Development and Application of InDel Markers Linked to Fruit-Shape and Peel-Colour Genes in Wax Gourd
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Experimental Methods
2.2.1. Screening of InDel Marker Loci
2.2.2. Design and Synthesis of InDel Primers
2.2.3. DNA Extraction, PCR Amplification, and 8% Nondenaturing Polyacrylamide Gels Electrophoresis
2.2.4. Construction of Physical Maps of InDel Markers
3. Results
3.1. Physical Map Analysis of InDel Markers
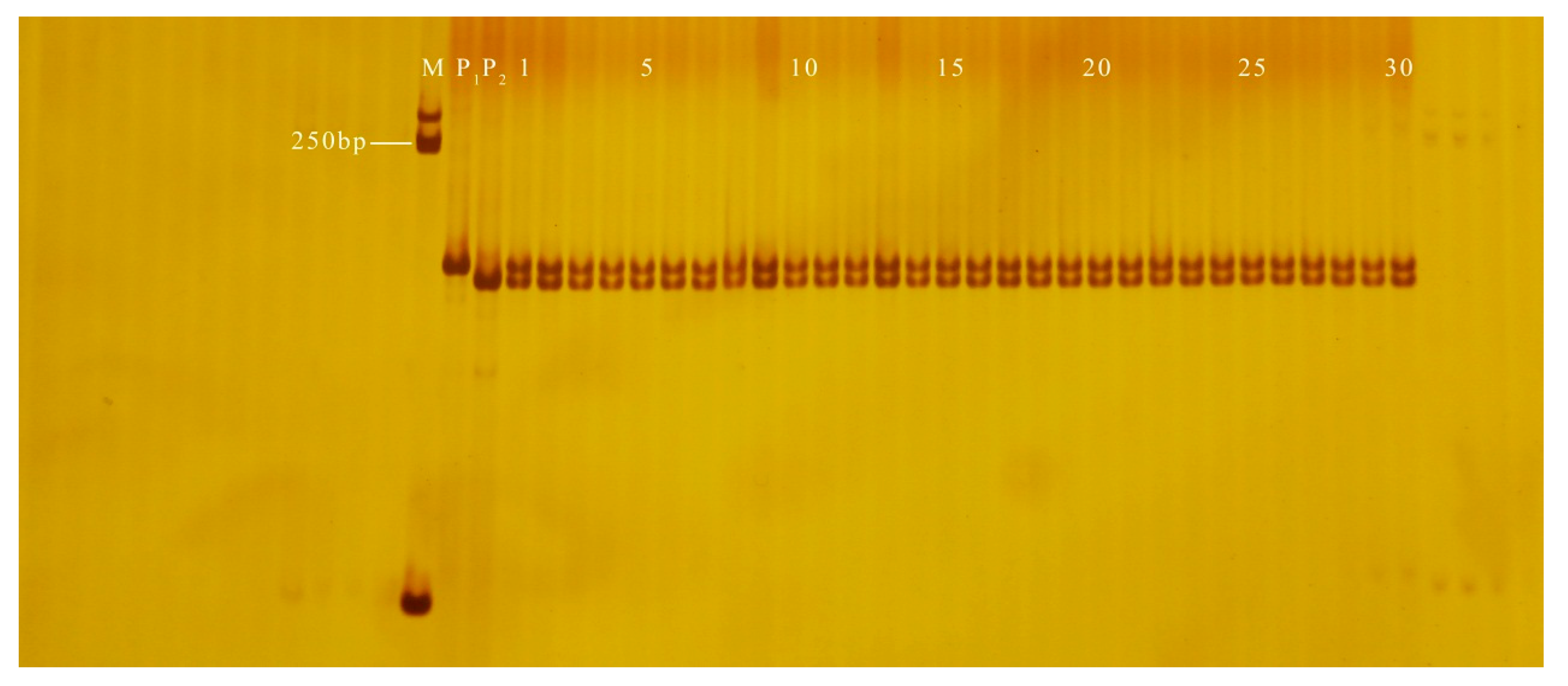
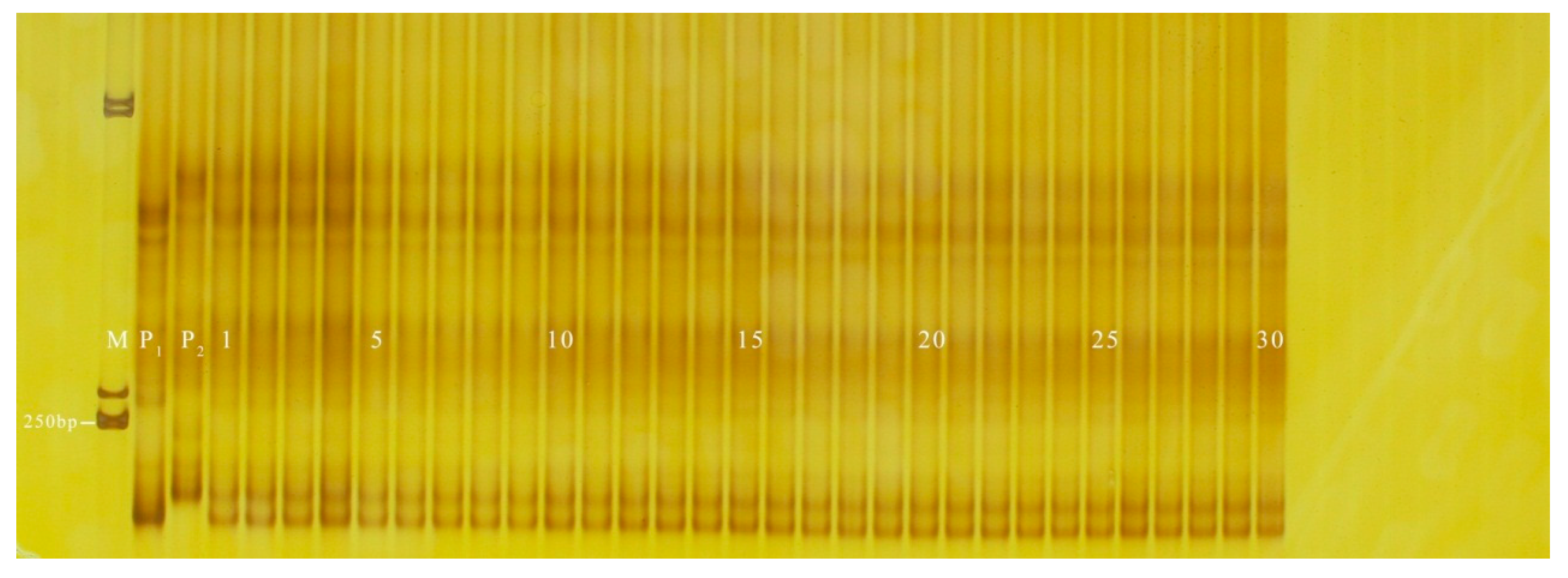
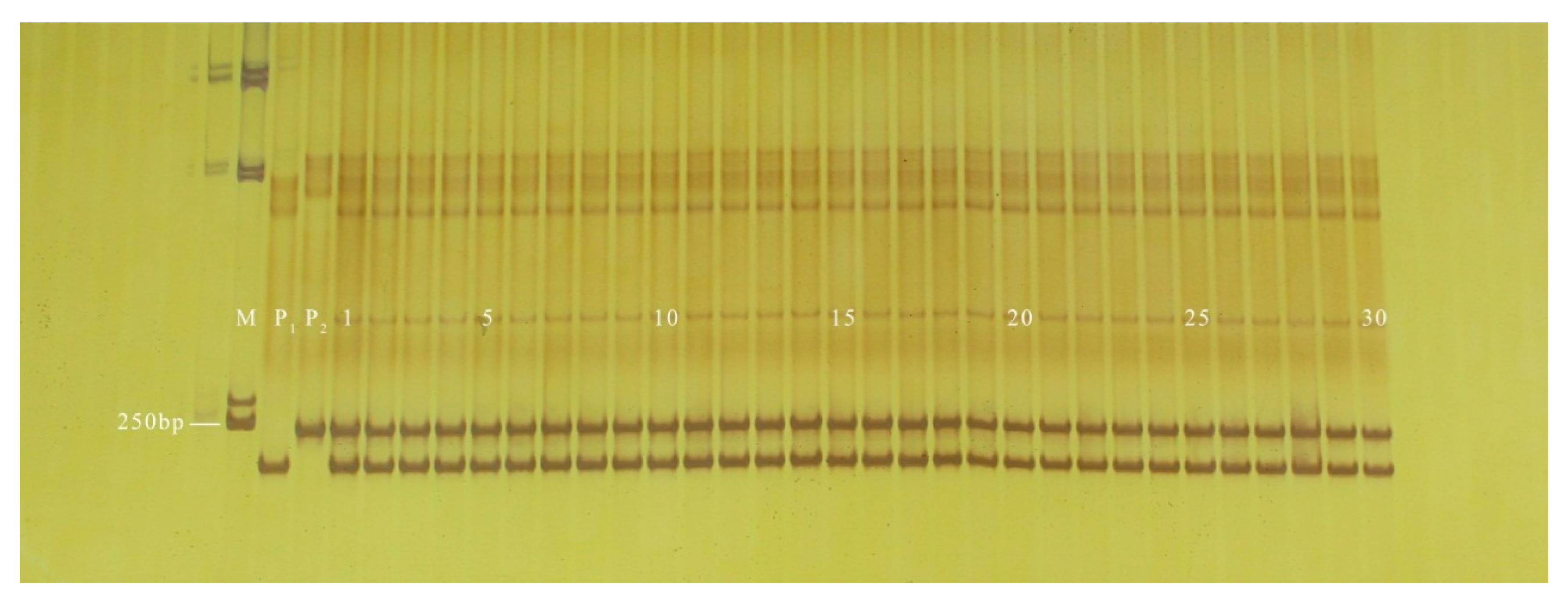
3.2. Validation of InDel Markers
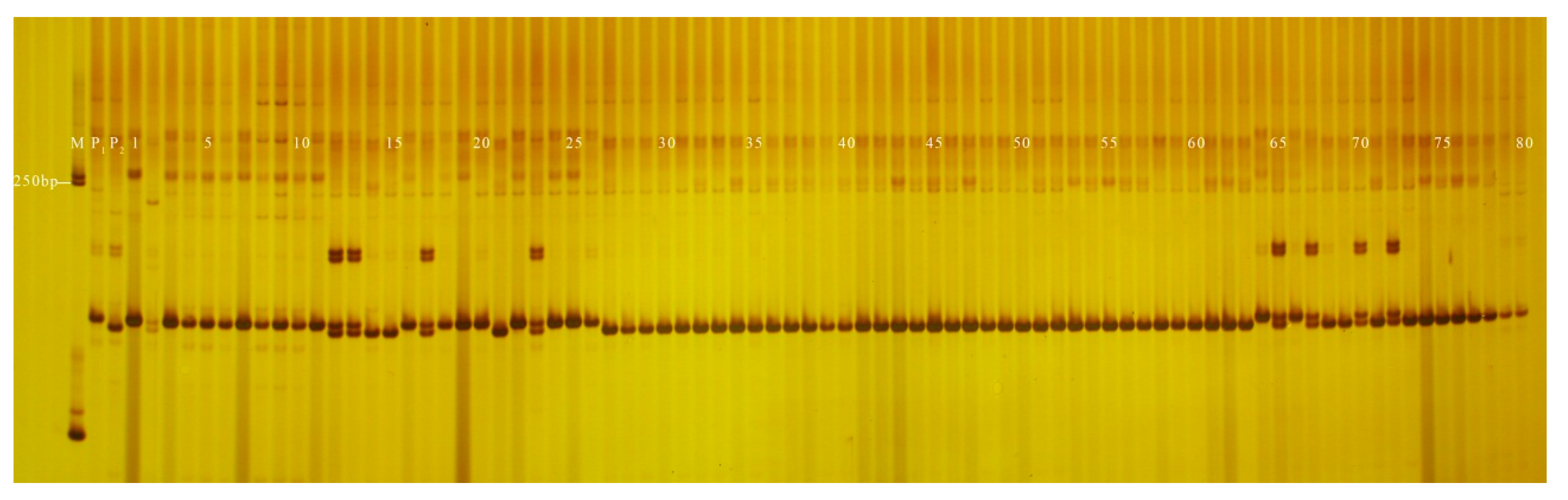
3.3. Application of InDel Markers in the Determination of Hybrid Purity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gu, M.; Fan, S.J.; Liu, G.G.; Guo, L.; Ding, X.B.; Lu, Y.; Zhang, Y.; Ji, G.; Huang, C. Extract of wax gourd peel prevents high-fat diet-induced hyperlipidemia in C57BL/6 mice via the inhibition of the PPARγ pathway. Evid.-Based Compl. Alt. 2013, 2013, 342561. [Google Scholar] [CrossRef]
- Resmi, J.; Sreelathakumary, I. Character association and path coefficient studies in ash gourd [Benincasa hispida (Thunb.) Congn.]. Agric. Sci. Digest. 2012, 32, 251–255. [Google Scholar]
- Zhu, D.D.; Liu, Z.G.; Bao, Z.Y.; Jiao, X.X.; Wang, X.Y. Analysis of ISSR markers linked to the fruit colour trait gene in chieh-qua. Genomics Appl. Biol. 2016, 35, 2781–2787. [Google Scholar]
- Chen, J.Y.; Chen, Q.M.; Liu, Z.G.; Wang, C.L.; Ma, L.L.; Gou, J.Q.; Cheng, Z.K. Seed genetic purity testing of F1 Benincasa hispida var. chieh-qua hybrids using SSR molecular marker analysis. Seed Sci. Technol. 2020, 3, 345–353. [Google Scholar] [CrossRef]
- Du, X.; Tian, S.B.; Zhang, H.M.; Liu, N. Genetic diversity of 36 chieh-qua resources based on SCoT. Mol. Plant Breed. 2021, 1–8. [Google Scholar]
- Jiao, X.X. Construction of Core Collections of Wax Gourd; Guangxi University: Nanning, China, 2018. [Google Scholar]
- Li, Z.L.; Qiao, Y.C.; Lin, J.Y.; Li, G.G. Studies on molecular markers to Fusarium Wilt resistance gene in chieh-qua. Genom. Appl. Biol. 2015, 34, 1946–1949. [Google Scholar]
- Qiao, Y.C.; Lin, J.Y.; Xie, W.P.; Xie, L.F.; Li, L.F. Genetic relationship of chieh-qua and wax-gourd on the morphological phenotypes and SRAP markers. J. Plant Genet. Resour. 2014, 15, 1150–1155. [Google Scholar]
- Ye, X.R.; Liu, J.T.; Li, Y.P.; Zhu, H.S.; Wen, Q.F. Identification of wax gourd by using EST-SSR markers-based MCID method. J. Nucl. Agr. Sci. 2021, 35, 780–788. [Google Scholar]
- Weber, J.L.; David, D.; Heil, J.; Fan, Y.; Zhao, C.F.; Marth, G. Human diallelic insertion/deletion polymorphisms. Am. J. Hum. Genet. 2002, 71, 854–862. [Google Scholar] [CrossRef]
- Wang, D.K.; Zhang, M.J.; Xu, N.N.; Yang, S.; Dou, J.L.; Liu, D.M.; Zhu, L.; Zhu, H.Y.; Hu, J.B.; Ma, C.S. Fine mapping a ClGS gene controlling dark-green stripe rind in watermelon. Sci. Hortic. 2022, 291, 110583. [Google Scholar] [CrossRef]
- Xiao, Z.L.; Kong, C.C.; Han, F.Q.; Yang, L.M.; Zhuang, M.; Zhang, Y.Y.; Wang, Y.; Ji, J.L.; Li, Z.S.; Fang, Z.Y. Two user-friendly molecular markers developed for the identification of hybrid lethality genes in Brassica oleracea. Agronomy 2021, 5, 982. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, T.; Li, L.; Xu, J.; Qin, X.D.; Zhang, T.L.; Cui, L.; Lou, Q.F.; Li, J.; Chen, J. Identification of a stable major-effect QTL(Parth 2.1)controlling parthenocarpy in cucumber and associated candidate gene analysis via whole genome re-sequencing. BMC Plant Biol. 2016, 16, 182. [Google Scholar] [CrossRef]
- Liu, X.; Geng, X.; Zhang, H.; Shen, H.; Yang, W. Association and genetic identification of loci for four fruit traits in tomato using InDel markers. Front. Plant Sci. 2017, 8, 1269. [Google Scholar] [CrossRef]
- Chen, R.; Chang, L.C.; Cai, X.; Wu, J.; Liang, J.L.; Lin, R.M.; Song, Y.; Wang, X.W. Development of InDel markers for Brassica rapa based on a high-resolution melting curve. Hortic. Plant J. 2021, 7, 31–37. [Google Scholar] [CrossRef]
- Liu, B.; Wang, Y.; Zhai, W.; Deng, J.; Wang, H.; Cui, Y.; Cheng, F.; Wang, X.W.; Wu, J. Development of InDel markers for Brassica rapa based on whole-genome re-sequencing. Theor. Appl. Genet. 2013, 126, 231–239. [Google Scholar] [CrossRef]
- Cheng, Z.K.; Liu, Z.G.; Xu, Y.C.; Ma, L.L.; Chen, J.Y.; Gou, J.Q.; Su, L.W.; Wu, W.T.; Chen, Y.; Yu, W.J.; et al. Fine mapping and identification of the candidate gene BFS for fruit shape in wax gourd (Benincasa hispida). Theor. Appl. Genet. 2021, 134, 3983–3995. [Google Scholar] [CrossRef]
- Ma, L.L.; Liu, Z.G.; Cheng, Z.K.; Gou, J.Q.; Chen, J.Y.; Yu, W.J.; Wang, P. Identification and application of BhAPRR2 controlling peel colour in wax gourd (Benincasa hispida). Front. Plant Sci. 2021, 12, 716772. [Google Scholar] [CrossRef]
- Mckenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Ren, L.; Zhu, B.Q.; Zhang, Y.B.; Wang, H.Y.; Li, C.Y.; Su, Y.H.; Ba, C.F. The research of applying primer premier 5.0 to design PCR primer. J. Jinzhou Med. Coll. 2004, 25, 43–46. [Google Scholar]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Yan, P.; Zhang, J.N.; Chen, Y. Purity identification of F1 hybrid seeds in watermelon and melon based on RAPD markers. J. Gansu Agr. Uni. 2007, 42, 43–46. [Google Scholar]
- Bassam, B.J.; Caetano-Anolles, G.; Gresshoff, P.M. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal. Biochem. 1991, 196, 80–83. [Google Scholar] [CrossRef]
- Neff, M.M.; Turk, E.; Kalishman, M. Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 2002, 18, 613–615. [Google Scholar] [CrossRef]
- Cui, J.Y.; Miao, H.; Ding, L.H.; Wehner, T.C.; Liu, P.N.; Wang, Y.; Zhang, S.P.; Gu, X.F. A new glabrous gene (csgl3) identified in trichome development in cucumber (Cucumis sativus L.). PLoS ONE. 2016, 11, e148422. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, S.B.; Goode, D.L.; Kvikstad, E.; Albers, C.A.; Zhang, Z.D.; Mu, X.J.; Ananda, G.; Howie, B.; Karczewski, K.J.; Smith, K.S.; et al. The origin, evolution, and functional impact of short insertion–deletion variants identified in 179 human genomes. Genome Res. 2013, 23, 749–761. [Google Scholar] [CrossRef]
- Wu, W.T.; Wang, P.; Huang, X.C.; Su, L.W.; Lv, H.X.; Gou, J.Q.; Cheng, Z.K.; Ma, L.L.; Yu, W.J.; Liu, Z.G. Fine mapping and functional analysis of major regulatory genes of soluble solids content in wax gourd (Benincasa hispida). Int. J. Mol. Sci. 2022, 23, 6999. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhang, B.B.; Ma, N.; Liu, X.; Qin, M.F.; Zhang, Y.; Wang, K.; Guo, N.; Zuo, K.F.; Liu, X.; et al. Quantitative trait locus mapping and identification of candidate genes controlling flowering time in Brassica napus L. Front. Plant Sci. 2021, 11, 626205. [Google Scholar] [CrossRef]
- Islam, M.R.; Hossain, M.R.; Jesse, D.M.I.; Jung, H.J.; Kim, H.T.; Park, J.I.; Nou, I.S. Development of molecular marker linked with bacterial fruit blotch resistance in melon (Cucumis melo L.). Genes. 2020, 11, 220. [Google Scholar] [CrossRef] [Green Version]
- Wei, C.; Chen, X.; Wang, Z.; Liu, Q.; Li, H.; Zhang, Y.; Ma, J.; Yang, J.; Zhang, X. Genetic mapping of the LOBED LEAF 1 (ClLL1) gene to a 127.6-kb region in watermelon (Citrullus lanatus L.). PLoS ONE. 2017, 12, e180741. [Google Scholar] [CrossRef]
- Dou, J.L.; Zhao, S.J.; Lu, X.Q.; He, N.; Zhang, L.; Ali, A.; Kuang, H.H.; Liu, W.G. Genetic mapping reveals a candidate gene (ClFS1) for fruit shape in watermelon (Citrullus lanatus L.). Theor. Appl. Genet. 2018, 131, 947–958. [Google Scholar] [CrossRef]
- Pan, Y.P.; Liang, X.J.; Gao, M.L.; Liu, H.Q.; Meng, H.W.; Weng, Y.Q.; Cheng, Z.H. Round fruit shape in WI7239 cucumber is controlled by two interacting quantitative trait loci with one putatively encoding a tomato SUN homolog. Theor. Appl. Genet. 2017, 130, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Lian, Q.; Fu, Q.S.; Xu, Y.Y.; Hu, Z.C.; Zheng, J.; Zhang, A.A.; He, Y.H.; Wang, C.S.; Xu, C.Q.; Chen, B.X.; et al. QTLs and candidate genes analyses for fruit size under domestication and differentiation in melon (Cucumis melo L.) based on high resolution maps. Bmc Plant Biol. 2021, 21, 126. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Kim, J.E.; Kim, H.T.; Lee, G.; Kim, B.; Lee, J.M. Genetic mapping of the c1 locus by GBS-based BSA-seq revealed Pseudo-Response Regulator 2 as a candidate gene controlling pepper fruit colour. Theor. Appl. Genet. 2020, 133, 1897–1910. [Google Scholar] [CrossRef]
- Liao, Y.; Sun, B.J.; Sun, G.W.; Liu, H.C.; Li, Z.L.; Xing, L.Z.; Wang, G.P.; Chen, R.Y. AFLP and SCAR markers associated with peel colour in eggplant (Solanum melongena ). Agric. Sci. China 2009, 8, 1466–1474. [Google Scholar] [CrossRef]
- Kim, B.; Kim, N.; Kang, J.; Choi, Y.; Sim, S.; Ran Min, S.; Park, Y. Single nucleotide polymorphisms linked to the SlMYB12 Gene that controls fruit peel colour in domesticated tomatoes (Solanum lycopersicum L.). Hortic. Sci. Technol. 2015, 33, 566–574. [Google Scholar]
- Hao, N.; Du, Y.L.; Li, H.Y.; Wang, C.; Wang, C.; Gong, S.Y.; Zhou, S.M.; Wu, T. CsMYB36 is involved in the formation of yellow green peel in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2018, 131, 1659–1669. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.A.; Zheng, J.; Chen, X.M.; Shi, X.Y.; Wang, H.S.; Fu, Q.S. Comprehensive analysis of transcriptome and metabolome reveals the flavonoid metabolic pathway is associated with fruit peel colouration of melon. Molecules 2021, 26, 2830. [Google Scholar] [CrossRef]
- Li, B.B.; Zhao, S.J.; Dou, J.L.; Ali, A.; Gebremeskel, H.; Gao, L.; He, N.; Lu, X.Q.; Liu, W.G. Genetic mapping and development of molecular markers for a candidate gene locus controlling rind colour in watermelon. Theor. Appl. Genet. 2019, 132, 2741–2753. [Google Scholar] [CrossRef]
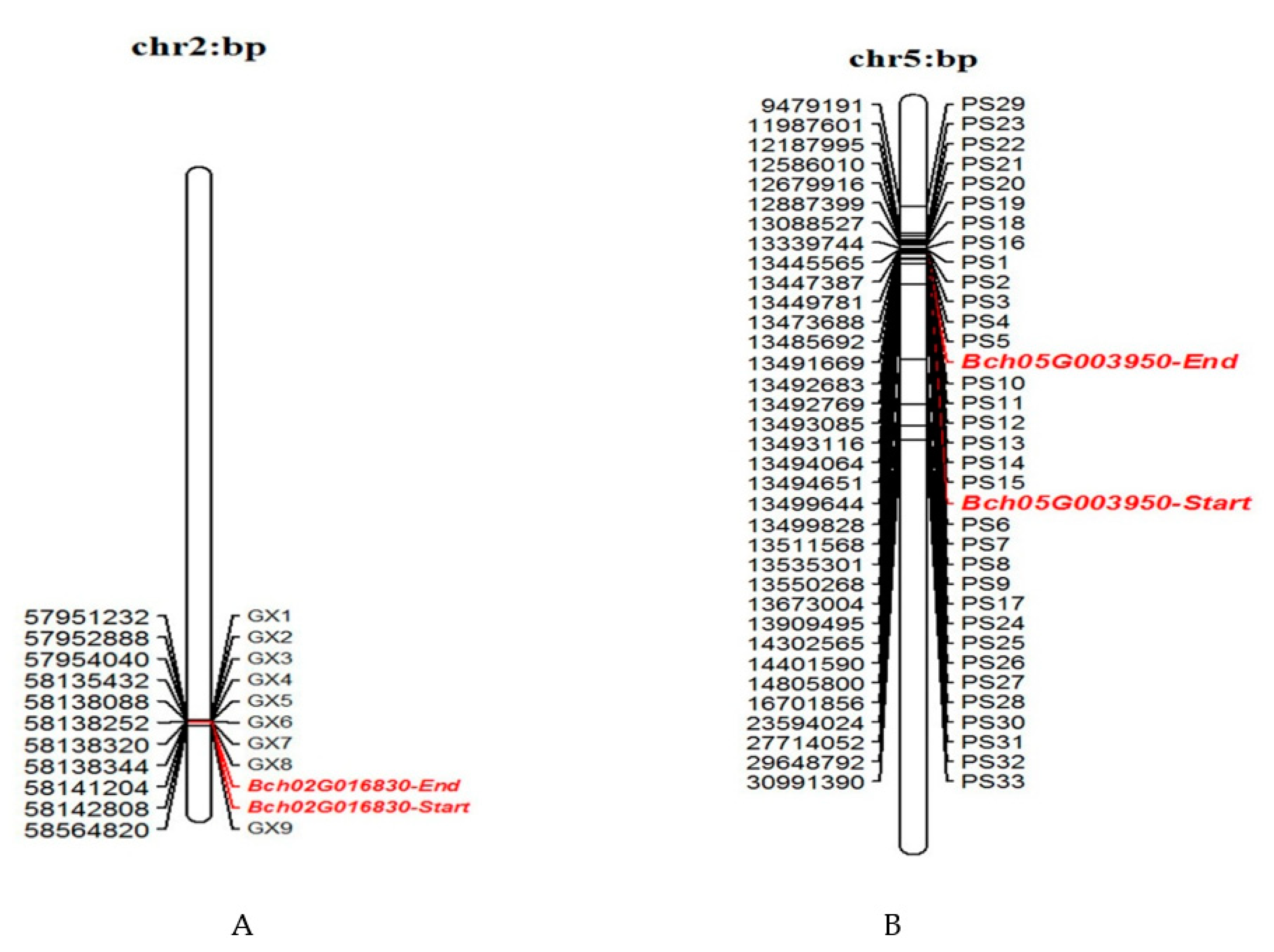

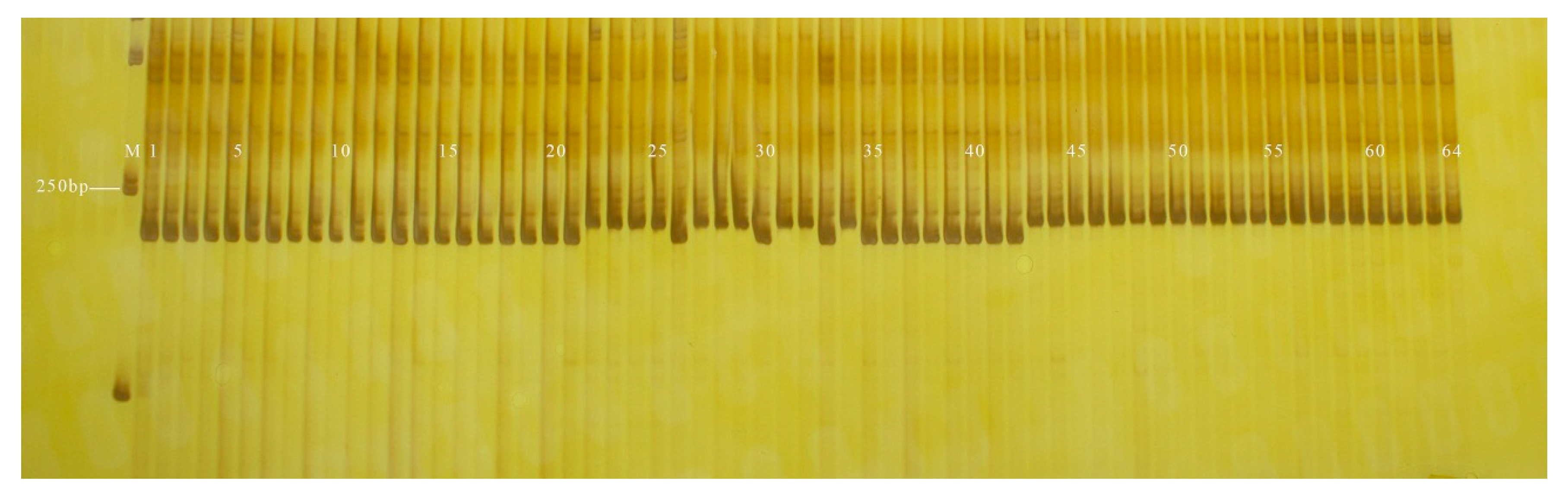


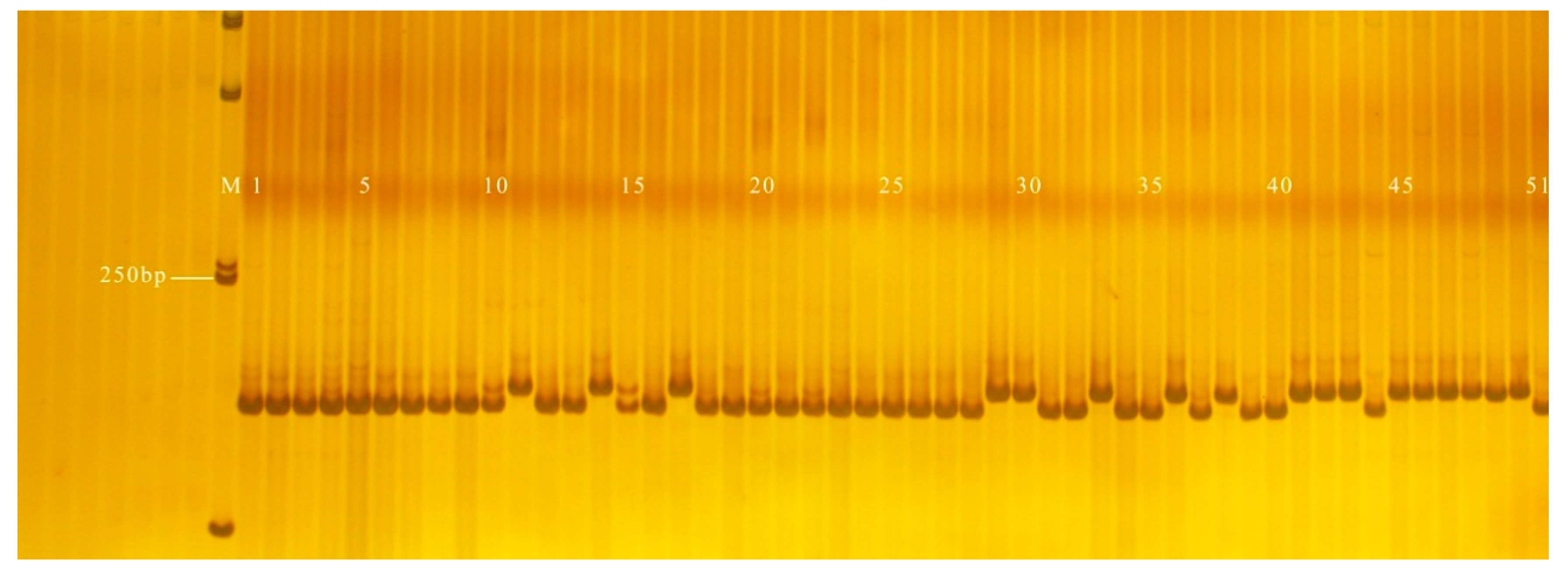


| InDel Markers | Upstream Primer Sequence (5′–3′) | Downstream Primer Sequence (5′–3′) | 120 Inbred Lines Coincidence Rate (%) |
|---|---|---|---|
| GX1 | GACTTCTTGTTGGGGGTTGA | CATATCATTGACTTGATTGGGC | 85.83 |
| GX2 | TGTTTTCTTTTTAACAAATGTCGG | GCTGTAATAATCAACCACGGC | 86.66 |
| GX3 | TCCTTAAAAGTGCTAGTCAATCCA | AAAAAGATTCATCGACATCTCTCC | 87.50 |
| GX4 | CAGAAGGGGAAAAACCTTGG | TGCTTGAAAAGTTAGACCAAACC | 88.33 |
| GX5 | GAGTGGTTGTCAAATATTTCGATTT | GAATTTGAAAACAGAAGGAATTAGAG | 90.00 |
| GX6 | TTGAAGCAGCAACCATACCA | GTGGTAGCCCCCACTTTTCT | 89.16 |
| GX7 | TTGAAGCAGCAACCATACCA | AGTGGTAGCCCCCACTTTTC | 91.66 |
| GX8 | TGAAGCAGCAACCATACCAA | GTGGTAGCCCCCACTTTTCT | 90.83 |
| GX9 | TCATAGATTTGATTTCAGGGCA | CAGTATCATTGAATTCGCACG | 84.16 |
| InDel Markers | Upstream Primer Sequence (5′–3′) | Downstream Primer Sequence (5′–3′) | 120 Inbred Lines Coincidence Rate (%) |
|---|---|---|---|
| PS1 | TGACTCCCACAGCAATGAAC | CCTCAATCTTCAAACACAAGCA | 100 |
| PS2 | CTCTGCAGGCCATCCTCTAT | AATGGAAAATTTCGTGCGTC | 100 |
| PS3 | GTCGGAGAATCAAGCTTTGC | TTCCATGCATAACCTGGAAAT | 100 |
| PS4 | TGCCTCCTTTTATCGCATGA | AATACGCCTCCCCCAATTTA | 100 |
| PS5 | TGCTACGAATTTGTAAATGGTGA | TGGTGGATTTGGAACATCTG | 100 |
| PS6 | TGTGATGTTTTCTTCAGCCAA | TGTTTGAACGGACAATTACATCTT | 100 |
| PS7 | TCCCTTTATTAGTTTCTCCCATGA | TTTCTAAAATTGAGTCCAAATCACA | 100 |
| PS8 | GTGTGCTGATTTGGTTCCCT | AGCCAAGCACGCTTAACATT | 100 |
| PS9 | GGAAACACCTCGATCTGGAA | CCTCCGTGTTGACTCCCTAA | 100 |
| PS10 | TGATTAAACCCAATTTGAAAACA | AACAACTGGAGAAATTTGGAGG | 100 |
| PS11 | CCTCCAAATTTCTCCAGTTG | GCATCTTTCTTAATTACAATGGTTGA | 100 |
| PS12 | CCAAAAACCAAAAACGAAATG | CCCATCCATTTATTTACGATGA | 100 |
| PS13 | CCAAAAACCAAAAACGAAATG | TCCCATCCATTTATTTACGATG | 100 |
| PS14 | TCTATTGCACTGACAAGTGTTTGA | CAAAATTTTTAAAGCTATTGTCCTTC | 100 |
| PS15 | TCATTACTTCAACCAATCACTCC | GCAACAAGGAATTCAGCCAT | 100 |
| PS16 | TTACTTTTCCTCATCCAATTTCTAA | GCATTGCACGTGTTATATAAAGTCT | 100 |
| PS17 | CCGATTCATGTGACCTTGAC | GCAGACAATCCACAAACCAC | 100 |
| PS18 | CAAAACCGTTACTAAACCAGACC | TGAAGGTGTCTGTGCTTTTCT | 100 |
| PS19 | GACATGGCTAAGTCAAGGGC | TGACGAAAAAGAAAATTTTGGAA | 100 |
| PS20 | TGGCAGAACCCAAATTTATTG | AAGGGGAGGGGATTGATAAAG | 100 |
| PS21 | TTCTCTACGCTGAGCCGTTT | GGGGAGGCAAACCAAATAAT | 100 |
| PS22 | TGGTGTGAAGGAAGGTGGTT | TTTTGGGGCAAAGTCTAACCT | 100 |
| PS23 | AAAAGAAATAACTCTTGAAAATGTTTG | CCAATTGCCTTTTGCATTTT | 100 |
| PS24 | CCACCATCTGTTAACTGCCA | GCATGCACATGCTTTCTTGT | 100 |
| PS25 | AACATCCAAAATTTGCACCA | CCTCATCTTCCAACAACTGTCA | 100 |
| PS26 | GGTTAAAAGATAAGCGGTTTGATT | AACCTCCCTCCACTCCCTTA | 100 |
| PS27 | TTCCTAGCCAGTTTGTCATTCA | AAAGCCATCATCTCTATTCCTCA | 100 |
| PS28 | AGGGTGAAATCCCGAAGAAG | TGATAGTTACCCCCGTTCGT | 90.00 |
| PS29 | AAAACCAAGCCGACTTTTGA | TTTGGTAACCATTTCATCTTTGG | 86.66 |
| PS30 | GCAATTTCAGACGGTGGTTT | TCCCTTGCCTTTCTGCTTTA | 65.00 |
| PS31 | TCAAAAGGCTCAAAACCCAC | TGTTGCTGCATTTCCATTGT | 60.00 |
| PS32 | CATGGTCAACGATGTGGAAG | AGAGTGGGTGGAAAGCGTTA | 56.66 |
| PS33 | GTGCAAGCTTATGCCATTGA | AAAGGTCAAACAAATGAGTGTTCA | 55.83 |
| InDel Markers | Coincidence Rate of F2 (96 Plants) (%) | Coincidence Rate of F5 (440 Plants) (%) |
|---|---|---|
| GX8 | 85.42 | |
| GX9 | 84.09 | |
| PS12 | 95.83 | |
| PS13 | 95.68 |
| Hybrid | InDel Assessment Average (%) | GOT Assessment Average (%) | Purity Deviation Range (%) |
|---|---|---|---|
| ‘Lvxianzi 2’ wax gourd | 98.3 | 97.0 | 1.3 |
| ‘Fenxianzi 11’ wax gourd | 100.0 | 100.0 | 0.0 |
| ‘Chunfeng 818’ wax gourd | 99.0 | 98.3 | 0.7 |
| ‘Fenxianzi 1’ wax gourd | 98.0 | 98.6 | 0.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, X.; Wu, W.; Su, L.; Lv, H.; Cheng, Z.; Yang, W.; Nong, L.; Liu, T.; Chen, Y.; Wang, P.; et al. Development and Application of InDel Markers Linked to Fruit-Shape and Peel-Colour Genes in Wax Gourd. Genes 2022, 13, 1567. https://doi.org/10.3390/genes13091567
Huang X, Wu W, Su L, Lv H, Cheng Z, Yang W, Nong L, Liu T, Chen Y, Wang P, et al. Development and Application of InDel Markers Linked to Fruit-Shape and Peel-Colour Genes in Wax Gourd. Genes. 2022; 13(9):1567. https://doi.org/10.3390/genes13091567
Chicago/Turabian StyleHuang, Xiaochun, Wenting Wu, Liwen Su, Haixuan Lv, Zhikui Cheng, Wenrui Yang, Lifeng Nong, Ting Liu, Yong Chen, Peng Wang, and et al. 2022. "Development and Application of InDel Markers Linked to Fruit-Shape and Peel-Colour Genes in Wax Gourd" Genes 13, no. 9: 1567. https://doi.org/10.3390/genes13091567
APA StyleHuang, X., Wu, W., Su, L., Lv, H., Cheng, Z., Yang, W., Nong, L., Liu, T., Chen, Y., Wang, P., & Liu, Z. (2022). Development and Application of InDel Markers Linked to Fruit-Shape and Peel-Colour Genes in Wax Gourd. Genes, 13(9), 1567. https://doi.org/10.3390/genes13091567




