Coyotes in New York City Carry Variable Genomic Dog Ancestry and Influence Their Interactions with Humans
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Genomic DNA Preparation
2.2. RAD Sequencing and Bioinformatic Processing
2.3. SNP Discovery and Filtering
2.4. Sex Inference
2.5. Inference of Autosomal Canid Ancestry
2.6. Human-Directed Hypersociability Genotypes
3. Results
3.1. Sequencing and Sex Inference
3.2. Dog Ancestry Detected in Coyotes in the New York Metro Area
3.3. Family Group Contains F1 and F2 Dog–Coyote Hybrids
3.4. Human-Directed Hypersociability Genotypes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gehrt, S.D.; Brown, J.L.; Anchor, C. Is the urban coyote a misanthropic synanthrope? The case from Chicago. Cities Environ. 2011, 4, 3. [Google Scholar] [CrossRef]
- Hody, J.W.; Kays, R. Mapping the expansion of coyotes (Canis latrans) across North and Central America. ZooKeys 2018, 759, 81–97. [Google Scholar] [CrossRef] [PubMed]
- Hinton, J.W.; Heppenheimer, E.; West, K.M.; Caudill, D.; Karlin, M.L.; Kilgo, J.C.; Mayer, J.J.; Miller, K.V.; Walch, M.; vonHoldt, B.; et al. Geographic patterns in morphometric and genetic variation for coyote populations with emphasis on southeastern coyotes. Ecol. Evol. 2019, 9, 3389–3404. [Google Scholar] [CrossRef] [PubMed]
- Way, J.G.; Rutledge, L.; Wheeldon, T.; White, B.N. Genetic characterization of eastern “coyotes” in eastern Massachusetts. Northeast. Nat. 2010, 17, 189–204. [Google Scholar] [CrossRef]
- Kays, R.; Curtis, A.; Kirchman, J.J. Rapid adaptive evolution of northeastern coyotes via hybridization with wolves. Biol. Lett. 2010, 6, 89–93. [Google Scholar] [CrossRef]
- Lehman, N.; Eisenhawer, A.; Hansen, K.; Mech, L.D.; Peterson, R.O.; Gogan, P.J.; Wayne, R.K. Introgression of coyote mitochondrial DNA into sympatric North American gray wolf populations. Evolution 1991, 45, 104–119. [Google Scholar] [CrossRef]
- vonHoldt, B.M.; Pollinger, J.P.; Earl, D.A.; Knowles, J.C.; Boyko, A.R.; Parker, H.; Geffen, E.; Pilot, M.; Jedrzejewski, W.; Jedrzejewska, B.; et al. A genome-wide perspective on the evolutionary history of enigmatic wolf-like canids. Genome Res. 2011, 21, 1294–1305. [Google Scholar] [CrossRef]
- Benson, J.F.; Patterson, B.R.; Wheeldon, T.J. Spatial genetic and morphologic structure of wolves and coyotes in relation to environmental heterogeneity in a C anis hybrid zone. Mol. Ecol. 2012, 21, 5934–5954. [Google Scholar] [CrossRef]
- Wheeldon, T.J.; Rutledge, L.Y.; Patterson, B.R.; White, B.N.; Wilson, P.J. Y-chromosome evidence supports asymmetric dog introgression into eastern coyotes. Ecol. Evol. 2013, 3, 3005–3020. [Google Scholar] [CrossRef]
- Monzón, J.; Kays, R.; Dykhuizen, D.E. Assessment of coyote–wolf–dog admixture using ancestry-informative diagnostic SNPs. Mol. Ecol. 2014, 23, 182–197. [Google Scholar] [CrossRef] [PubMed]
- Bohling, J.H.; Waits, L.P. Factors influencing red wolf–coyote hybridization in eastern North Carolina, USA. Biol. Conserv. 2015, 184, 108–116. [Google Scholar] [CrossRef]
- vonHoldt, B.M.; Cahill, J.A.; Fan, Z.; Gronau, I.; Robinson, J.; Pollinger, J.P.; Shapiro, B.; Wall, J.; Wayne, R.K. Whole-genome sequence analysis shows that two endemic species of North American wolf are admixtures of the coyote and gray wolf. Sci. Adv. 2016, 2, e1501714. [Google Scholar] [CrossRef] [PubMed]
- vonHoldt, B.M.; Aardema, M.L. Updating the bibliography of interbreeding among Canis in North America. J. Hered. 2020, 111, 249–262. [Google Scholar] [CrossRef]
- Gese, E.M.; Knowlton, F.F.; Adams, J.R.; Beck, K.; Fuller, T.K.; Murray, D.L.; Steury, T.D.; Stoskopf, M.K.; Waddell, W.T.; Waits, L.P. Managing hybridization of a recovering endangered species: The red wolf Canis rufus as a case study. Curr. Zool. 2015, 61, 191–205. [Google Scholar] [CrossRef]
- Thornton, D.H.; Murray, D.L. Influence of hybridization on niche shifts in expanding coyote populations. Divers. Distrib. 2014, 20, 1355–1364. [Google Scholar] [CrossRef]
- Heppenheimer, E.; Brzeski, K.E.; Hinton, J.W.; Patterson, B.R.; Rutledge, L.Y.; DeCandia, A.L.; Wheeldon, T.; Fain, S.R.; Hohenlohe, P.A.; Kays, R. High genomic diversity and candidate genes under selection associated with range expansion in eastern coyote (Canis latrans) populations. Ecol. Evol. 2018, 8, 12641–12655. [Google Scholar] [CrossRef] [PubMed]
- Gehrt, S.D.; Anchor, C.; White, L.A. Home Range and Landscape Use of Coyotes in a Metropolitan Landscape: Conflict or Coexistence? J. Mammal. 2009, 90, 1045–1057. [Google Scholar] [CrossRef]
- Weckel, M.; Bogan, D.A.; Burke, R.L.; Nagy, C.; Siemer, W.F.; Green, T.; Mitchell, N. Coyotes go “bridge and tunnel”: A narrow opportunity to study the socio-ecological impacts of coyote range expansion on Long Island, NY pre-and post-arrival. Cities Environ. 2015, 8, 5. [Google Scholar]
- Toomey, A.; Weckel, M.; Nagy, C.; Gormezano, L.; Silver, S. The last frontier: Eastern coyotes in New York City. Wildl. Prof. 2012, 6, 54–57. [Google Scholar]
- Henger, C.S.; Herrera, G.A.; Nagy, C.M.; Weckel, M.E.; Gormezano, L.J.; Wultsch, C.; Munshi-South, J. Genetic diversity and relatedness of a recently established population of eastern coyotes (Canis latrans) in New York City. Urban Ecosyst. 2019, 23, 319–330. [Google Scholar] [CrossRef]
- DeCandia, A.L.; Henger, C.S.; Krause, A.; Gormezano, L.J.; Weckel, M.E.; Nagy, C.M.; Munshi-South, J.; vonHoldt, B.M. Genetics of urban colonization: Neutral and adaptive variation in coyotes (Canis latrans) inhabiting the New York metropolitan area. J. Urban Ecol. 2019, 5, juz002. [Google Scholar] [CrossRef]
- Bekoff, M. Canis latrans. Mamm. Species 1977, 79, 1–9. [Google Scholar] [CrossRef]
- MacCracken, J.G.; Hansen, R.M. Coyote feeding strategies in southeastern Idaho: Optimal foraging by an opportunistic predator? J. Wildl. Manage. 1987, 51, 278–285. [Google Scholar] [CrossRef]
- Windberg, L.A.; Mitchell, C.D. Winter diets of coyotes in relation to prey abundance in southern Texas. J. Mammal. 1990, 71, 439–447. [Google Scholar] [CrossRef]
- Arjo, W.M.; Pletscher, D.H.; Ream, R.R. Dietary overlap between wolves and coyotes in northwestern Montana. J. Mammal. 2002, 83, 754–766. [Google Scholar] [CrossRef]
- Nagy, C.M.; Koestner, C.; Clemente, S.; Weckel, M. Occupancy and breeding status of coyotes in New York City parks, 2011 to 2014. Urban Nat. 2016, 9, 1–16. [Google Scholar]
- Duncan, N.; Asher, O.; Weckel, M.; Nagy, C.; Henger, C.; Yau, F.; Gormanzano, L. Baseline diet of an urban carnivore on an expanding range front. J. Urban Ecol. 2020, 6, juaa021. [Google Scholar] [CrossRef]
- Larson, R.N.; Brown, J.L.; Karels, T.; Riley, S.P. Effects of urbanization on resource use and individual specialization in coyotes (Canis latrans) in southern California. PLoS ONE 2020, 15, e0228881. [Google Scholar] [CrossRef]
- Bradfield, A.A.; Nagy, C.M.; Weckel, M.; Lahti, D.C.; Habig, B. Predictors of Mammalian Diversity in the New York Metropolitan Area. Front. Ecol. Evol. 2022, 10, 903211. [Google Scholar] [CrossRef]
- Henger, C.; Hargous, E.; Nagy, C.; Weckel, M.; Wultsch, C.; Krampis, K.; Duncan, N.; Gormanzano, L.; Munshi-South, J. DNA metabarcoding reveals that coyotes in New York City consume wide variety of native prey species and human food. PeerJ 2021, 59, 37–50. [Google Scholar] [CrossRef]
- Mowry, C.B.; Wilson, L.A.; vonHoldt, B.M. Interface of Human/Wildlife Interactions: An Example of a Bold Coyote (Canis latrans) in Atlanta, GA, USA. Diversity 2021, 13, 372. [Google Scholar] [CrossRef]
- Ali, O.A.; O’Rourke, S.M.; Amish, S.J.; Meek, M.H.; Luikart, G.; Jeffres, C.; Miller, M.R. RAD capture (Rapture): Flexible and efficient sequence-based genotyping. Genetics 2016, 202, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Catchen, J.; Hohenlohe, P.A.; Bassham, S.; Amores, A.; Cresko, W.A. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef]
- Rochette, N.C.; Rivera-Colón, A.G.; Catchen, J.M. Stacks 2: Analytical methods for paired-end sequencing improve RADseq-based population genomics. Mol. Ecol. 2019, 28, 4737–4754. [Google Scholar] [CrossRef] [PubMed]
- Lindblad-Toh, K.; Wade, C.M.; Mikkelsen, T.S.; Karlsson, E.K.; Jaffe, D.B.; Kamal, M.; Clamp, M.; Chang, J.L.; Kulbokas, E.J.; Zody, M.C. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 2005, 438, 803–819. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, s13742-015-0047-8. [Google Scholar] [CrossRef]
- Pew, J.; Muir, P.H.; Wang, J.; Frasier, T.R. related: An R package for analysing pairwise relatedness from codominant molecular markers. Mol. Ecol. Resour. 2015, 15, 557–561. [Google Scholar] [CrossRef]
- vonHoldt, B.M.; DeCandia, A.L.; Heppenheimer, E.; Janowitz-Koch, I.; Shi, R.; Zhou, H.; German, C.A.; Brzeski, K.E.; Cassidy, K.A.; Stahler, D.R. Heritability of interpack aggression in a wild pedigreed population of North American grey wolves. Mol. Ecol. 2020, 29, 1764–1775. [Google Scholar] [CrossRef] [PubMed]
- Milligan, B.G. Maximum-likelihood estimation of relatedness. Genetics 2003, 163, 1153–1167. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Davis, B.W.; Raudsepp, T.; Wilkerson, A.J.P.; Mason, V.C.; Ferguson-Smith, M.; O’Brien, P.C.; Waters, P.D.; Murphy, W.J. Comparative analysis of mammalian Y chromosomes illuminates ancestral structure and lineage-specific evolution. Genome Res. 2013, 23, 1486–1495. [Google Scholar] [CrossRef] [PubMed]
- Raudsepp, T.; Das, P.; Avila, F.; Chowdhary, B. The pseudoautosomal region and sex chromosome aneuploidies in domestic species. Sex. Dev. 2012, 6, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Heppenheimer, E.; Brzeski, K.E.; Wooten, R.; Waddell, W.; Rutledge, L.Y.; Chamberlain, M.J.; Stahler, D.R.; Hinton, J.W.; VonHoldt, B.M. Rediscovery of red wolf ghost alleles in a canid population along the American Gulf Coast. Genes 2018, 9, 618. [Google Scholar] [CrossRef] [PubMed]
- Heppenheimer, E.; Harrigan, R.J.; Rutledge, L.Y.; Koepfli, K.-P.; DeCandia, A.L.; Brzeski, K.E.; Benson, J.F.; Wheeldon, T.; Patterson, B.R.; Kays, R. Population Genomic Analysis of North American Eastern Wolves (Canis lycaon) Supports Their Conservation Priority Status. Genes 2018, 9, 606. [Google Scholar] [CrossRef]
- Heppenheimer, E.; Brzeski, K.E.; Hinton, J.W.; Chamberlain, M.J.; Robinson, J.; Wayne, R.K.; vonHoldt, B.M. A Genome-Wide Perspective on the Persistence of Red Wolf Ancestry in Southeastern Canids. J. Hered. 2020, 111, 277–286. [Google Scholar] [CrossRef]
- Guan, Y. Detecting structure of haplotypes and local ancestry. Genetics 2014, 196, 625–642. [Google Scholar] [CrossRef]
- vonHoldt, B.M.; Shuldiner, E.; Koch, I.J.; Kartzinel, R.Y.; Hogan, A.; Brubaker, L.; Wanser, S.; Stahler, D.; Wynne, C.D.L.; Ostrander, E.A.; et al. Structural variants in genes associated with human Williams-Beuren syndrome underlie stereotypical hypersociability in domestic dogs. Sci. Adv. 2017, 3, e1700398. [Google Scholar] [CrossRef]
- Tandon, D.; Ressler, K.; Petticord, D.; Papa, A.; Jiranek, J.; Wilkinson, R.; Kartzinel, R.Y.; Ostrander, E.A.; Burney, N.; Borden, C.; et al. Homozygosity for Mobile Element Insertions Associated with WBSCR17 Could Predict Success in Assistance Dog Training Programs. Genes 2019, 10, 439. [Google Scholar] [CrossRef]
- Karolchik, D.; Hinrichs, A.S.; Furey, T.S.; Roskin, K.M.; Sugnet, C.W.; Haussler, D.; Kent, W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004, 32, D493–D496. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Harrison, D.J. Dispersal characteristics of juvenile coyotes in Maine. J. Wildl. Manag. 1992, 56, 128–138. [Google Scholar] [CrossRef]
- Nagy, C.M.; Weckel, M.E.; Monzón, J.; Duncan, N.; Rosenthal, M.R. Initial colonization of Long Island, New York by the eastern coyote, Canis latrans (Carnivora, Canidae), including first record of breeding. Check List 2017, 13, 901–907. [Google Scholar] [CrossRef]
- Carrete, M.; Martínez-Padilla, J.; Rodríguez-Martínez, S.; Rebolo-Ifrán, N.; Palma, A.; Tella, J.L. Heritability of fear of humans in urban and rural populations of a bird species. Sci. Rep. 2016, 6, 31060. [Google Scholar] [CrossRef] [PubMed]
- Cook, M.O.; Weaver, M.J.; Hutton, P.; McGraw, K.J. The effects of urbanization and human disturbance on problem solving in juvenile house finches (Haemorhous mexicanus). Behav. Ecol. Sociobiol. 2017, 71, 85. [Google Scholar] [CrossRef]
- Martin, J.G.; Réale, D. Temperament, risk assessment and habituation to novelty in eastern chipmunks, Tamias striatus. Anim. Behav. 2008, 75, 309–318. [Google Scholar] [CrossRef]
- Uchida, K.; Suzuki, K.; Shimamoto, T.; Yanagawa, H.; Koizumi, I. Seasonal variation of flight initiation distance in Eurasian red squirrels in urban versus rural habitat. J. Zool. 2016, 298, 225–231. [Google Scholar] [CrossRef]
- Vincze, E.; Papp, S.; Preiszner, B.; Seress, G.; Bókony, V.; Liker, A. Habituation to human disturbance is faster in urban than rural house sparrows. Behav. Ecol. 2016, 27, 1304–1313. [Google Scholar] [CrossRef]
- Schell, C.J.; Young, J.K.; Lonsdorf, E.V.; Santymire, R.M.; Mateo, J.M. Parental habituation to human disturbance over time reduces fear of humans in coyote offspring. Ecol. Evol. 2018, 8, 12965–12980. [Google Scholar] [CrossRef]
- Thompson, C.A.; Malcolm, J.R.; Patterson, B.R. Individual and temporal variation in use of residential areas by urban coyotes. Front. Ecol. Evol. 2021, 9, 687504. [Google Scholar] [CrossRef]
- Atwood, T.C.; Weeks, H.P.; Gehring, T.M. Spatial ecology of coyotes along a suburban-to-rural gradient. J. Wildl. Manag. 2004, 68, 1000–1009. [Google Scholar] [CrossRef]
- Gese, E.M.; Morey, P.S.; Gehrt, S.D. Influence of the urban matrix on space use of coyotes in the Chicago metropolitan area. J. Ethol. 2012, 30, 413–425. [Google Scholar] [CrossRef]
- Young, J.K.; Hammill, E.; Breck, S.W. Interactions with humans shape coyote responses to hazing. Sci. Rep. 2019, 9, 20046. [Google Scholar] [CrossRef]
- Anderson, T.M.; vonHoldt, B.M.; Candille, S.I.; Musiani, M.; Greco, C.; Stahler, D.R.; Smith, D.W.; Padhukasahasram, B.; Randi, E.; Leonard, J.A. Molecular and evolutionary history of melanism in North American gray wolves. Science 2009, 323, 1339–1343. [Google Scholar] [CrossRef]
- Gompper, M.E. Top Carnivores in the Suburbs? Ecological and Conservation Issues Raised by Colonization of North-eastern North America by Coyotes: The expansion of the coyote’s geographical range may broadly influence community structure, and rising coyote densities in the suburbs may alter how the general public views wildlife. Bioscience 2002, 52, 185–190. [Google Scholar] [CrossRef]
- Breck, S.W.; Poessel, S.A.; Mahoney, P.; Young, J.K. The intrepid urban coyote: A comparison of bold and exploratory behavior in coyotes from urban and rural environments. Sci. Rep. 2019, 9, 2104. [Google Scholar] [CrossRef]
- Dietz, L.; Arnold, A.-M.K.; Goerlich-Jansson, V.C.; Vinke, C.M. The importance of early life experiences for the development of behavioural disorders in domestic dogs. Behavior 2018, 155, 83. [Google Scholar] [CrossRef]


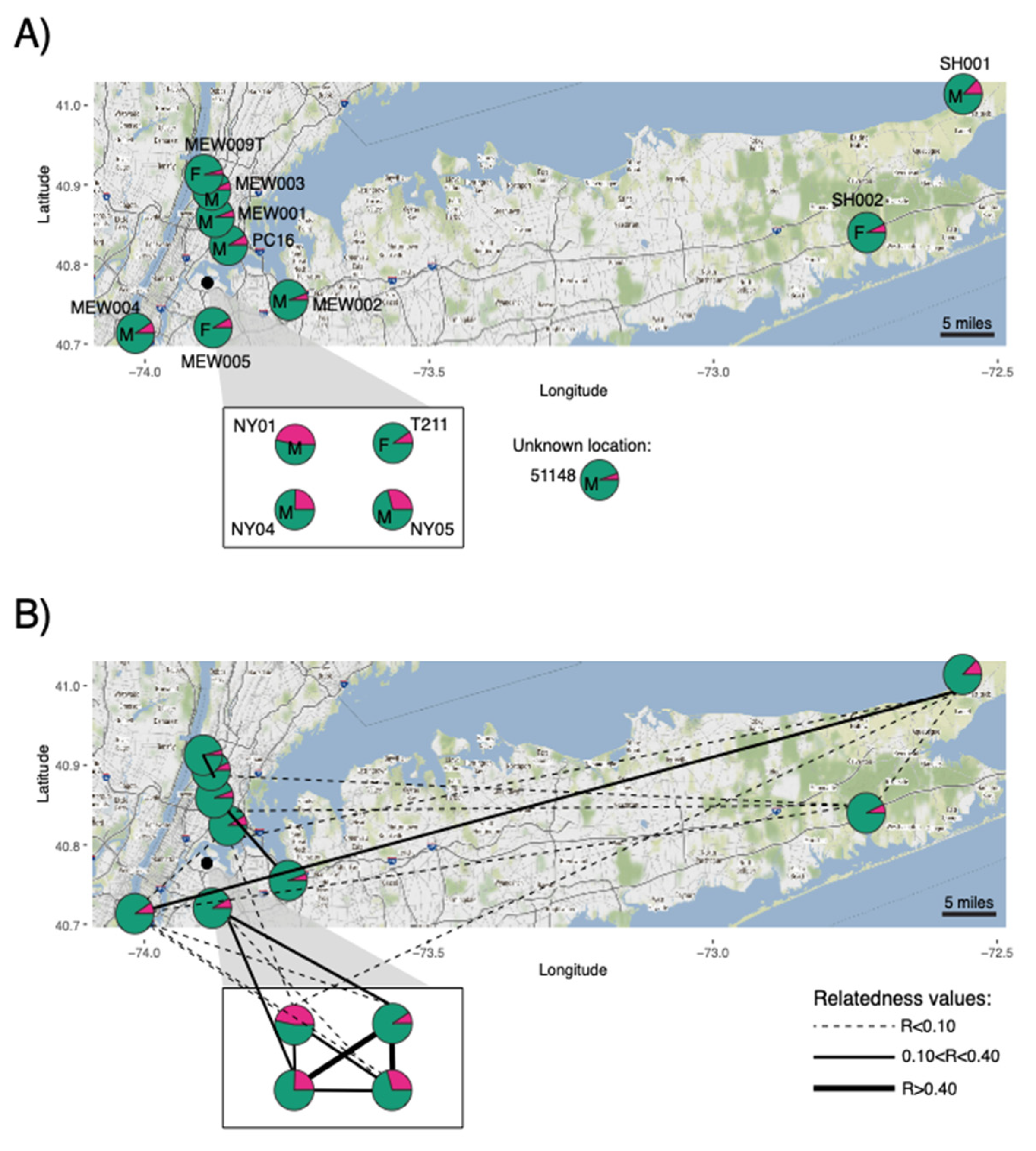
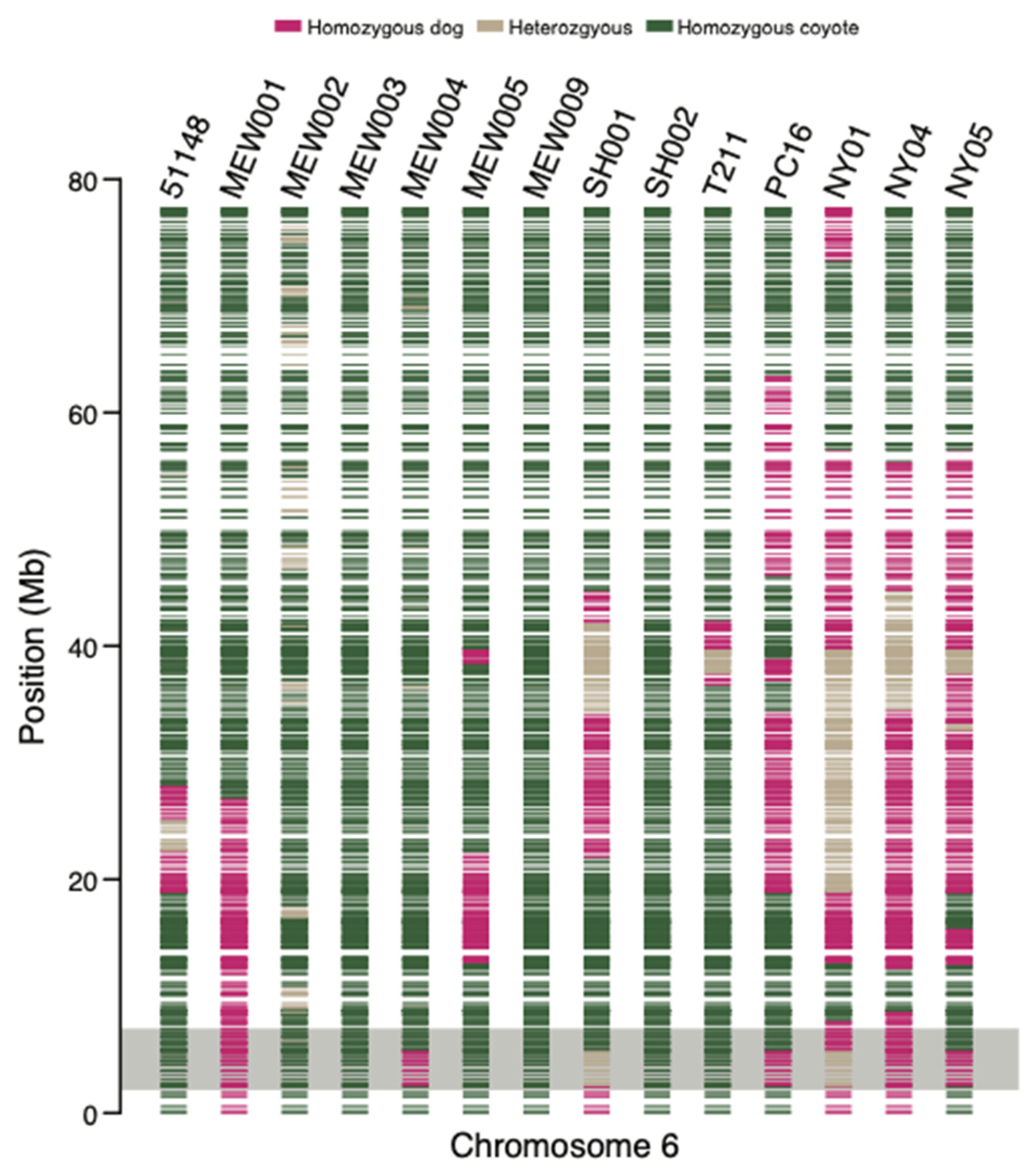
| Sample ID | Cfa | Cla |
|---|---|---|
| PC16 | 0.084 | 0.917 |
| MEW005 | 0.081 | 0.919 |
| NY04 | 0.245 | 0.755 |
| MEW001 | 0.058 | 0.942 |
| T211 | 0.094 | 0.907 |
| MEW009B | 0.048 | 0.952 |
| MEW004 | 0.092 | 0.908 |
| SH002 | 0.070 | 0.930 |
| NY01 | 0.465 | 0.535 |
| NY05 | 0.292 | 0.708 |
| MEW009T | 0.043 | 0.957 |
| MEW003 | 0.067 | 0.934 |
| SH001 | 0.125 | 0.875 |
| MEW002 | 0.056 | 0.944 |
| 51148 | 0.056 | 0.944 |
| Sample ID 1 | Sample ID 2 | Dyadml r |
|---|---|---|
| T211 | NY05 | 0.50 |
| NY04 | T211 | 0.47 |
| NY04 | NY05 | 0.40 |
| NY01 | NY05 | 0.39 |
| NY04 | NY01 | 0.35 |
| T211 | NY01 | 0.0 |
| Sample ID | Cfa6.6 | Cfa6.7 | Cfa6.66 |
|---|---|---|---|
| MEW007 | 0 | 0 | 0 |
| PC16 | 0 | 0 | 0 |
| MEW008 | 0 | 0 | 0 |
| MEW005 | 0 | 0 | 0 |
| NY04 * | 1 | 0 | 1 |
| MEW001 | 1 | 1 | 0 |
| T211 * | 0 | 0 | 0 |
| MEW009 | 0 | 0 | 0 |
| MEW004 | 0 | 0 | 0 |
| SH002 | 0 | 0 | 0 |
| NY01 * | 1 | 0 | 1 |
| NY05 * | 0 | 0 | 0 |
| MEW003 | 0 | 0 | 0 |
| SH001 | 0 | 0 | 0 |
| MEW002 | 0 | 0 | - |
| 51148 | 0 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caragiulo, A.; Gaughran, S.J.; Duncan, N.; Nagy, C.; Weckel, M.; vonHoldt, B.M. Coyotes in New York City Carry Variable Genomic Dog Ancestry and Influence Their Interactions with Humans. Genes 2022, 13, 1661. https://doi.org/10.3390/genes13091661
Caragiulo A, Gaughran SJ, Duncan N, Nagy C, Weckel M, vonHoldt BM. Coyotes in New York City Carry Variable Genomic Dog Ancestry and Influence Their Interactions with Humans. Genes. 2022; 13(9):1661. https://doi.org/10.3390/genes13091661
Chicago/Turabian StyleCaragiulo, Anthony, Stephen J. Gaughran, Neil Duncan, Christopher Nagy, Mark Weckel, and Bridgett M. vonHoldt. 2022. "Coyotes in New York City Carry Variable Genomic Dog Ancestry and Influence Their Interactions with Humans" Genes 13, no. 9: 1661. https://doi.org/10.3390/genes13091661
APA StyleCaragiulo, A., Gaughran, S. J., Duncan, N., Nagy, C., Weckel, M., & vonHoldt, B. M. (2022). Coyotes in New York City Carry Variable Genomic Dog Ancestry and Influence Their Interactions with Humans. Genes, 13(9), 1661. https://doi.org/10.3390/genes13091661





