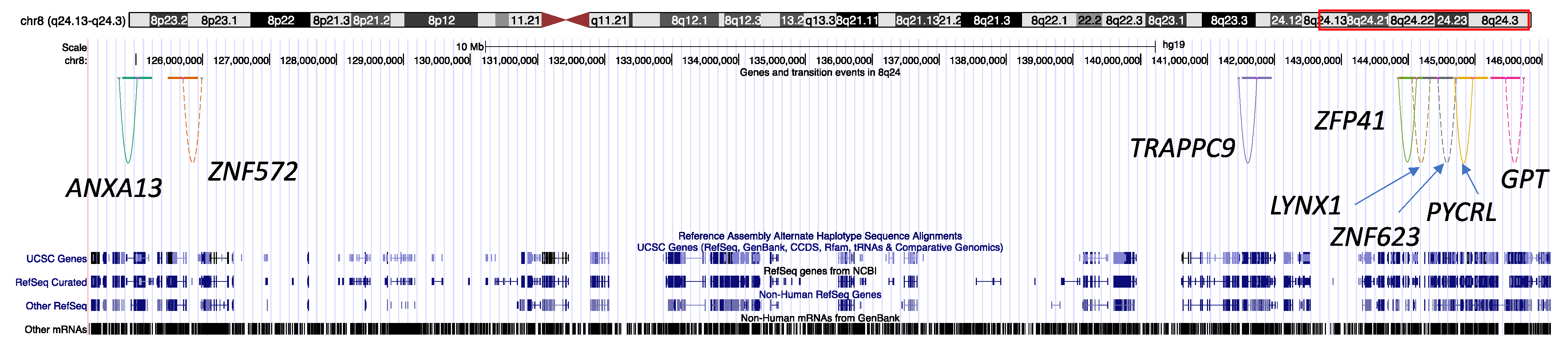
3.4. Gene Expression Levels in Chromosome 8q24 Associated with Local Ancestral Transition between Africans and Europeans
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Benton, M.L.; Abraham, A.; LaBella, A.L.; Abbot, P.; Rokas, A.; Capra, J.A. The influence of evolutionary history on human health and disease. Nat. Rev. Genet. 2021, 22, 269–283. [Google Scholar] [CrossRef]
- Maples, B.K.; Gravel, S.; Kenny, E.E.; Bustamante, C.D. RFMix: A discriminative modeling approach for rapid and robust local-ancestry inference. Am. J. Hum. Genet. 2013, 93, 278–288. [Google Scholar] [CrossRef]
- Kong, A.; Thorleifsson, G.; Gudbjartsson, D.F.; Masson, G.; Sigurdsson, A.; Jonasdottir, A.; Walters, G.B.; Jonasdottir, A.; Gylfason, A.; Kristinsson, K.T.; et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature 2010, 467, 1099–1103. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, P.F.; Balding, D.J. Admixture provides new insights into recombination. Nat. Genet. 2011, 43, 819–820. [Google Scholar] [CrossRef]
- Hindorff, L.A.; Sethupathy, P.; Junkins, H.A.; Ramos, E.M.; Mehta, J.P.; Collins, F.S.; Manolio, T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA 2009, 106, 9362–9367. [Google Scholar] [CrossRef] [PubMed]
- Fish, A.E.; Crawford, D.C.; Capra, J.A.; Bush, W.S. Local ancestry transitions modify snp-trait associations. Acific Symp. Biocomput. 2018, 23, 424–435. [Google Scholar]
- Montgomery, S.B.; Dermitzakis, E.T. From expression QTLs to personalized transcriptomics. Nat. Rev. Genet. 2011, 12, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Hentze, M.W.; Castello, A.; Schwarzl, T.; Preiss, T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018, 19, 327–341. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.R.; Kanai, M.; Kamatani, Y.; Okada, Y.; Neale, B.M.; Daly, M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019, 51, 584–591. [Google Scholar] [CrossRef]
- Mills, M.C.; Rahal, C. The GWAS Diversity Monitor tracks diversity by disease in real time. Nat. Genet. 2020, 52, 242–243. [Google Scholar] [CrossRef]
- Sollis, E.; Mosaku, A.; Abid, A.; Buniello, A.; Cerezo, M.; Gil, L.; Groza, T.; Gunes, O.; Hall, P.; Hayhurst, J.; et al. The NHGRI-EBI GWAS Catalog: Knowledgebase and deposition resource. Nucleic Acids Res. 2022. [Google Scholar] [CrossRef] [PubMed]
- Consortium, G.T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar]
- Lappalainen, T.; Sammeth, M.; Friedlander, M.R.; Hoen, P.A.; Monlong, J.; Rivas, M.A.; Gonzalez-Porta, M.; Kurbatova, N.; Griebel, T.; Ferreira, P.G.; et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 2013, 501, 506–511. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Perera, M.A.; Gamazon, E.R. On Using Local Ancestry to Characterize the Genetic Architecture of Human Traits: Genetic Regulation of Gene Expression in Multiethnic or Admixed Populations. Am. J. Hum. Genet. 2019, 104, 1097–1115. [Google Scholar] [CrossRef] [PubMed]
- Baran, Y.; Pasaniuc, B.; Sankararaman, S.; Torgerson, D.G.; Gignoux, C.; Eng, C.; Rodriguez-Cintron, W.; Chapela, R.; Ford, J.G.; Avila, P.C.; et al. Fast and accurate inference of local ancestry in Latino populations. Bioinformatics 2012, 28, 1359–1367. [Google Scholar] [CrossRef]
- Consortium, G.T. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015, 348, 648–660. [Google Scholar] [CrossRef]
- Consortium, G.T. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020, 369, 1318–1330. [Google Scholar] [CrossRef]
- Genomes Project, C.; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar]
- Harrow, J.; Frankish, A.; Gonzalez, J.M.; Tapanari, E.; Diekhans, M.; Kokocinski, F.; Aken, B.L.; Barrell, D.; Zadissa, A.; Searle, S.; et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012, 22, 1760–1774. [Google Scholar] [CrossRef]
- Frankish, A.; Diekhans, M.; Ferreira, A.M.; Johnson, R.; Jungreis, I.; Loveland, J.; Mudge, J.M.; Sisu, C.; Wright, J.; Armstrong, J.; et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019, 47, D766–D773. [Google Scholar] [CrossRef]
- Jovov, B.; Araujo-Perez, F.; Sigel, C.S.; Stratford, J.K.; McCoy, A.N.; Yeh, J.J.; Keku, T. Differential gene expression between African American and European American colorectal cancer patients. PLoS ONE 2012, 7, e30168. [Google Scholar] [CrossRef] [PubMed]
- Freedman, M.L.; Haiman, C.A.; Patterson, N.; McDonald, G.J.; Tandon, A.; Waliszewska, A.; Penney, K.; Steen, R.G.; Ardlie, K.; John, E.M.; et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc. Natl. Acad. Sci. USA 2006, 103, 14068–14073. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, S.; Wang, H.; Yang, W.; Li, F.; Yang, F.; Yu, D.; Ramsey, F.V.; Tuszyski, G.P.; Hu, W. Elevated NIBP/TRAPPC9 mediates tumorigenesis of cancer cells through NFkappaB signaling. Oncotarget 2015, 6, 6160–6178. [Google Scholar] [CrossRef]
- Liu, W.; Hancock, C.N.; Fischer, J.W.; Harman, M.; Phang, J.M. Proline biosynthesis augments tumor cell growth and aerobic glycolysis: Involvement of pyridine nucleotides. Sci. Rep. 2015, 5, 17206. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Majumder, P.K.; Ross, K.; Shim, Y.; Golub, T.R.; Loda, M.; Sellers, W.R. Identification of prostate cancer modifier pathways using parental strain expression mapping. Proc. Natl. Acad. Sci. USA 2007, 104, 17771–17776. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D. Gene--environment-wide association studies: Emerging approaches. Nat. Rev. Genet. 2010, 11, 259–272. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zeng, J.; Zhang, F.; Zhu, Z.; Qi, T.; Zheng, Z.; Lloyd-Jones, L.R.; Marioni, R.E.; Martin, N.G.; Montgomery, G.W.; et al. Integrative analysis of omics summary data reveals putative mechanisms underlying complex traits. Nat. Commun. 2018, 9, 918. [Google Scholar] [CrossRef] [PubMed]
- Gamazon, E.R.; Wheeler, H.E.; Shah, K.P.; Mozaffari, S.V.; Aquino-Michaels, K.; Carroll, R.J.; Eyler, A.E.; Denny, J.C.; Consortium, G.T.; Nicolae, D.L.; et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015, 47, 1091–1098. [Google Scholar] [CrossRef] [PubMed]
- Gusev, A.; Ko, A.; Shi, H.; Bhatia, G.; Chung, W.; Penninx, B.W.; Jansen, R.; de Geus, E.J.; Boomsma, D.I.; Wright, F.A.; et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016, 48, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Aguet, F.; Barbeira, A.N.; Ardlie, K.; Im, H.K. A scalable unified framework of total and allele-specific counts for cis-QTL, fine-mapping, and prediction. Nat. Commun. 2021, 12, 1424. [Google Scholar] [CrossRef]
- Pai, A.A.; Pritchard, J.K.; Gilad, Y. The genetic and mechanistic basis for variation in gene regulation. PLoS Genet. 2015, 11, e1004857. [Google Scholar] [CrossRef] [PubMed]
- Reich, D.E.; Cargill, M.; Bolk, S.; Ireland, J.; Sabeti, P.C.; Richter, D.J.; Lavery, T.; Kouyoumjian, R.; Farhadian, S.F.; Ward, R.; et al. Linkage disequilibrium in the human genome. Nature 2001, 411, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Yan, D.; Cooper, R.S.; Luke, A.; Ikeda, M.A.; Chang, Y.P.; Weder, A.; Chakravarti, A. Linkage disequilibrium and haplotype diversity in the genes of the renin-angiotensin system: Findings from the family blood pressure program. Genome Res. 2003, 13, 173–181. [Google Scholar] [CrossRef]
- Hinch, A.G.; Tandon, A.; Patterson, N.; Song, Y.; Rohland, N.; Palmer, C.D.; Chen, G.K.; Wang, K.; Buxbaum, S.G.; Akylbekova, E.L.; et al. The landscape of recombination in African Americans. Nature 2011, 476, 170–175. [Google Scholar] [CrossRef]
- Uren, C.; Hoal, E.G.; Moller, M. Putting RFMix and ADMIXTURE to the test in a complex admixed population. BMC Genet. 2020, 21, 40. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Urs, M.J.; Sanchez-Gonzalez, I.; Olayioye, M.A.; Herde, M.; Witte, C.P. m(6)A RNA Degradation Products Are Catabolized by an Evolutionarily Conserved N(6)-Methyl-AMP Deaminase in Plant and Mammalian Cells. Plant Cell 2018, 30, 1511–1522. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Zhang, D.; Nie, H.; Shen, S.; Li, Y.; Li, S. Structure of Arabidopsis thaliana N(6)-methyl-AMP deaminase ADAL with bound GMP and IMP and implications for N(6)-methyl-AMP recognition and processing. RNA Biol. 2019, 16, 1504–1512. [Google Scholar] [CrossRef] [PubMed]
- Zhabotynsky, V.; Huang, L.; Little, P.; Hu, Y.J.; Pardo-Manuel de Villena, F.; Zou, F.; Sun, W. eQTL mapping using allele-specific count data is computationally feasible, powerful, and provides individual-specific estimates of genetic effects. PLoS Genet. 2022, 18, e1010076. [Google Scholar] [CrossRef]
- Dreau, A.; Venu, V.; Avdievich, E.; Gaspar, L.; Jones, F.C. Genome-wide recombination map construction from single individuals using linked-read sequencing. Nat. Commun. 2019, 10, 4309. [Google Scholar] [CrossRef]
- Bentley, A.R.; Callier, S.; Rotimi, C.N. Diversity and inclusion in genomic research: Why the uneven progress? J. Community Genet. 2017, 8, 255–266. [Google Scholar] [CrossRef]
- Paigen, K.; Petkov, P.M. PRDM9 and Its Role in Genetic Recombination. Trends Genet. 2018, 34, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dai, M.; Cai, D.; Shi, Z. Proteome and transcriptome profile analysis reveals regulatory and stress-responsive networks in the russet fruit skin of sand pear. Hortic. Res. 2020, 7, 16. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Shao, J.; Zhou, Z.; Davis, R.E. Genotype-specific suppression of multiple defense pathways in apple root during infection by Pythium ultimum. Hortic. Res. 2019, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- Georges, M.; Charlier, C.; Hayes, B. Harnessing genomic information for livestock improvement. Nat. Rev. Genet. 2019, 20, 135–156. [Google Scholar] [CrossRef]

| Model | Distance from Transcription Start Site | Tissue Type | Gene | False Discovery Rate |
|---|---|---|---|---|
| Dominant | Less than 5 kbps | Small Intestine, Terminal Ileum | SLC17A9 | 0.047 |
| 5~50 kbps | Brain, Cerebellar Hemisphere | HLA-DMA | 0.018 | |
| 50~500 kbps | Adipose, Subcutaneous | ADAL | 0.00029 | |
| C10orf107 | 0.0049 | |||
| HLA-DQB2 | 0.016 | |||
| Artery, Aorta | ADAL | 8.4 × 10−6 | ||
| PSORS1C2 | 0.007 | |||
| Artery, Tibial | ADAL | 6.6 × 10−7 | ||
| Brain Cerebellum | HLA-A | 0.01 | ||
| Breast, Mammary Tissue | ADAL | 0.0035 | ||
| Colon, Transverse | ADAL | 0.00083 | ||
| Esophagus, Muscularis | C10orf107 | 0.02 | ||
| ADAL | 0.02 | |||
| Heart, Atrial Appendage | C10orf107 | 0.00068 | ||
| PLEK2 | 0.04 | |||
| ALOX12 | 0.04 | |||
| Lung | PCDHGA6 | 0.029 | ||
| PSORS1C2 | 0.029 | |||
| STEAP2 | 0.03 | |||
| Muscle, Skeletal | HLA-DQB2 | 0.025 | ||
| COL8A2 | 0.025 | |||
| Nerve, Tibial | ADAL | 2.4 × 10−6 | ||
| Ovary | ADAL | 0.0029 | ||
| Skin, Not Sun Exposed Suprapubic | ADAL | 0.035 | ||
| Skin, Sun Exposed Lower leg | ADAL | 0.00068 | ||
| Spleen | ADAL | 0.00055 | ||
| Stomach | ADAL | 0.00074 | ||
| Thyroid | WDR87 | 5.5 × 10−5 | ||
| ADAL | 0.00012 | |||
| ZSCAN31 | 0.0053 | |||
| Whole Blood | MISP3 | 0.033 | ||
| Recessive | Less than 5 kbps | Uterus | SH3GLB1 | 0.043 |
| Additive | Less than 5 kbps | Small Intestine, Terminal Ileum | SLC17A9 | 0.047 |
| 5~50 kbps | Brain, Cerebellar Hemisphere | HLA-DMA | 0.018 | |
| 50~500 kbps | Adipose, Subcutaneous | HLA-DQB2 | 6.6 × 10−6 | |
| ADAL | 0.00014 | |||
| C10orf107 | 0.0033 | |||
| Adipose, Visceral Omentum | HLA-DQB2 | 0.0092 | ||
| Artery, Aorta | ADAL | 2.1 × 10−5 | ||
| Artery, Tibial | ADAL | 6.6 × 10−7 | ||
| Breast, Mammary Tissue | ADAL | 0.0074 | ||
| Colon, Transverse | ADAL | 0.00083 | ||
| Esophagus, Muscularis | C10orf107 | 0.02 | ||
| Heart, Atrial Appendage | HLA-DQB2 | 3.3 × 10−5 | ||
| C10orf107 | 0.00034 | |||
| Heart, Left Ventricle | HLA-DRB5 | 0.015 | ||
| Lung | PCDHGA6 | 0.032 | ||
| TLDC1 | 0.032 | |||
| Muscle, Skeletal | HLA-DQB2 | 3.1 × 10−7 | ||
| Nerve, Tibial | ADAL | 3.7 × 10−6 | ||
| Ovary | ADAL | 0.0053 | ||
| Skin, Not Sun Exposed Suprapubic | ZNF347 | 0.022 | ||
| ADAL | 0.022 | |||
| Skin, Sun Exposed Lower leg | ADAL | 0.00068 | ||
| Spleen | ADAL | 0.00055 | ||
| Stomach | ADAL | 0.00074 | ||
| Thyroid | WDR87 | 5.9 × 10−5 | ||
| ADAL | 0.00012 | |||
| ZSCAN31 | 0.0014 | |||
| Vagina | CTNNA2 | 0.029 | ||
| Whole Blood | ZFP57 | 0.0029 |
| Model | Distance from Transcription Start Site | Tissue Type | Gene | False Discovery Rate |
|---|---|---|---|---|
| Dominant | 5~50 kbps | Skin, Not Sun Exposed Suprapubic | ANXA13 | 0.015 |
| TRAPPC9 | 0.025 | |||
| 50~500 kbps | Brain, Spinal Cord Cervical C1 | GPT | 0.035 | |
| Whole Blood | ZNF572 | 0.041 | ||
| Recessive | Less than 5 kbps | Brain, Anterior Cingulate Cortex | ZNF623 | 0.032 |
| Brain, Frontal Cortex BA9 | ZNF623 | 0.03 | ||
| Colon, Transverse | LYNX1 | 0.0015 | ||
| 5~50 kbps | Brain, Caudate Basal Ganglia | PYCRL | 0.02 | |
| Brain, Cerebellar Hemisphere | PYCRL | 0.037 | ||
| TRAPPC9 | 0.037 | |||
| Brain, Hypothalamus | PYCRL | 0.024 | ||
| TRAPPC9 | 0.024 | |||
| Skin, Not Sun Exposed Suprapubic | TRAPPC9 | 0.049 | ||
| Additive | 5~50 kbps | Skin, Not Sun Exposed Suprapubic | ANXA13 | 0.015 |
| TRAPPC9 | 0.025 | |||
| 50~500 kbps | Brain, Spinal Cord Cervical C1 | GPT | 0.031 | |
| Skin, Sun Exposed Lower Leg | ZFP41 | 0.025 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, I.-H.; Kong, S.W. ADGR: Admixture-Informed Differential Gene Regulation. Genes 2023, 14, 147. https://doi.org/10.3390/genes14010147
Lee I-H, Kong SW. ADGR: Admixture-Informed Differential Gene Regulation. Genes. 2023; 14(1):147. https://doi.org/10.3390/genes14010147
Chicago/Turabian StyleLee, In-Hee, and Sek Won Kong. 2023. "ADGR: Admixture-Informed Differential Gene Regulation" Genes 14, no. 1: 147. https://doi.org/10.3390/genes14010147
APA StyleLee, I.-H., & Kong, S. W. (2023). ADGR: Admixture-Informed Differential Gene Regulation. Genes, 14(1), 147. https://doi.org/10.3390/genes14010147





