C-to-U RNA Editing: A Site Directed RNA Editing Tool for Restoration of Genetic Code
Abstract
:1. Introduction
2. RNA Editing
3. C-to-U RNA Editing
4. Artificial C-to-U RNA Editing
5. Enzymes (Editors) for Artificial C-to-U RNA Editing
6. C-to-U RNA Editing in Mammals
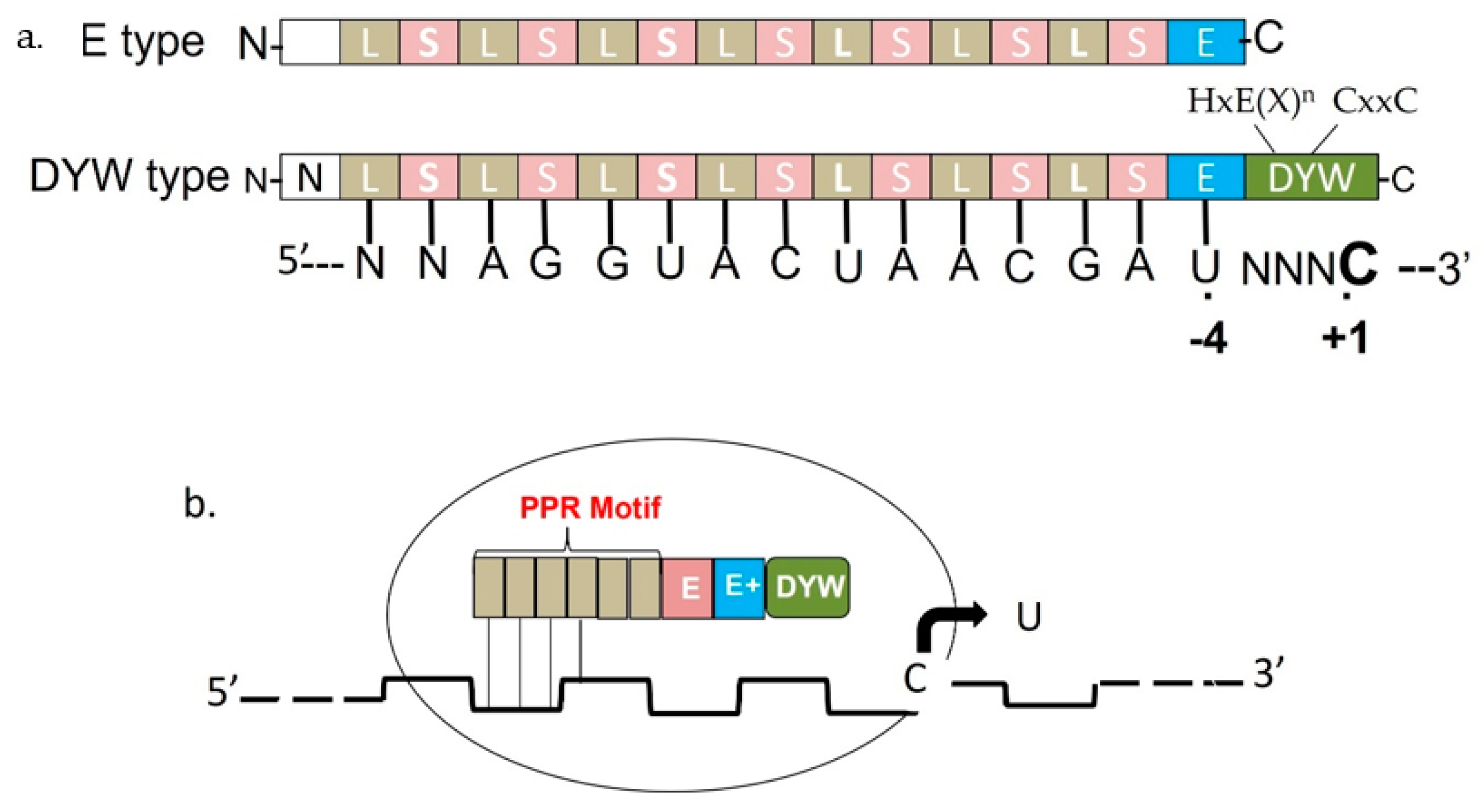
7. C-to-U RNA Editing in Plants
8. Human Diseases Related to C-to-U Editing
9. Future Perspectives of RNA Editing in Diagnosis and Treatment
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wolf, J.; Gerber, A.P.; Keller, W. tadA, an essential tRNA-specific adenosine deaminase from Escherichia coli. EMBO J. 2002, 21, 3841–3851. [Google Scholar] [CrossRef] [PubMed]
- Brennicke, A.; Marchfelder, A.; Binder, S. RNA editing. FEMS Microbiol. Rev. 1999, 23, 297–316. [Google Scholar] [CrossRef] [PubMed]
- John, D.; Weirick, T.; Dimmeler, S.; Uchida, S. RNAEditor: Easy detection of RNA editing events and the introduction of editing islands. Brief. Bioinform. 2017, 18, 993–1001. [Google Scholar] [CrossRef]
- Bahn, J.H.; Lee, J.H.; Li, G.; Greer, C.; Peng, G.; Xiao, X. Accurate identification of A-to-I RNA editing in human by transcriptome sequencing. Genome Res. 2012, 22, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Nishikura, K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010, 79, 321–349. [Google Scholar] [CrossRef] [PubMed]
- Zanlungo, S.; Béqu, D.; Quiñones, V.; Araya, A.; Jordana, X. RNA editing of a pocytochrome b (cob) transcripts in mitochondria from two genera of plants. Curr. Genet. 1993, 24, 344–348. [Google Scholar] [CrossRef]
- Gott, J.M.; Emeson, R.B. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 2000, 34, 499–531. [Google Scholar] [CrossRef]
- Chen, S.H.; Habib, G.; Yang, C.Y.; Gu, Z.W.; Lee, B.R.; Weng, S.A.; Silberman, S.R.; Cai, S.J.; Deslypere, J.P.; Rosseneu, M.; et al. Apolipoprotein B-48 is the product of a messenger RNA with an organ-specific inframe stop codon. Science 1987, 238, 363–366. [Google Scholar] [CrossRef]
- Powell, L.M.; Wallis, S.C.; Pease, R.J.; Edwards, Y.H.; Knott, T.J.; Scott, J. A novel form of tissue-specific RNA processing produces a polipoprotein-B48 in intestine. Cell 1987, 50, 831–840. [Google Scholar] [CrossRef]
- Freyer, R.; Kiefer-Meyer, M.C.; Kössel, H. Occurrence of plastid RNA editing in all major lineages of land plants. Proc. Natl. Acad. Sci. USA 1997, 94, 6285–6290. [Google Scholar] [CrossRef] [Green Version]
- Navaratnam, N.; Bhattacharya, S.; Fujino, T.; Patel, D.; Jarmuz, A.L.; Scott, J. Evolutionary origins of apoB mRNA editing: Catalysis by a cytidine deaminase that has acquired a novel RNA-binding motif at its active site. Cell 1995, 81, 187–195. [Google Scholar] [CrossRef]
- Blanc, V.; Davidson, N.O. APOBEC-1-mediated RNA editing. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010, 2, 594–602. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Patnaik, S.K.; Taggart, R.T.; Kannisto, E.D.; Enriquez, S.M.; Gollnick, P.; Baysal, B.E. APOBEC3A cytidine deaminase induces RNA editing in monocytes and macrophages. Nat. Commun. 2015, 6, 6881. [Google Scholar] [CrossRef]
- Sharma, S.; Patnaik, S.K.; Taggart, R.T.; Baysal, B.F. The double-domain cytidine deaminase APOBEC3G is a cellular site-specific RNA editing enzyme. Sci. Rep. 2016, 6, 39100. [Google Scholar] [CrossRef] [PubMed]
- Patnaik, S.K.; Kannisto, E. APOBEC3B is a new RNA editing enzyme. In Proceedings of the RNA 2016, Annual Meeting of RNA Society, Kyoto, Japan, 28 June–2 July 2016. [Google Scholar]
- Moris, A.; Murray, S.; Cardinaud, S. AID and APOBECs span the gap between innate and adaptive immunity. Front. Microbiol. 2014, 5, 534. [Google Scholar] [CrossRef] [PubMed]
- Bhakta, S.; Sakari, M.; Tsukahara, T. RNA editing of BFP, a point mutant of GFP, using artificial APOBEC1 deaminase to restore the genetic code. Sci. Rep. 2020, 10, 17304. [Google Scholar] [CrossRef] [PubMed]
- Gerber, A.; Keller, W. RNA editing by base deamination: More enzymes, more targets, new mysteries. Trends Biochem. Sci. 2001, 26, 376–384. [Google Scholar] [CrossRef]
- Roberts, S.A.; Lawrence, M.S.; Klimczak, L.J.; Grimm, S.A.; Fargo, D.; Stojanov, P.; Kiezun, A.; Kryukov, G.V.; Carter, S.L.; Saksena, G.; et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 2013, 45, 970–976. [Google Scholar] [CrossRef]
- Swanton, C.; McGranahan, N.; Starrett, G.J.; Harris, R.S. APOBEC Enzymes: Mutagenic fuel for cancer evolution and heterogeneity. Cancer Discov. 2015, 5, 704–712. [Google Scholar] [CrossRef]
- Kung, C.; Maggi, L.B., Jr.; Weber, J.D. The Role of RNA Editing in Cancer Development and Metabolic Disorders. Front. Endocrinol. 2018, 9, 762. [Google Scholar] [CrossRef] [Green Version]
- Chan, K.; Roberts, S.A.; Klimczak, L.J.; Sterling, J.F.; Saini, N.; Malc, E.P.; Kim, J.; Kwiatkowski, D.J.; Fargo, D.C.; Mieczkowski, P.A.; et al. An APOBEC3A hyper-mutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat. Genet. 2015, 47, 1067–1072. [Google Scholar] [CrossRef] [PubMed]
- Burns, M.B.; Lackey, L.; Carpenter, M.A.; Rathore, A.; Land, A.M.; Leonard, B.; Refsland, E.W.; Kotandeniya, D.; Tretyakova, N.; Nikas, J.B.; et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 2013, 494, 366–370. [Google Scholar] [CrossRef] [PubMed]
- Knisbacher, B.A.; Gerber, D.; Levanon, E.Y. DNA Editing by APOBECs: A genomic preserver and transformer. Trends Genet. 2016, 32, 16–28. [Google Scholar] [CrossRef] [PubMed]
- Davidson, N.O.; Shelness, G.S. APOLIPOPROTEIN B: mRNA editing, lipoprotein assembly, and presecretory degradation. Annu. Rev. Nutr. 2000, 20, 169–193. [Google Scholar] [CrossRef] [PubMed]
- Prohaska, K.M.; Bennett, R.P.; Salter, J.D.; Smith, H.C. The multifaceted roles of RNA binding in APOBEC cytidine deaminase functions. Wiley Interdiscip. Rev. RNA 2014, 5, 493–508. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Patnaik, S.K.; Kemer, Z.; Baysal, B.E. Transient overexpression of exogenous APOBEC3A causes C to U RNA editing of thousands of genes. RNA Biol. 2017, 14, 603–610. [Google Scholar] [CrossRef]
- Sharma, S.; Wang, J.; Alqassim, E.; Portwood, S.; Cortes Gomez, E.; Maguire, O.; Basse, P.H.; Wang, E.S.; Segal, B.H.; Baysal, B.E. Mitochondrial hypoxic stress induces widespread RNA editing by APOBEC3G in natural killer cells. Genome Biol. 2019, 20, 37. [Google Scholar] [CrossRef]
- Marek-Trzonkowska, N.; Piekarska, K.; Filipowicz, N.; Piotrowski, A.; Gucwa, M.; Vogt, K.; Sawitzki, B.; Siebert, J.; Trzonkowski, P. Mild hypothermia provides Treg stability. Sci. Rep. 2017, 7, 11915. [Google Scholar] [CrossRef]
- Takeda, E.; Tsuji-Kawahara, S.; Sakamoto, M.; Langlois, M.A.; Neuberger, M.S.; Rada, C.; Miyazawa, M. Mouse APOBEC3 restricts friend leukemia virus infection and pathogenesis in vivo. J. Virol. 2008, 82, 10998–11008. [Google Scholar] [CrossRef] [PubMed]
- Lai, F.; Drakas, R.; Nishikura, K. Mutagenic analysis of double-stranded RNA adenosine deaminase, a candidate enzyme for RNA editing of glutamate-gated ion channel transcripts. J. Biol. Chem. 1995, 270, 17098–17105. [Google Scholar] [CrossRef] [Green Version]
- McGrath, E.; Shin, H.; Zhang, L.; Phue, J.N.; Wu, W.W.; Shen, R.F.; Jang, Y.Y.; Revollo, J.; Ye, Z. Targeting specificity of APOBEC-based cytosine base editor in human iPSCs determined by whole genome sequencing. Nat. Commun. 2019, 10, 5353. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.C.; Bennett, R.P.; Kizilyer, A.; McDougall, W.M.; Prohaska, K.M. Functions and regulation of the APOBEC family of proteins. Semin. Cell Dev. Biol. 2012, 23, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Bazak, L.; Haviv, A.; Barak, M.; Jacob-Hirsch, J.; Deng, P.; Zhang, R.; Isaacs, F.J.; Rechavi, G.; Li, J.B.; Eisenberg, E.; et al. A to I RNA editing occurs at over a hundred million genomic sites located in a majority of human genes. Genome Res. 2013, 24, 365–376. [Google Scholar] [CrossRef]
- Ramaswami, G.; Lin, W.; Piskol, R.; Tan, M.H.; Davis, C.; Li, J.B. Accurate identification of human alu and non-alu RNA editing sites. Nat. Methods 2012, 9, 579–581. [Google Scholar] [CrossRef]
- Li, J.B.; Church, G.M. Deciphering the functions and regulation of brain-enriched A to I RNA editing. Nat. Neurosci. 2013, 16, 1518–1522. [Google Scholar] [CrossRef] [PubMed]
- Rosenber, B.R.; Hamilton, C.E.; Mwangi, M.M.; Dewell, S.; Papavasiliou, F.N. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3′ UTRs. Nat. Struct. Mol. Biol. 2011, 18, 230–236. [Google Scholar] [CrossRef]
- Blanc, V.; Park, E.; Schaefer, S.; Miller, M.; Lin, Y.; Kennedy, S.; Billing, A.M.; Hamidane, H.B.; Graumann, J.; Mortazavi, A.; et al. Genome-wide identification and functional analysis of Apobec-1-mediated C to U RNA editing in mouse small intestine and liver. Genome Biol. 2014, 15, R79. [Google Scholar] [CrossRef] [PubMed]
- Harjanto, D.; Papamarkou, T.; Oates, C.J.; Rayon-Estrada, V.; Papavasiliou, F.N.; Papavasiliou, A. RNA editinggenerates cellular subsets with diverse sequence within populations. Nat. Commun. 2016, 7, 12145. [Google Scholar] [CrossRef]
- Blanc, V.; Davidson, N.O. Mouse and other rodent models of C to U RNA editing. Methods Mol. Biol. 2011, 718, 121–135. [Google Scholar]
- Navaratnam, N.; Patel, D.; Shah, R.R.; Greeve, J.C.; Powell, L.M.; Knott, T.J.; Scott, J. An additional editing site is present in apolipoprotein B mRNA. Nucleic Acids Res. 1991, 19, 1741–1744. [Google Scholar] [CrossRef] [PubMed]
- Backus, J.W.; Smith, H.C. Three distinct RNA sequence elements are required for efficient apolipoprotein B (apoB) RNA editing in vitro. Nucleic Acids Res. 1992, 20, 6007–6014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Backus, J.W.; Smith, H.C. Apolipoprotein B mRNA sequences 3′ of the editing site are necessary and sufficient for editing and editosome assembly. Nucleic Acids Res. 1991, 19, 6781–6786. [Google Scholar] [CrossRef] [PubMed]
- Hersberger, M.; Innerarity, T.L. Two efficiency elements flanking the editing site of cytidine 6666 in the apolipoprotein B mRNA support mooring-dependent editing. J. Biol. Chem. 1998, 273, 9435–9442. [Google Scholar] [CrossRef] [PubMed]
- Nakamuta, M.; Tsai, A.; Chan, L.; Davidson, N.O.; Teng, B.B. Sequence elements required for apolipoprotein B mRNA editing enhancement activity from chicken enterocytes. Biochem. Biophys. Res. Commun. 1999, 254, 744–750. [Google Scholar] [CrossRef]
- Shah, R.R.; Knott, T.J.; Legros, J.E.; Navaratnam, N.; Greeve, J.C.; Scott, J. Sequence requirements for the editing of apolipoprotein B mRNA. J. Biol. Chem. 1991, 266, 16301–16304. [Google Scholar] [CrossRef]
- Mehta, A.; Kinter, M.T.; Sherman, N.E.; Driscoll, D.M. Molecular cloning of apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA. Mol. Cell Biol. 2000, 20, 1846–1854. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Navaratnam, N.; Morrison, J.R.; Scott, J.; Taylor, W.R. Cytosine nucleoside/nucleotide deaminases and apolipoprotein B mRNA editing. Trends Biochem. Sci. 1994, 3, 105–106. [Google Scholar] [CrossRef]
- MacGinnitie, A.J.; Anant, S.; Davidson, N.O. Mutagenesis of apobec-1, the catalytic subunit of the mammalian apolipoprotein B mRNA editing enzyme, reveals distinct domains that mediate cytosine nucleoside deaminase, RNA binding, and RNA editing activity. J. Biol. Chem. 1995, 270, 14768–14775. [Google Scholar] [CrossRef]
- Wedekind, J.E.; Dance, G.S.; Sowden, M.P.; Smith, H.C. Messenger RNA editing in mammals: New members of the APOBEC family seeking roles in the family business. Trends Genet. 2003, 19, 207–216. [Google Scholar] [CrossRef]
- Blanc, V.; Henderson, J.O.; Newberry, E.P.; Kennedy, S.; Luo, J.; Davidson, N.O. Targeted deletion of the murine apobec-1 complementation factor (acf) gene results in embryonic lethality. Mol. Cell Biol. 2005, 25, 7260–7269. [Google Scholar] [CrossRef]
- Fossat, N.; Tourle, K.; Radziewic, T.; Barratt, K.; Leibhold, D.; Studdert, J.B.; Power, M.; Jones, V.; Loebel, D.A.F.; Tam, P.P. C to U RNA editing meditated by APOBEC1 requires RNA-binding protein RBM47. EMBO Rep. 2014, 15, 903–910. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Zhang, J. Human C-to-U Coding RNA Editing Is Largely Nonadaptive. Mol. Biol. Evol. 2018, 35, 963–969. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Lv, J.; Li, Y.; Mao, S.; Li, Z.; Jing, Z.; Sun, Y.; Zhang, X.; Shen, S.; Wang, X.; et al. Programmable C-to-U RNA editing using the human APOBEC3A deaminase. EMBO J. 2021, 40, e108209. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, M.; Verbitskly, D.; van der Merwe, J.A.; Zehrmann, A.; Brennicke, A. The process of RNA editing in plant mito-chondria. Mitochondrion 2008, 8, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, M.; Zehrmann, A.; Verbitskiy, D.; Hartel, B.; Brennicke, A. RNA editing in plants and its evolution. Annu. Rev. Genet. 2013, 47, 335–352. [Google Scholar] [CrossRef] [PubMed]
- Covello, P.S.; Gray, M.W. RNA editing in plant mitochondria. Nature 1989, 341, 662–666. [Google Scholar] [CrossRef] [PubMed]
- Gualberto, J.M.; Lamattina, L.; Bonnard, G.; Weil, J.H.; Grienenberger, J.M. RNA editing in wheat mitochondria results in the conversation of protein sequences. Nature 1989, 341, 660–662. [Google Scholar] [CrossRef]
- Hiesel, R.; Wissinger, B.; Schuster, W.; Brennicke, A. RNA editing in plant mitochondria. Science 1989, 246, 1632–1634. [Google Scholar] [CrossRef]
- Maier, M.R.; Neckermann, K.; Igloi, G.L.; Kössel, H. Complete sequence of the maize chloroplast genome: Gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J. Mol. Biol. 1995, 251, 614–628. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Rochaix, J.D.; Wassenegger, M.; Filipowicz, W. Plant, their organelles, viruses and transgenes reveal the mechanisms and relevance of post-transcriptional processes Leysin, VD, Switzerland, February 25–28, 1999. EMBO J. 1999, 18, 5153–5158. [Google Scholar] [CrossRef]
- Kotera, E.; Tasaka, M.; Shikanai, T. A pentatricopetide repeat protein is essential for RNA editing in chloroplasts. Nature 2005, 433, 326–330. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, Y.; Kozaki, A.; Ohmori, A.; Iguchi, H.; Nagano, Y. Chloroplast RNA editing required for functional acetyl-CoA carboxylase in plants. J. Biol. Chem. 2001, 276, 3937–3940. [Google Scholar] [CrossRef] [PubMed]
- Wintz, H.; Hanson, M.R. A termination codon is created by RNA editing in the petunia atp9 transcript. Curr. Genet. 1991, 19, 61–64. [Google Scholar] [CrossRef] [PubMed]
- Unseld, M.; Marienfeld, J.R.; Brandt, P.; Brennicke, A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 1997, 15, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Giegé, P.; Brennicke, A. RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl. Acad. Sci. USA 1999, 96, 15324–15329. [Google Scholar] [CrossRef] [PubMed]
- Marchfelder, A.; Binder, S. Plastid and plant mitochondrial RNA processing and RNA stability. In Molecular Biology and Biotechnology of Plant Organelles; Springer: Dordrecht, The Netherlands, 2004; pp. 261–294. [Google Scholar]
- Takenaka, M.; Verbitskiy, D.; Van Der Merwe, J.A.; Zehrmann, A.; Plessmann, U.; Urlaub, H.; Brennicke, A. In vitro RNA editing in plant mitochondria does not require added energy. FEBS Lett. 2007, 581, 2743–2747. [Google Scholar] [CrossRef]
- Chaudhuri, S.; Carrer, H.; Maliga, P. Site-specific factor involved in the editing of the psbL mRNA in tobacco plastids. EMBO J. 1995, 14, 2951–2957. [Google Scholar] [CrossRef]
- Bock, R.; Hermann, M.; Kössel, H. In vivo dissection of cis-acting determinants for plastid RNA editing. EMBO J. 1996, 15, 5052–5059. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, S.; Maliga, P. Sequences directing C to U editing of the plastid psbL mRNA are located within a 22 nucleotide segment spanning the editing site. EMBO J. 1996, 15, 5958–5964. [Google Scholar] [CrossRef]
- Bock, R.; Hermann, M.; Fuchs, M. Identification of critical nucleotide positions for plastid RNA editing site recognition. RNA 1997, 3, 1194–1200. [Google Scholar]
- Hermann, M.; Bock, R. Transfer of plastid RNA-editing activity to novel sites suggests a critical role for spacing in editing-site recognition. Proc. Natl. Acad. Sci. USA 1999, 96, 4856–4861. [Google Scholar] [CrossRef] [PubMed]
- Shikanai, T. RNA editing in plant organelles: Machinery, physiological function and evolution. Cell Mol. Life Sci. 2006, 63, 698–708. [Google Scholar] [CrossRef] [PubMed]
- Small, I.D.; Peeters, N. The PPR motif—A TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 2000, 25, 46–47. [Google Scholar] [CrossRef]
- Aubourg, S.; Boudet, N.; Kreis, M.; Lecharny, A. In Arabidopsis thaliana, 1% of the genome codes for a novel protein family unique to plants. Plant Mol. Biol. 2000, 42, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Lurin, C.; Andres, C.; Aubourg, S.; Bellaoui, M.; Bitton, F.; Bruyere, C.; Caboche, M.; Debast, C.; Gualberto, J.; Hoffmann, B.; et al. Genome-wide analysis of Arabidopsis pentatrico-peptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 2004, 16, 2089–2103. [Google Scholar] [CrossRef]
- Shikanai, T.; Endo, T.; Hashimoto, T.; Yamada, Y.; Asada, K.; Yokota, A. Directed disruption of the tobacco ndhB gene impairs cyclic electron flow around photosystem I. Proc. Natl. Acad. Sci. USA 1998, 95, 9705–9709. [Google Scholar] [CrossRef]
- Schmitz-Linneweber, C.; Small, I. Pentatricopeptide repeat proteins: A socket set for organelle gene expression. Trends Plant Sci. 2008, 13, 663–670. [Google Scholar] [CrossRef]
- Barkan, A.; Small, I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014, 65, 415–442. [Google Scholar] [CrossRef]
- Zhu, Q.; Dugardeyn, J.; Zhang, C.; Takenaka, M.; Kühn, K.; Craddock, C.; Smalle, J.; Karampelias, M.; Denecke, J.; Peters, J.; et al. SLO2, a mitochondrial pentatricopeptide repeat protein affecting several RNA editing sites, is required for energy metabolism. Plant J. 2012, 71, 836–849. [Google Scholar] [CrossRef]
- Okuda, K.; Chateigner-Boutin, A.L.; Nakamura, T.; Delannoy, E.; Sugita, M.; Myouga, F.; Motohashi, R.; Shinozaki, K.; Small, I.; Shikanai, T. Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. Plant Cell 2009, 21, 147–156. [Google Scholar] [CrossRef]
- Bentolila, S.; Oh, J.; Hanson, M.R.; Bukowski, R. Comprehensive high-resolution analysis of the role of an Arabidopsis gene family in RNA editing. PLoS Genet. 2013, 9, e1003584. [Google Scholar] [CrossRef] [PubMed]
- Bentolila, S.; Heller, W.P.; Sun, T.; Babina, A.M.; Friso, G.; van Wijk, K.J.; Hanson, M.R. RIP1, a member of an Arabidopsis protein family, interacts with the protein RARE1 and broadly affects RNA editing. Proc. Natl. Acad. Sci. USA 2012, 109, E1453–E1461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takenaka, M.; Zehrmann, A.; Verbitskiy, D.; Kugelmann, M.; Hartel, B.; Brennicke, A. Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proc. Natl. Acad. Sci. USA 2012, 109, 5104–5109. [Google Scholar] [CrossRef] [PubMed]
- Boussardon, C.; Avon, A.; Kindgren, P.; Bond, C.S.; Challenor, M.; Lurin, C.; Small, I. The cytidine deaminase signature HxE(x)n CxxC of DYW1 binds zinc and is necessary for RNA editing of ndhD-1. New Phyto. 2014, 203, 1090–1095. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Germain, A.; Giloteaux, L.; Hammani, K.; Barkan, A.; Hanson, M.R.; Bentolila, S. An RNA recognition motif containing protein is required for plastid RNA editing in Arabidopsis and maize. Proc. Natl. Acad. Sci. USA 2013, 110, E1169–E1178. [Google Scholar] [CrossRef]
- Shi, X.; Bentolila, S.; Hanson, M.R. Organelle RNA recognition motif-containing (ORRM) proteins are plastid and mitochondrial editing factors in Arabidopsis. Plant Signal Behav. 2016, 11, e1167299. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Herde, M.; Witte, C.P. Of the nine cytidine deaminase-like genes in Arabidopsis, eight are pseudogenes and only one is required to maintain pyrimidine homeostasis in vivo. Plant Physiol. 2016, 171, 799–809. [Google Scholar] [CrossRef]
- Ruchika; Tsukahara, T. The U to C RNA editing affects the mRNA stability of nuclear genes in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2021, 571, 110–117. [Google Scholar]
- Ruchika; Okudaira, C.; Sakari, M.; Tsukahara, T. Genome-wide identification of U to C RNA editing events for nuclear genes in Arabidopsis thaliana. Cells 2021, 10, 635. [Google Scholar]
- Qulsum, U.; Azad, M.T.A.; Tsukahara, T. Analysis of tissue-specific RNA editing events of genes involved in RNA editing in Arabidopsis thaliana. J. Plant Biol. 2019, 62, 351–358. [Google Scholar] [CrossRef]
- Meng, Y.; Chen, D.; Jin, Y.; Mao, C.; Wu, P.; Chen, M. RNA editing of nuclear transcripts in Arabidopsis thaliana. BMC Genom. 2010, 11, S12. [Google Scholar] [CrossRef] [PubMed]
- Koito, A.; Ikeda, T. Apolipoprotein B mRNA-editing, catalytic polypeptide cytidine deaminases and retroviral restriction. Wiley Interdiscip. Rev. RNA 2012, 3, 529–541. [Google Scholar] [CrossRef] [PubMed]
- Ichinose, M.; Sugita, M. RNA Editing and Its Molecular Mechanism in Plant Organelles. Genes 2017, 8, 5. [Google Scholar] [CrossRef]
- Prochnow, C.; Bransteitter, R.; Klein, M.G.; Goodman, M.F.; Chen, X.S. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature 2007, 445, 447–451. [Google Scholar] [CrossRef] [PubMed]
- Conticello, S.G.; Thomas, C.J.; Petersen-Mahrt, S.K.; Neuberger, M.S. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol. Biol. Evol. 2005, 22, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Bhakta, S.; Azad, M.T.A.; Tsukahara, T. Genetic code restoration by artificial RNA editing of Ochre stop codon with ADAR1 deaminase. PEDS 2018, 31, 471–478. [Google Scholar] [CrossRef]
- Bhakta, S.; Tsukahara, T. Artificial RNA editing with ADAR for gene therapy. Curr. Gene Ther. 2020, 20, 44–54. [Google Scholar] [CrossRef]
- Bhakta, S.; Tsukahara, T. Double MS2 guided restoration of genetic code in amber (TAG), opal (TGA) and ochre (TAA) stop codon. Enzym. Microb. Technol. 2021, 149, 109851. [Google Scholar] [CrossRef]
- Krishnan, A.; Iyer, L.M.; Holland, S.J.; Boehm, T.; Aravind, L. Diversifcation of AID/APOBEC-like deaminases in metazoa: Multiplicity of clades and widespread roles in immunity. Proc. Natl. Acad. Sci. USA 2018, 115, E3201–E3210. [Google Scholar] [CrossRef]
- Salter, J.D.; Bennett, R.P.; Smith, H.C. The APOBEC protein family: United by structure, divergent in function. Trends Biochem. Sci. 2016, 41, 578–594. [Google Scholar] [CrossRef]
- Iyer, L.M.; Zhang, D.; Rogozin, I.B.; Aravind, L. Evolution of the deaminase fold and multiple origins of eukaryotic editing and mutagenic nucleic acid deaminases from bacterial toxin systems. Nucleic Acids Res. 2011, 39, 9473–9497. [Google Scholar] [CrossRef] [PubMed]
- Randau, L.; Stanley, B.J.; Kohlway, A.; Mechta, S.; Xiong, Y.; Soll, D. A cytidine deaminase edits C to U in transfer RNAs in archaea. Science 2009, 324, 657–659. [Google Scholar] [CrossRef]
- Salter, J.D.; Smith, H.C. Modeling the embrace of a mutator: APOBEC selection of nucleic acid ligands. Trends Biochem. Sci. 2018, 43, 606–622. [Google Scholar] [CrossRef] [PubMed]
- Münk, C.; Willemsen, A.; Bravo, I.G. An ancient history of gene duplications, fusions and losses in the evolution of APOBEC3 mutators in mammals. BMC Evol. Biol. 2012, 12, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saraconi, G.; Severi, F.; Sala, C.; Mattiuz, G.; Conticello, S.G. The RNA editing enzyme APOBEC1 induces somatic mutations and a compatible mutational signature is present in esophageal adenocarcinomas. Genome Biol. 2014, 15, 417. [Google Scholar] [CrossRef]
- Eichler, S.A.; Kirischuk, S.; Jüttner, R.; Schafermeier, P.K.; Legendre, P.; Lehmann, T.N.; Gloveli, T.; Grantyn, R.; Meier, J.C. Glycinergic tonic inhibition of hippocampal neurons with depolarizing GABAergic transmission elicits histopathological signs of temporal lobe epilepsy. J. Cell Mol. Med. 2008, 12, 2848–2866. [Google Scholar] [CrossRef]
- Winkelmann, A.; Maggio, N.; Eller, J.; Caliskan, G.; Semtner, M.; Häussler, U.; Jüttner, R.; Dugladze, T.; Smolinsky, B.; Kowalczyk, S.; et al. Changes in neural network homeostasis trigger neuropsychiatric symptoms. J. Clin. Investig. 2014, 124, 696–711. [Google Scholar] [CrossRef]
- Çaliskan, G.; Müller, I.; Semtner, M.; Winkelmann, A.; Raza, A.S.; Hollnagel, J.O.; Rösler, A.; Heinemann, U.; Stork, O.; Meier, J.C. Identification of parvalbumin interneurons as cellular substrate of fear memory persistence. Cereb. Cortex 2016, 26, 2325–2340. [Google Scholar] [CrossRef]
- Kankowski, S.; Förstera, B.; Winkelmann, A.; Knauff, P.; Wanker, E.E.; You, X.A.; Semtner, M.; Hetsch, F.; Meier, J.C. A Novel RNA editing sensor tool and a specific agonist determine neuronal protein expression of RNA-edited glycine receptors and identify a genomic APOBEC1 dimorphism as a new genetic risk factor of epilepsy. Front. Mol. Neurosci. 2018, 10, 439. [Google Scholar] [CrossRef]
- Hirano, K.; Min, J.; Funahashi, T.; Baunoch, D.A.; Davidson, N.O. Characterization of the human apobec-1 gene: Expression in gastrointestinal tissues determined by alternative splicing with production of a novel truncated peptide. J. Lipid Res. 1997, 38, 847–859. [Google Scholar] [CrossRef]
- Grohmann, M.; Hammer, P.; Walther, M.; Paulmann, N.; Büttner, A.; Eisenmenger, W.; Baghai, T.C.; Schüle, C.; Rupprecht, R.; Bader, M.; et al. Alternative splicing and extensive RNA editing of human TPH2 transcripts. PLoS ONE 2010, 5, e8956. [Google Scholar] [CrossRef] [PubMed]
- Vanharanta, S.; Marney, C.B.; Shu, W.; Valiente, M.; Zou, Y.; Mele, A.; Darnell, R.B.; Massagué, J. Loss of the multifunctional RNA-binding protein RBM47 as a source of selectable metastatic traits in breast cancer. Elife 2014, 3, e02734. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, T.; Isogaya, K.; Sakai, S.; Morikawa, M.; Morishita, Y.; Ehata, S.; Miyazono, K.; Koinuma, D. RNA-binding motif protein 47 inhibits Nrf2 activity to suppress tumor growth in lung adenocarcinoma. Oncogene 2017, 35, 5000–5009. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, Y.; Nishitsuji, H.; Marusawa, H.; Ujino, S.; Takaku, H.; Shimotohno, K. The RNA-editing enzyme APOBEC1 requires heterogeneous nuclear ribonucleoprotein Q Isoform 6 for efficient interaction with interleukin-8 mRNA. J. Biol. Chem. 2014, 289, 26226–26238. [Google Scholar] [CrossRef] [Green Version]
- Chiche, L.; Jourde-Chiche, N.; Whalen, E.; Presnell, S.; Gersuk, V.; Dang, K.; Anguiano, E.; Quinn, C.; Burtey, S.; Berland, Y.; et al. Modular transcriptional repertoire analyses of adults with systemic lupus erythematosus reveal distinct type I and type II interferon signatures. Arthritis Rheumatol. 2014, 66, 1583–1595. [Google Scholar] [CrossRef]
- Hagberg, N.; Rönnblom, L. Systemic Lupus Erythematosus-A disease with a dysregulated type I interferon system. Scand. J. Immunol. 2015, 82, 199–207. [Google Scholar] [CrossRef]
- Roth, S.H.; Danan-Gotthold, M.; Ben-Izhak, M.; Rechavi, G.; Cohen, C.J.; Louzoun, Y.; Levanon, E.Y. Increased RNA editing may provide a source for autoantigens in Systemic Lupus Erythematosus. Cell Rep. 2018, 23, 50–57. [Google Scholar] [CrossRef]
- Sharma, S.; Baysal, B.E. Stem-loop structure preference for site-specific RNA editing by APOBEC3A and APOBEC3G. Peer J. 2017, 5, e4136. [Google Scholar] [CrossRef]
- Radanova, M.; Vasilev, V.; Dimitrov, T.; Deliyska, B.; Ikonomov, V.; Ivanova, D. Association of rs172378 C1q gene cluster polymorphism with lupus nephritis in Bulgarian patients. Lupus 2015, 24, 280–289. [Google Scholar] [CrossRef]
- Namjou, B.; Gray-McGuire, C.; Sestak, A.L.; Gilkeson, G.S.; Jacob, C.O.; Merrill, J.T.; James, J.A.; Wakeland, E.K.; Li, Q.Z.; Langefeld, C.D.; et al. Evaluation of C1q genomic region in minority racial groups of lupus. Genes Immun. 2009, 10, 517–524. [Google Scholar] [CrossRef]
- Peng, X.; Xu, X.; Wang, Y.; Hawke, D.H.; Yu, S.; Han, L.; Zhou, Z.; Mojumdar, K.; Jeong, K.J.; Labrie, M.; et al. A to I RNA editing contributes to proteomic diversity in cancer. Cancer Cell 2018, 33, 817–828.e7. [Google Scholar] [CrossRef] [PubMed]
- Chan, T.H.; Qamra, A.; Tan, K.T.; Guo, J.; Yang, H.; Qi, L.; Lin, J.S.; Ng, V.H.E.; Song, Y.; Hong, H.; et al. ADAR-mediated RNA editing predicts progression and prognosis of gastric cancer. Gastroenterology 2016, 151, 637–650.e10. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, Y.; Lin, C.H.; Chan, T.H.; Chow, R.K.; Song, Y.; Liu, M.; Yuan, Y.F.; Fu, L.; Kong, K.L.; et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat. Med. 2013, 19, 209–216. [Google Scholar] [CrossRef]
- Han, L.; Diao, L.; Yu, S.; Xu, X.; Li, J.; Zhang, R.; Yang, Y.; Werner, H.M.; Eterovic, A.K.; Yuan, Y.; et al. The genomic landscape and clinical relevance of A to I RNA editing in human cancers. Cancer Cell 2015, 28, 515–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sieuwerts, A.M.; Willis, S.; Burns, M.B.; Look, M.P.; Gelder, M.E.; Schlicker, A.; Heideman, M.R.; Jacobs, H.; Wessels, L.; Leyland-Jones, B.; et al. Elevated APOBEC3B correlates with poor outcomes for estrogen-receptor-positive breast cancers. Horm. Cancer 2014, 5, 405–413. [Google Scholar] [CrossRef]
- Landry, S.; Narvaiza, I.; Linfesty, D.C.; Weitzman, M.D. APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep. 2011, 12, 444–450. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.R.; Qiao, J.J.; Chan, T.H.; Zhu, Y.H.; Li, F.F.; Liu, H.; Fei, J.; Li, Y.; Guan, X.Y.; Chen, L. Adenosine-to-inosine RNA editing mediated by ADARs in esophageal squamous cell carcinoma. Cancer Res. 2014, 74, 840–851. [Google Scholar] [CrossRef]
- Shigeyasu, K.; Okugawa, Y.; Toden, S.; Miyoshi, J.; Toiyama, Y.; Nagasaka, T.; Takahashi, N.; Kusunoki, M.; Takayama, T.; Yamada, Y.; et al. AZIN1 RNA editing confers cancer stemness and enhances oncogenic potential in colorectal cancer. JCI Insight 2018, 3, e99976. [Google Scholar] [CrossRef]
- Law, E.K.; Sieuwerts, A.M.; LaPara, K.; Leonard, B.; Starrett, G.J.; Molan, A.M.; Temiz, N.A.; Vogel, R.I.; Meijer-van Gelder, M.E.; Sweep, F.C.; et al. The DNA cytosine deaminase APOBEC3B promotes tamoxifen resistance in ER-positive breast cancer. Sci. Adv. 2016, 2, e1601737. [Google Scholar] [CrossRef]
- Fritzell, K.; Xu, L.D.; Otrocka, M.; Andreasson, C.; Ohman, M. Sensitive ADAR editing reporter in cancer cells enables high-throughput screening of small molecule libraries. Nucleic Acids Res. 2018, 47, e22. [Google Scholar] [CrossRef]
- Olson, M.E.; Harris, R.S.; Harki, D.A. APOBEC enzymes as targets for virus and cancer therapy. Cell Chem. Biol. 2018, 25, 36–49. [Google Scholar] [CrossRef]
- Mizrahi, R.A.; Schirle, N.T.; Beal, P.A. Potent and selective inhibition of A to I RNA editing with 2’-O-methyl/locked nucleic acid-containing antisense oligoribonucleotides. ACS Chem. Biol. 2013, 8, 832–839. [Google Scholar] [CrossRef] [PubMed]
- Montiel-Gonzalez, M.F.; Vallecillo-Viejo, I.; Yudowski, G.A.; Rosenthal, J.J. Correction of mutations within the cystic fbrosis transmembrane conductance regulator by site-directed RNA editing. Proc. Natl. Acad. Sci. USA 2013, 110, 18285–18290. [Google Scholar] [CrossRef] [PubMed]
- Hanswillemenke, A.; Kuzdere, T.; Vogel, P.; Jekely, G.; Staforst, T. Site-directed RNA editing in vivo can be triggered by the light-driven assembly of an artifcial riboprotein. J. Am. Chem. Soc. 2015, 137, 15875–15881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simpson, L.; Sbicego, S.; Aphasizhev, R. Uridine insertion/deletion RNA editing in trypanosome mitochondria: A complex business. RNA 2003, 9, 265–276. [Google Scholar] [CrossRef]
- Rubio, M.A.T.; Ragone, F.L.; Gaston, K.W.; Ibba, M.; Alfonzo, J.D. C to U Editing Stimulates A to I Editing in the Anticodon Loop of a Cytoplasmic Threonyl tRNA in Trypanosoma brucei. J. Biol. Chem. 2006, 281, 115–120. [Google Scholar] [CrossRef]
- Maier, R.M.; Zeltz, P.; Kössel, H.; Bonnard, G.; Gualberto, J.M.; Grienenberger, J.M. RNA editing in plant mitochondria and chloroplasts. Plant Mol. Biol. 1996, 32, 343–365. [Google Scholar] [CrossRef]
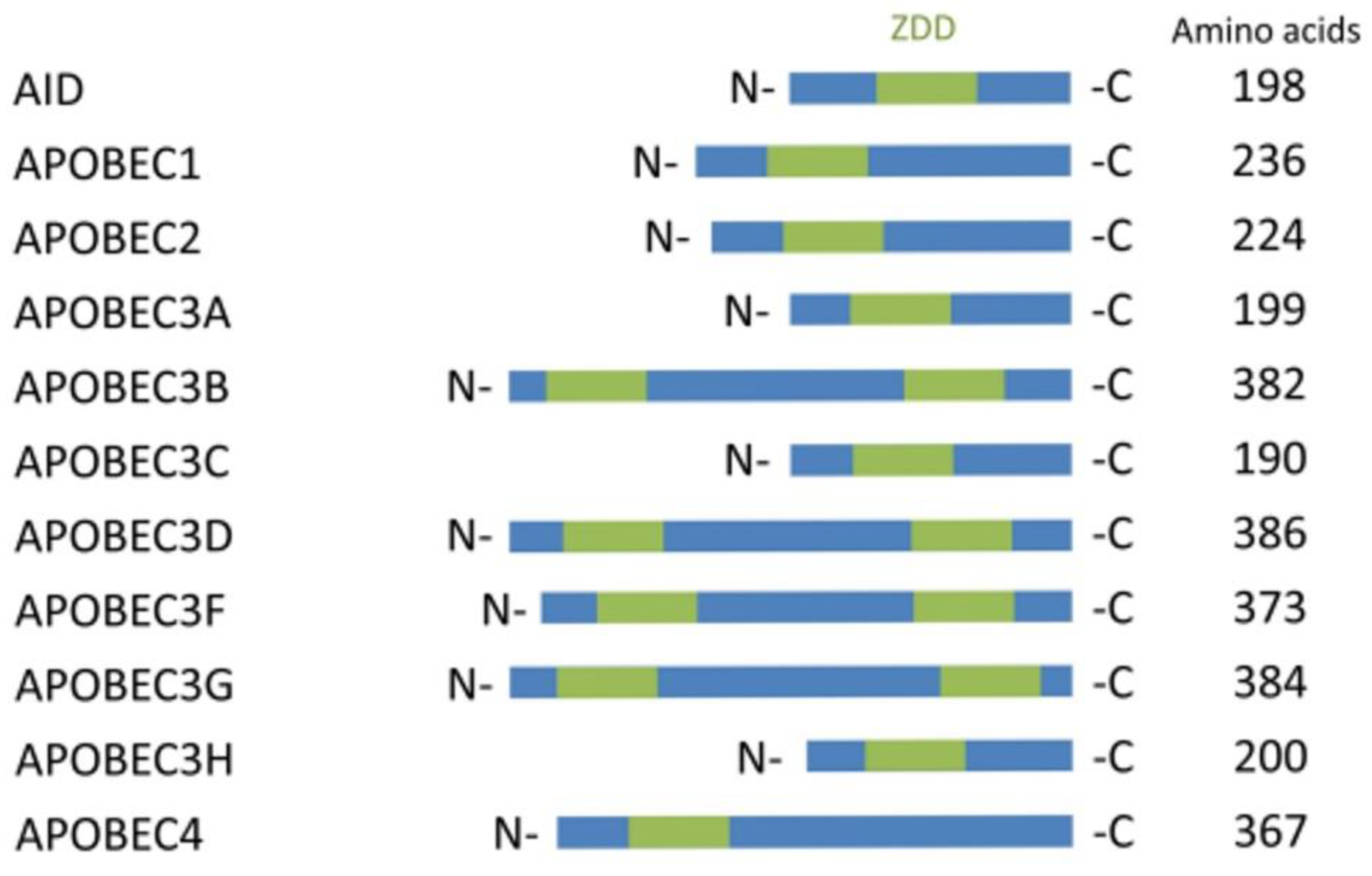
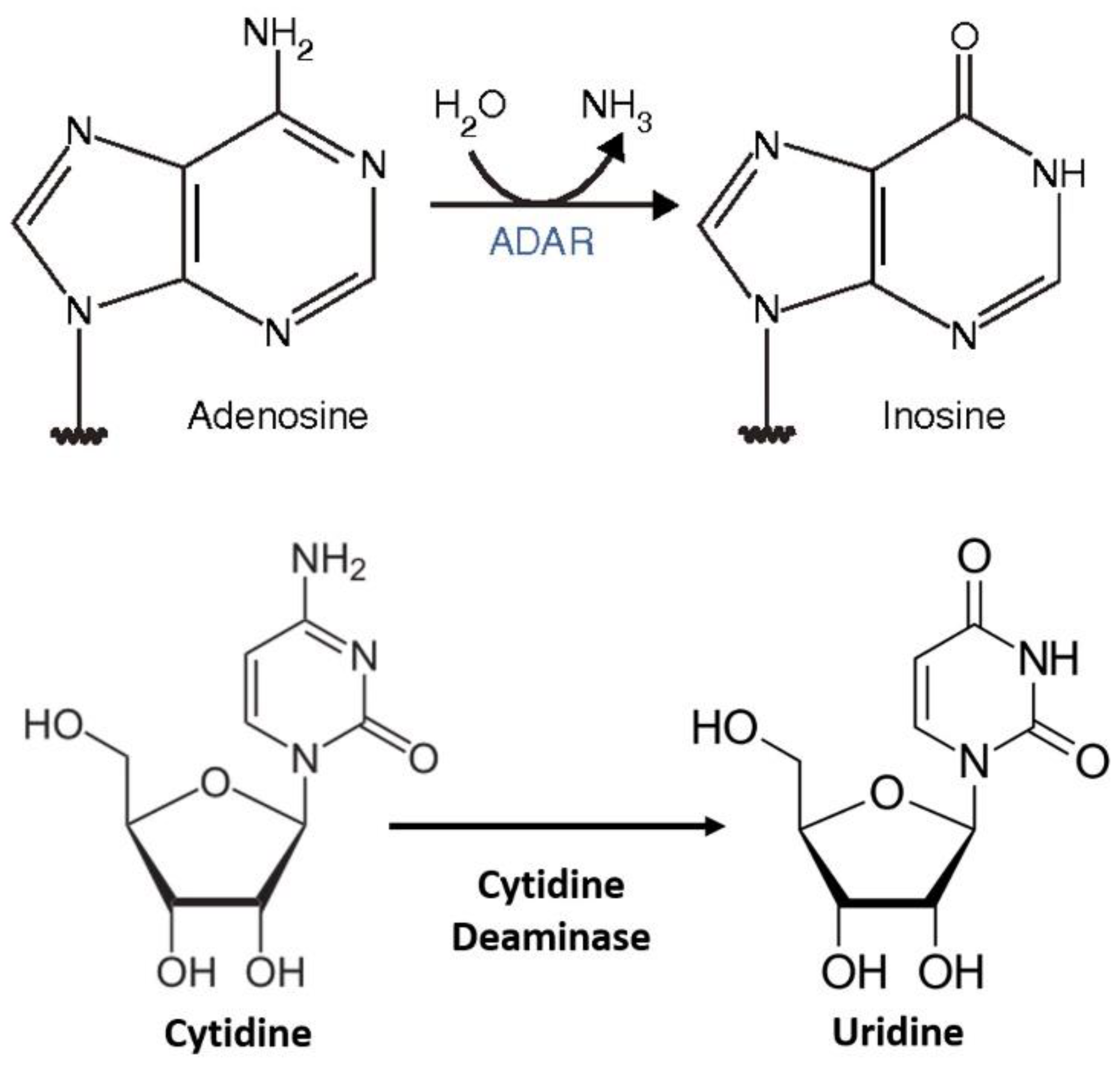
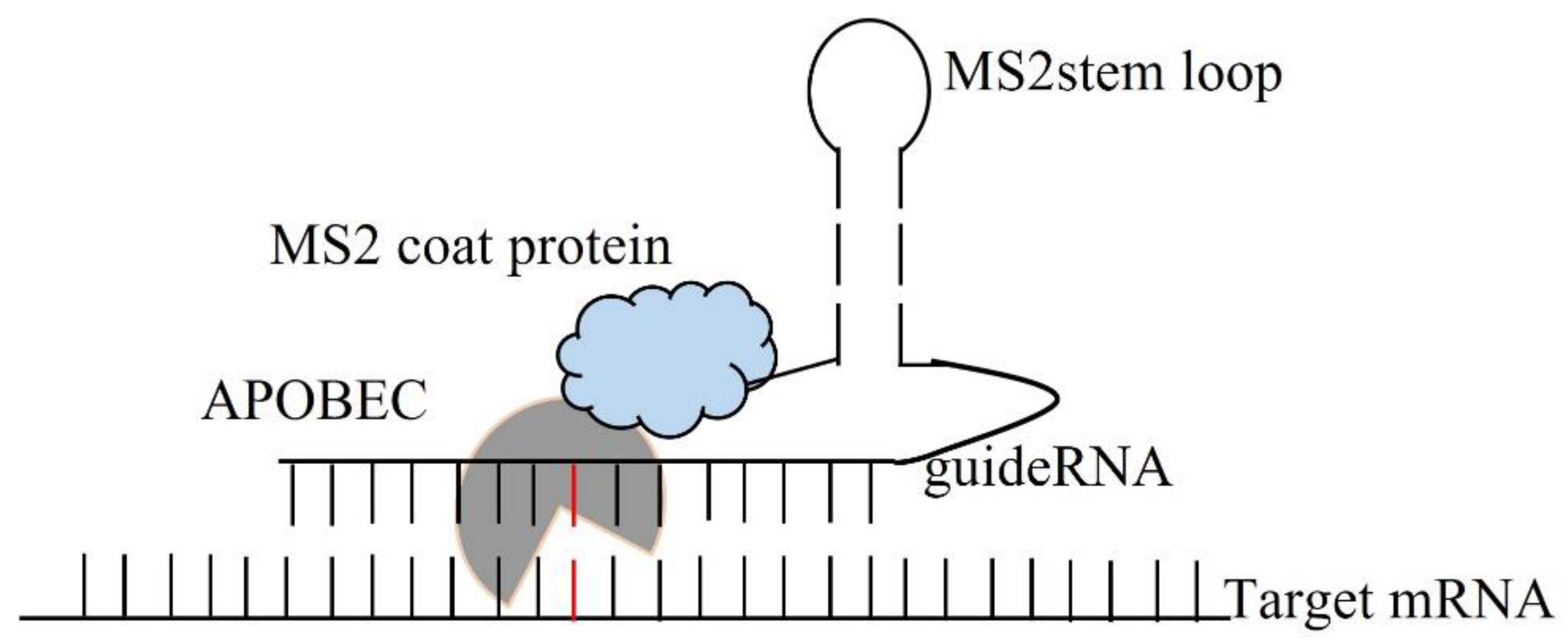
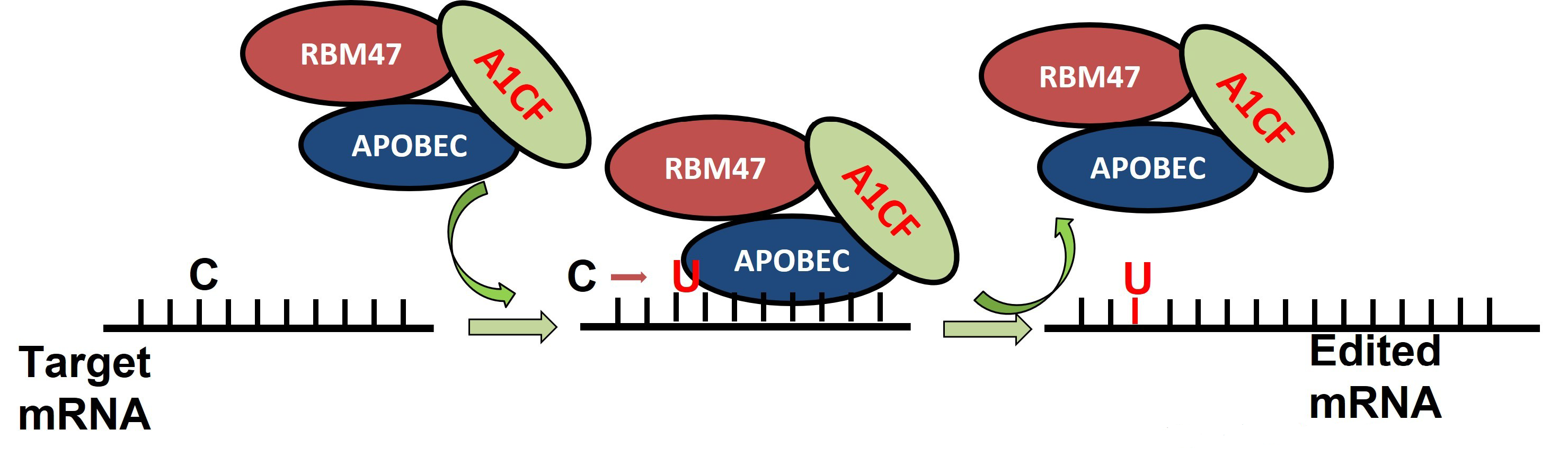
| No. | Disease State | Gene Symbol | Base Change | Amino Acid | Codon |
|---|---|---|---|---|---|
| 1 | ADA deficiency | ADA | CTG-CCG | Leu-Pro | 107 |
| 2 | APRT Deficiency | ART | ATG-ACG | Met-Thr | 136 |
| 3 | Amyloid prealbumin | PALB | GTG-GCG | Val-Ala | 30 |
| 4 | Antithrombin III def. | AT3 | TTC-TCC | Phe-Ser | 402 |
| 5 | Antitrypsin ∝ 1 def. | PI | CTC-CCG | Leu-Pro | 41 |
| 6 | Antitrypsin ∝1 def. | PI | CTC-GCG | Val-Ala | 213 |
| 7 | Elliptocytosis | SPTA | CTC-CCG | Leu-Pro | 207 |
| 8 | Epidermolysis bull | KRT14 | CTG-CCG | Leu-Pro | 384 |
| 9 | G6PD Deficiency | G6PD | CTG-CCG | Leu-pro | 968 |
| 10 | Galactosaemia | GALT | CTG-CCG | Leu-Pro | 195 |
| 11 | Gangliosidosis GM1 | GLB1 | ATC-ACC | Ile-Thr | 51 |
| 12 | HPRT deficiency | HPRT | ATT-ACT | Ile-Thr | 182 |
| 13 | Haemolytic anaemia | PGK | CTG-CCG | Leu-Pro | 88 |
| 14 | Haemophilia A | F8 | TTC-TCC | Phe-Ser | 293 |
| 15 | Haemophilia A | F8 | TTG-TCG | Leu-Ser | 2166 |
| 16 | Insulin Resistance | INSR | CTG-CCG | Leu-Pro | 233 |
| 17 | Laron dwarfism | GHR | TTT-TCT | Phe-Ser | 96 |
| 18 | Leukocyte adhes. Def. | LFA1 | CTA-CCA | Leu-Pro | 149 |
| 19 | Lipoprt. lipase def. | LPL | ATT-ACT | Ile-Thr | 194 |
| 20 | MCAD deficiency | MCAD | ATA-ACA | Ile-thr | 375 |
| 21 | Methaemoglobin | DIA1 | CTG-CCG | Leu-Pro | 148 |
| 22 | Neurofibromatos is (1) | NF1 | CTC-CCG | Leu-Pro | |
| 23 | OTC deficiency | OTC | CTA-CCA | Leu-Pro | 45 |
| 24 | OTC deficiency | OTC | CTT-CCT | Leu Pro | 111 |
| 25 | Phenylketonuria | PAH | TTG-TCG | Leu-Ser | 48 |
| 26 | Phenylketonuria | PAH | TTG-TCG | Leu-Ser | 255 |
| 27 | Phenylketonuria | PAH | CTG-CCG | Leu-Pro | 311 |
| 28 | Pompe disease | GAA | ATG-ACG | Met-Thr | 318 |
| 29 | Retinitis pigmentosa | RDS | CTG-CCG | Leu-Pro | 185 |
| 30 | Ster.18-hydrox. Def. | CYP18 | GTG-GCG | Val-Ala | 386 |
| 31 | Thalassaemia ∝ | HBA2 | ATG-ACG | Met-Thr | −1 |
| 32 | Thalassaemia ∝ | HBA2 | CTG-CCG | Leu-Pro | 125 |
| 33 | Thalassaemia ∝ | HBB | CTG-CCG | Leu-Pro | 110 |
| 34 | Thalassaemia ∝ | HBD | CTG-CCG | Leu-Pro | 141 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhakta, S.; Tsukahara, T. C-to-U RNA Editing: A Site Directed RNA Editing Tool for Restoration of Genetic Code. Genes 2022, 13, 1636. https://doi.org/10.3390/genes13091636
Bhakta S, Tsukahara T. C-to-U RNA Editing: A Site Directed RNA Editing Tool for Restoration of Genetic Code. Genes. 2022; 13(9):1636. https://doi.org/10.3390/genes13091636
Chicago/Turabian StyleBhakta, Sonali, and Toshifumi Tsukahara. 2022. "C-to-U RNA Editing: A Site Directed RNA Editing Tool for Restoration of Genetic Code" Genes 13, no. 9: 1636. https://doi.org/10.3390/genes13091636
APA StyleBhakta, S., & Tsukahara, T. (2022). C-to-U RNA Editing: A Site Directed RNA Editing Tool for Restoration of Genetic Code. Genes, 13(9), 1636. https://doi.org/10.3390/genes13091636









