Transcriptomic Analysis Reveals the Correlation between End-of-Day Far Red Light and Chilling Stress in Setaria viridis
Abstract
:1. Introduction
2. Materials and Methods
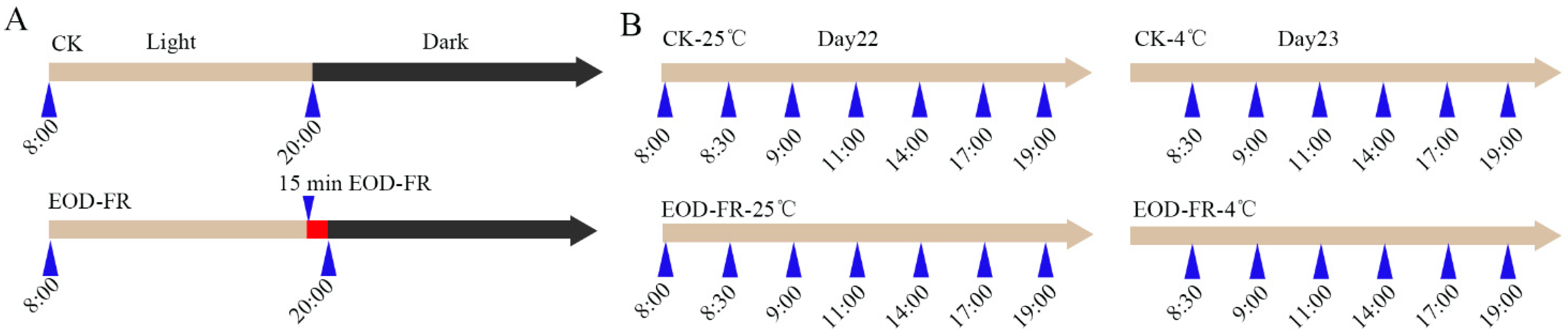
2.1. Plant Material for RNA-Seq Experiments
2.2. Total RNA Isolation
2.3. RNA-Seq Library Construction, Sequencing and Analysis
2.4. Gene Network Construction and Visualization
3. Results
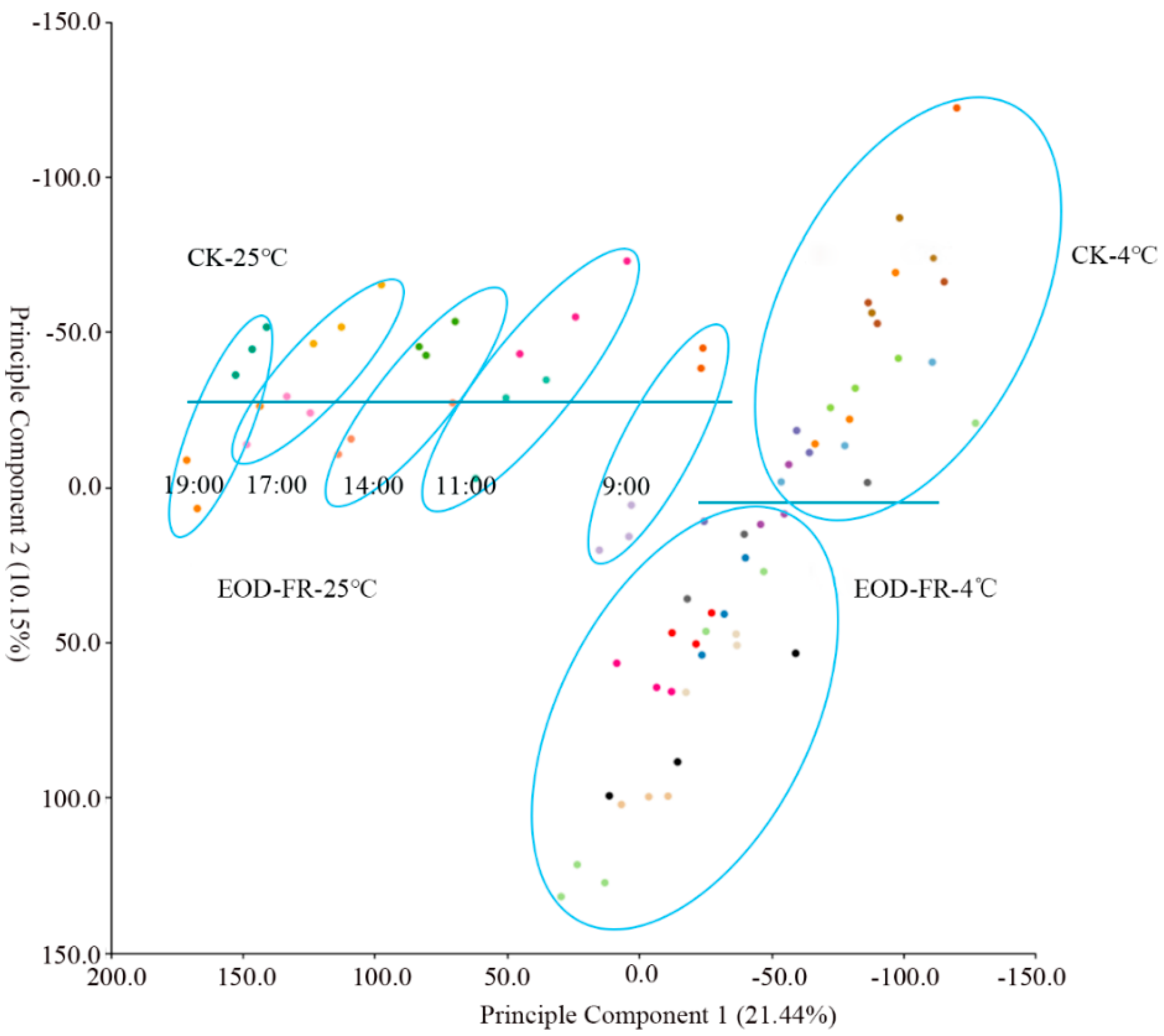
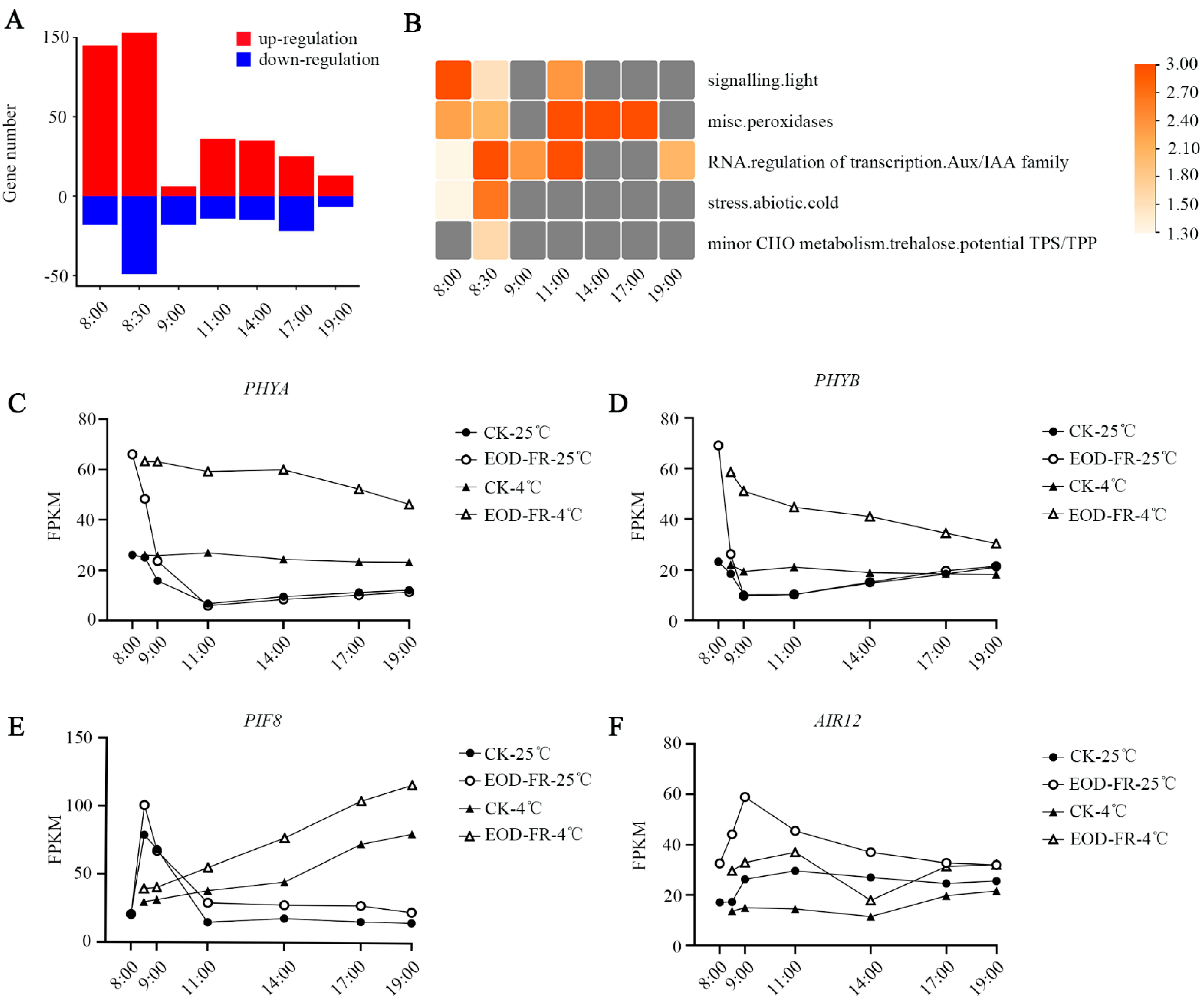
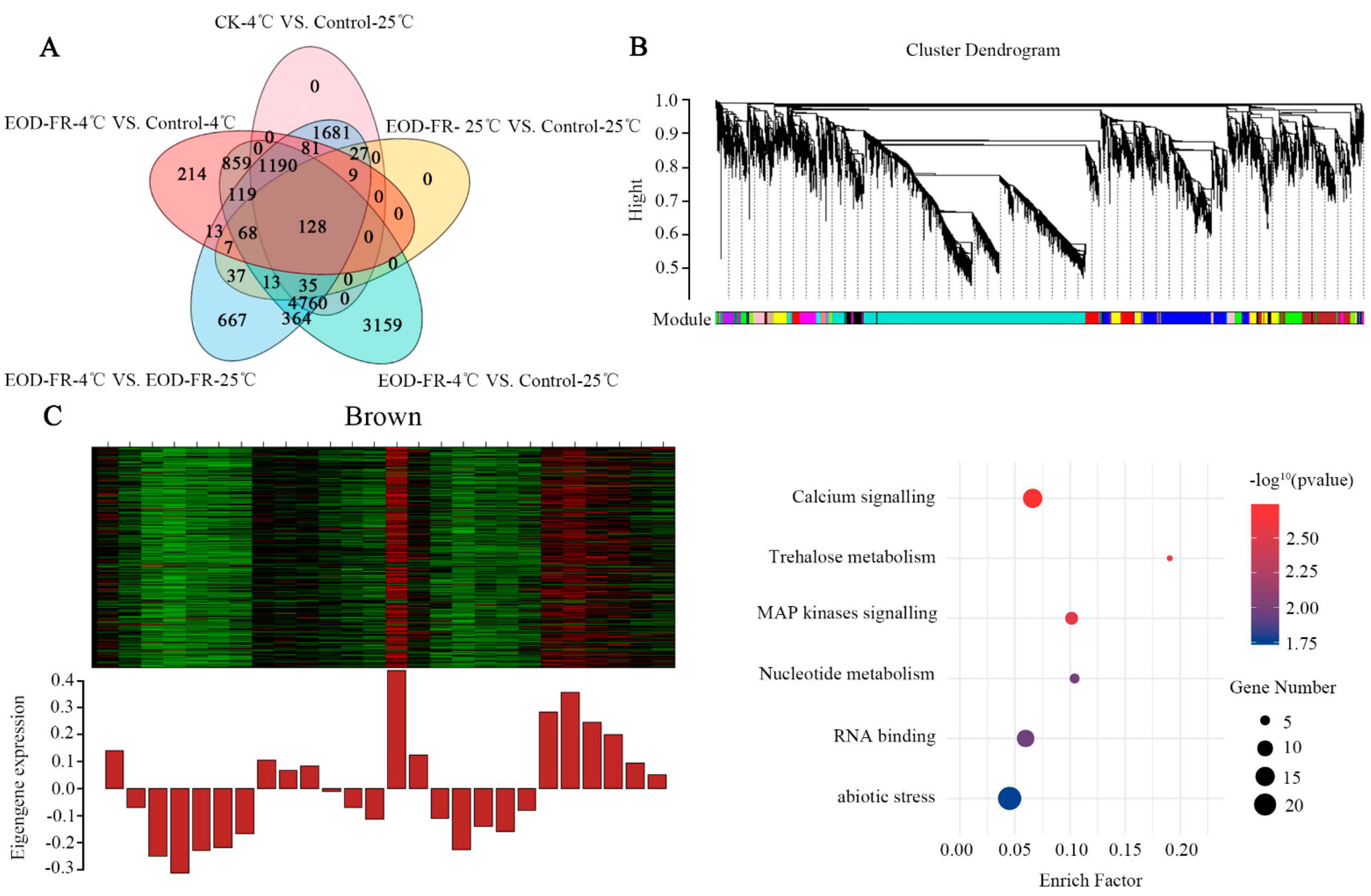
3.1. Transcriptome Atlas under EOD-FR and Cold Treatment
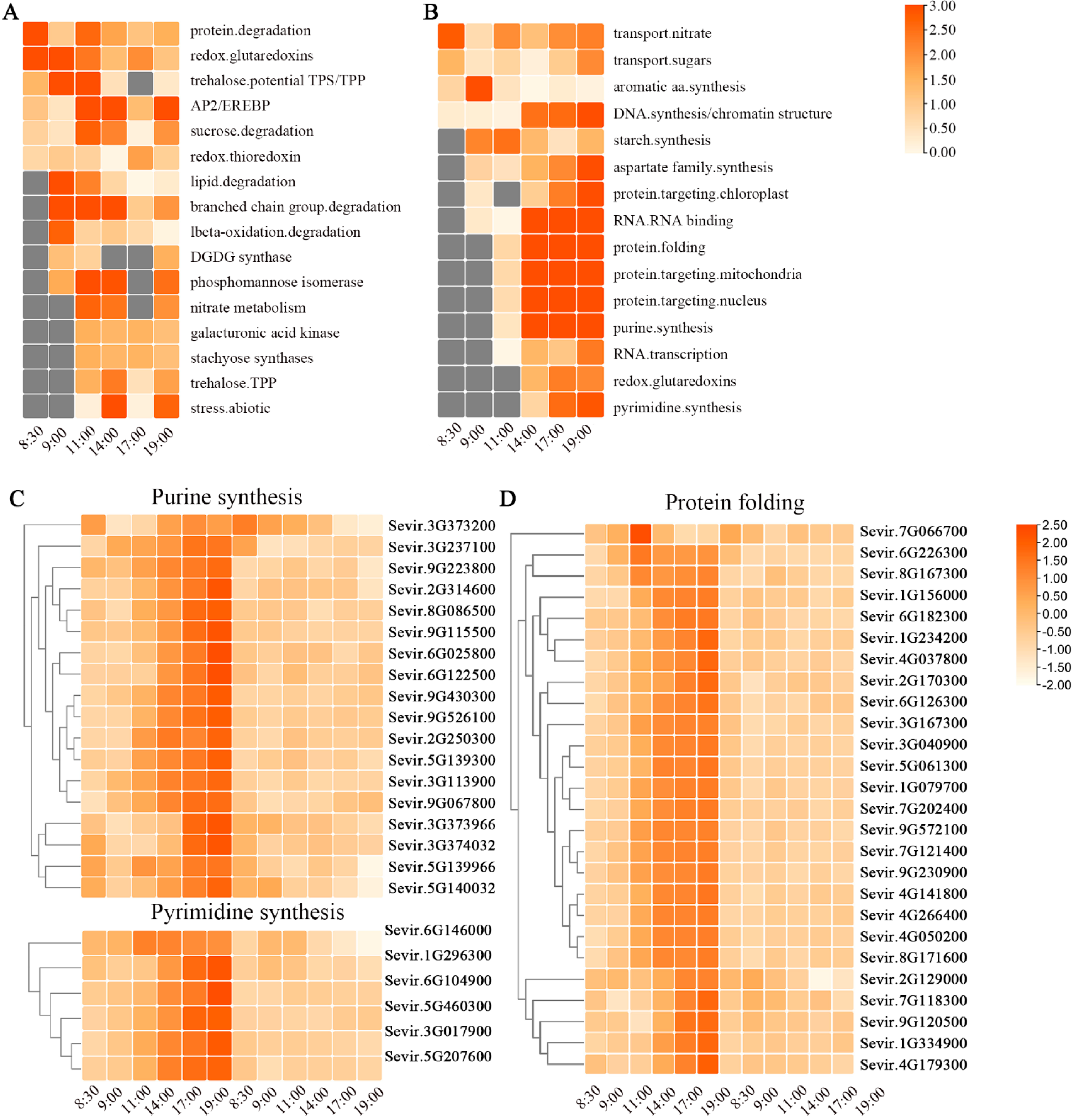
3.2. Functional Category Enrichment under Chilling Stress
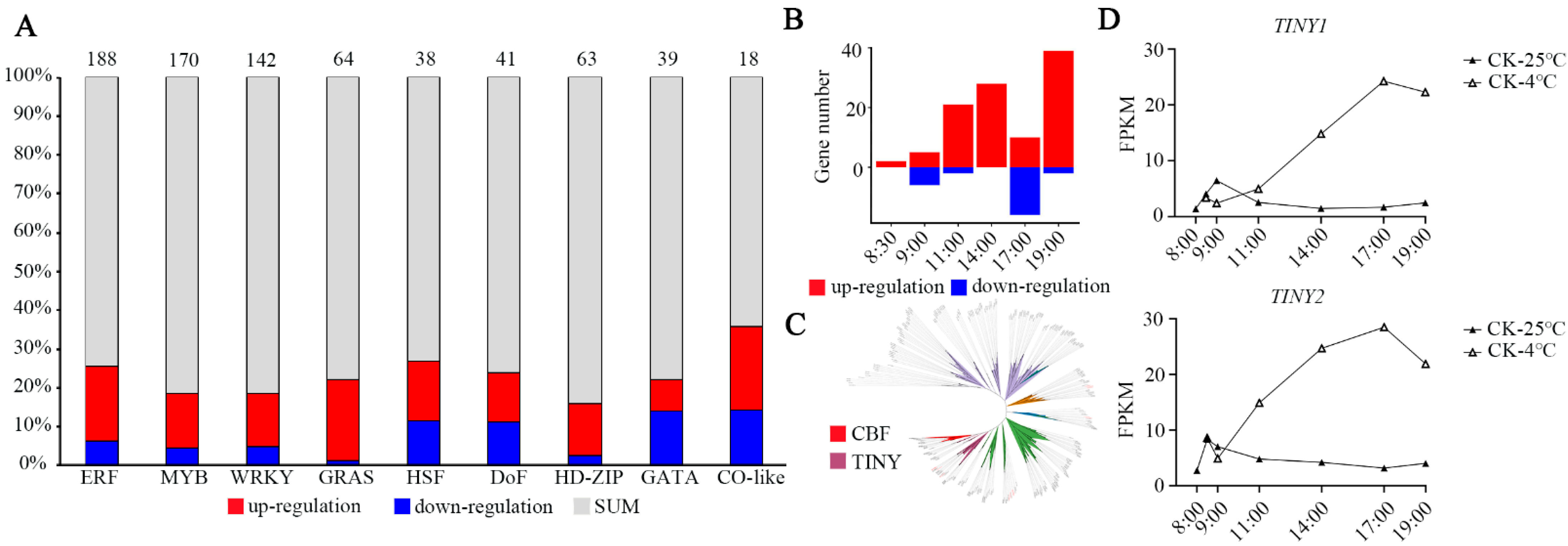
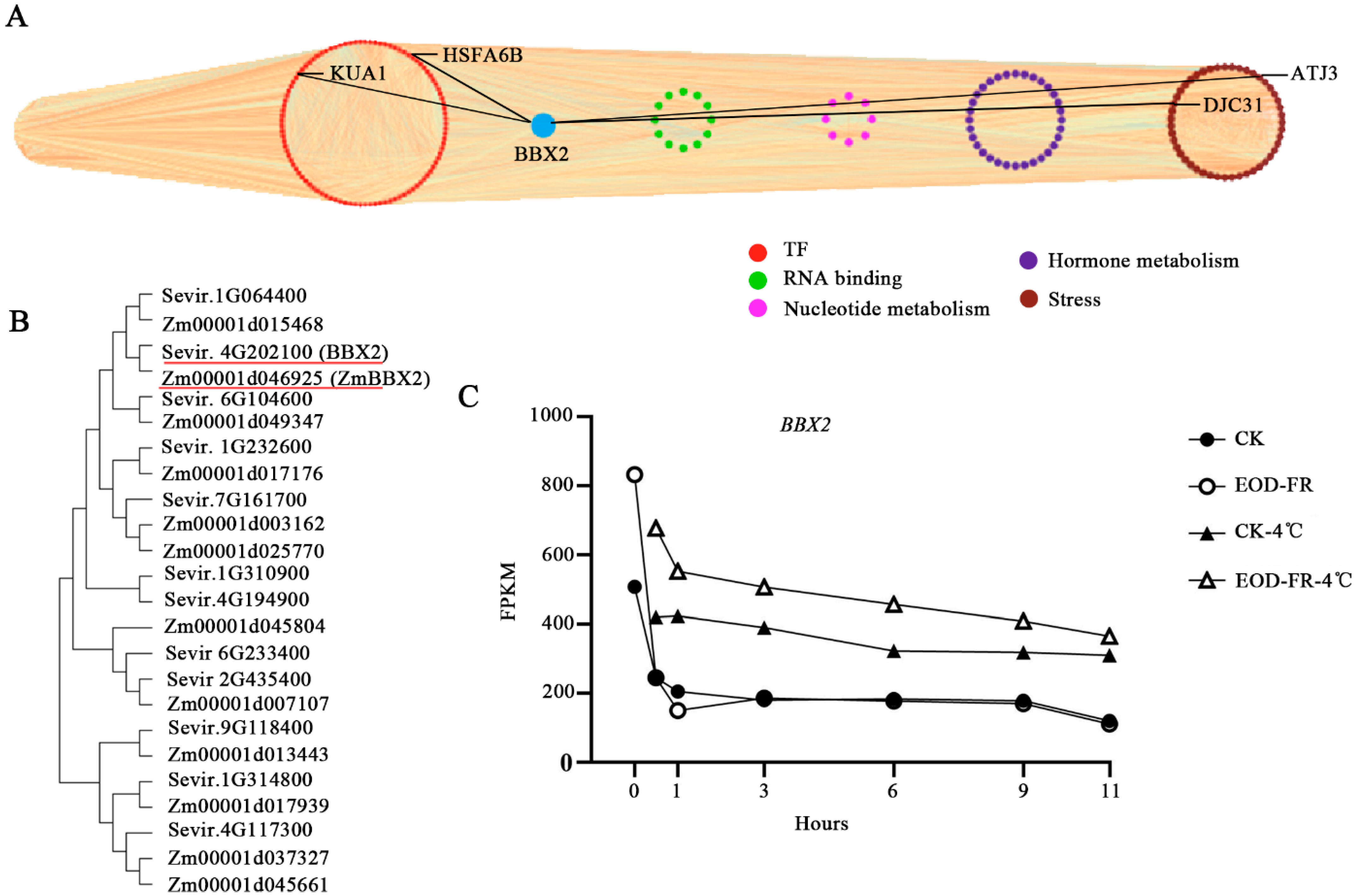
3.3. Changes in Transcription Factors during Chilling Stress
3.4. Functional Characterization of DEGs under EOD-FR
3.5. Transcriptional Dynamics under EOD-FR and Cold Stress
3.6. Identification of TFs Expressed at High Levels in Five Modules
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ruelland, E.; Vaultier, M.-N.l.; Zachowski, A.; Hurry, V. Cold signalling and cold acclimation in plants. Adv. Bot. Res. 2009, 49, 35–150. [Google Scholar]
- Thomashow, M.F. PLANT COLD ACCLIMATION: Freezing tolerance genes and tegulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.H.; Hong, X.; Agarwal, M.; Zhu, J.K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef]
- Thomashow, M.F. So what’s new in the field of plant cold acclimation? Lots! Plant Physiol. 2001, 125, 89–93. [Google Scholar] [CrossRef]
- Novillo, F.; Medina, J.; Salinas, J. Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc. Natl. Acad. Sci. USA 2007, 104, 21002–21007. [Google Scholar] [CrossRef]
- Novillo, F.; Alonso, J.M.; Ecker, J.R.; Salinas, J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 3985–3990. [Google Scholar] [CrossRef]
- Fowler, S.; Thomashow, M.F. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 2002, 14, 1675–1690. [Google Scholar] [CrossRef]
- Kreps, J.A.; Wu, Y.; Chang, H.S.; Zhu, T.; Wang, X.; Harper, J.F. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002, 130, 2129–2141. [Google Scholar] [CrossRef]
- Uno, Y.; Furihata, T.; Abe, H.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 2000, 97, 11632–11637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.J.; Kim, Y.K.; Park, J.Y.; Kim, J. Light signalling mediated by phytochrome plays an important role in cold-induced gene expression through the C-repeat/dehydration responsive element (C/DRE) in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 2002, 29, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.T.; Zarka, D.G.; Van Buskirk, H.A.; Fowler, S.G.; Thomashow, M.F. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. Cell Mol. Biol. 2005, 41, 195–211. [Google Scholar] [CrossRef] [PubMed]
- Pruneda-Paz, J.L.; Kay, S.A. An expanding universe of circadian networks in higher plants. Trends Plant Sci. 2010, 15, 259–265. [Google Scholar] [CrossRef]
- Salomé, P.A.; McClung, C.R. PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 2005, 17, 791–803. [Google Scholar] [CrossRef]
- Maibam, P.; Nawkar, G.M.; Park, J.H.; Sahi, V.P.; Lee, S.Y.; Kang, C.H. The influence of light quality, circadian rhythm, and photoperiod on the CBF-mediated freezing tolerance. Int. J. Mol. Sci. 2013, 14, 11527–11543. [Google Scholar] [CrossRef]
- Franklin, K.A.; Whitelam, G.C. Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nat. Genet. 2007, 39, 1410–1413. [Google Scholar] [CrossRef]
- Wang, F.; Guo, Z.; Li, H.; Wang, M.; Onac, E. Phytochrome A and B function antagonistically to regulate cold tolerance via abscisic acid-dependent jasmonate signaling. Plant Physiol. 2016, 170, 459–471. [Google Scholar] [CrossRef]
- Wang, F.; Wu, N.; Zhang, L.; Ahammed, G.J. Light signaling-dependent regulation of photoinhibition and photoprotection in tomato. Plant Physiol. 2018, 176, 1311–1326. [Google Scholar] [CrossRef]
- Jung, J.H.; Domijan, M.; Klose, C.; Biswas, S.; Ezer, D.; Gao, M.; Khattak, A.K.; Box, M.S.; Charoensawan, V.; Cortijo, S.; et al. Phytochromes function as thermosensors in Arabidopsis. Science 2016, 354, 886–889. [Google Scholar] [CrossRef]
- He, Y.; Li, Y.; Cui, L.; Xie, L.; Zheng, C.; Zhou, G.; Zhou, J.; Xie, X. Phytochrome B Negatively affects cold tolerance by regulating OsDREB1 gene expression through phytochrome interacting factor-like protein OsPIL16 in Rice. Front. Plant Sci. 2016, 7, 1963. [Google Scholar] [CrossRef] [Green Version]
- Pooam, M.; Dixon, N.; Hilvert, M.; Misko, P.; Waters, K.; Jourdan, N.; Drahy, S.; Mills, S.; Engle, D.; Link, J.; et al. Effect of temperature on the Arabidopsis cryptochrome photocycle. Physiol. Plant. 2021, 172, 1653–1661. [Google Scholar] [CrossRef]
- Kohnen, M.V.; Schmid-Siegert, E.; Trevisan, M.; Petrolati, L.A.; Sénéchal, F.; Müller-Moulé, P.; Maloof, J.; Xenarios, I.; Fankhauser, C. Neighbor detection induces organ-specific transcriptomes, revealing patterns underlying hypocotyl-specific growth. Plant Cell 2016, 28, 2889–2904. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Jeong, J.; Kang, M.Y.; Kim, J.; Paek, N.C.; Choi, G. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 2014, 5, 4636. [Google Scholar] [CrossRef]
- Kidokoro, S.; Maruyama, K.; Nakashima, K.; Imura, Y.; Narusaka, Y.; Shinwari, Z.K.; Osakabe, Y.; Fujita, Y.; Mizoi, J.; Shinozaki, K.; et al. The phytochrome-interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol. 2009, 151, 2046–2057. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Huq, E.; Rodríguez-Concepción, M. Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 2010, 107, 11626–11631. [Google Scholar] [CrossRef]
- Shor, E.; Potavskaya, R.; Kurtz, A.; Paik, I.; Huq, E. PIF-mediated sucrose regulation of the circadian oscillator is light quality and temperature dependent. Genes 2018, 9, 628. [Google Scholar] [CrossRef]
- He, Z.; Zhao, T.; Yin, Z.; Liu, J.; Cheng, Y.; Xu, J. The phytochrome-interacting transcription factor CsPIF8 contributes to cold tolerance in citrus by regulating superoxide dismutase expression. Plant Sci. Int. J. Exp. Plant Biol. 2020, 298, 110584. [Google Scholar] [CrossRef]
- Wang, F.; Chen, X.; Dong, S.; Jiang, X.; Wang, L.; Yu, J. Crosstalk of PIF4 and DELLA modulates CBF transcript and hormone homeostasis in cold response in tomato. Plant Biotechnol. J. 2020, 18, 1041–1055. [Google Scholar] [CrossRef]
- Jiang, B.; Shi, Y.; Zhang, X.; Xin, X.; Qi, L.; Guo, H.; Li, J.; Yang, S. PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E6695–E6702. [Google Scholar] [CrossRef]
- Li, Y.; Shi, Y.; Li, M.; Fu, D.; Wu, S.; Li, J.; Gong, Z.; Liu, H.; Yang, S. The CRY2–COP1–HY5–BBX7/8 module regulates blue light-dependent cold acclimation in Arabidopsis. Plant Cell 2021, 11, 3555–3573. [Google Scholar] [CrossRef]
- Dong, M.A.; Farré, E.M.; Thomashow, M.F. Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 7241–7246. [Google Scholar] [CrossRef]
- Keily, J.; MacGregor, D.R.; Smith, R.W.; Millar, A.J.; Halliday, K.J.; Penfield, S. Model selection reveals control of cold signalling by evening-phased components of the plant circadian clock. Plant J. Cell Mol. Biol. 2013, 76, 247–257. [Google Scholar] [CrossRef]
- Li, P.; Ponnala, L.; Gandotra, N.; Wang, L.; Si, Y.; Tausta, S.L.; Kebrom, T.H.; Provart, N.; Patel, R.; Myers, C.R.; et al. The developmental dynamics of the maize leaf transcriptome. Nat. Genet. 2010, 42, 1060–1067. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Han, Q.; Qi, J.; Hao, G.; Zhang, C.; Wang, C.; Dirk, L.M.A.; Downie, A.B.; Zhao, T. ZmDREB1A regulates RAFFINOSE SYNTHASE controlling raffinose accumulation and plant chilling stress tolerance in maize. Plant Cell Physiol. 2020, 61, 331–341. [Google Scholar] [CrossRef]
- Lunn, J.E.; Delorge, I.; Figueroa, C.M.; Van Dijck, P.; Stitt, M. Trehalose metabolism in plants. Plant J. Cell Mol. Biol. 2014, 79, 544–567. [Google Scholar] [CrossRef]
- Ge, L.F.; Chao, D.Y.; Shi, M.; Zhu, M.Z.; Gao, J.P.; Lin, H.X. Overexpression of the trehalose-6-phosphate phosphatase gene OsTPP1 confers stress tolerance in rice and results in the activation of stress responsive genes. Planta 2008, 228, 191–201. [Google Scholar] [CrossRef]
- Burgos, A.; Szymanski, J.; Seiwert, B.; Degenkolbe, T.; Hannah, M.A.; Giavalisco, P.; Willmitzer, L. Analysis of short-term changes in the Arabidopsis thaliana glycerolipidome in response to temperature and light. Plant J. Cell Mol. Biol. 2011, 66, 656–668. [Google Scholar] [CrossRef]
- Sharma, R.; Singh, G.; Bhattacharya, S.; Singh, A. Comparative transcriptome meta-analysis of Arabidopsis thaliana under drought and cold stress. PLoS ONE 2018, 13, e0203266. [Google Scholar] [CrossRef]
- Abogadallah, G.M.; Nada, R.M.; Malinowski, R.; Quick, P. Overexpression of HARDY, an AP2/ERF gene from Arabidopsis, improves drought and salt tolerance by reducing transpiration and sodium uptake in transgenic Trifolium alexandrinum L. Planta 2011, 233, 1265–1276. [Google Scholar] [CrossRef]
- Park, S.; Lee, C.M.; Doherty, C.J.; Gilmour, S.J.; Kim, Y.; Thomashow, M.F. Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J. Cell Mol. Biol. 2015, 82, 193–207. [Google Scholar] [CrossRef]
- Olate, E.; Jiménez-Gómez, J.M.; Holuigue, L.; Salinas, J. NPR1 mediates a novel regulatory pathway in cold acclimation by interacting with HSFA1 factors. Nat. Plants 2018, 4, 811–823. [Google Scholar] [CrossRef]
- Xin, Z.; Browse, J. Eskimo1 mutants of Arabidopsis are constitutively freezing-tolerant. Proc. Natl. Acad. Sci. USA 1998, 95, 7799–7804. [Google Scholar] [CrossRef]
- Kim, B.H.; Kim, S.Y.; Nam, K.H. Genes encoding plant-specific class III peroxidases are responsible for increased cold tolerance of the brassinosteroid-insensitive 1 mutant. Mol. Cells 2012, 34, 539–548. [Google Scholar] [CrossRef]
- Wang, Q.; Shi, H.; Huang, R.; Ye, R.; Luo, Y.; Guo, Z. AIR12 confers cold tolerance through regulation of the CBF cold response pathway and ascorbate homeostasis. Plant Cell Environ. 2021, 44, 1522–1533. [Google Scholar] [CrossRef]
- Soitamo, A.J.; Piippo, M.; Allahverdiyeva, Y.; Battchikova, N.; Aro, E.M. Light has a specific role in modulating Arabidopsis gene expression at low temperature. BMC Plant biol. 2008, 8, 13. [Google Scholar] [CrossRef] [Green Version]
- Avila, L.M.; Obeidat, W.; Earl, H.; Niu, X.; Hargreaves, W.; Lukens, L. Shared and genetically distinct Zea mays transcriptome responses to ongoing and past low temperature exposure. Plant Cell Environ. 2018, 19, 761. [Google Scholar] [CrossRef]
- Onaga, G.; Wydra, K. Advances in Plant Tolerance to Biotic Stresses. Plant Genom. 2016, 29, 167–227. [Google Scholar] [CrossRef]
- Datta, S.; Johansson, H.; Hettiarachchi, C.; Irigoyen, M.L.; Desai, M.; Rubio, V.; Holm, M. LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell 2008, 20, 2324–2338. [Google Scholar] [CrossRef]
- Weller, J.L.; Hecht, V.; Vander Schoor, J.K.; Davidson, S.E.; Ross, J.J. Light regulation of gibberellin biosynthesis in pea is mediated through the COP1/HY5 pathway. Plant Cell 2009, 21, 800–813. [Google Scholar] [CrossRef]
- Crocco, C.D.; Holm, M.; Yanovsky, M.J.; Botto, J.F. Function of B-BOX under shade. Plant Signal. Behav. 2011, 6, 101–104. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Botto, J.F. The BBX family of plant transcription factors. Trends Plant Sci. 2014, 19, 460–470. [Google Scholar] [CrossRef]
- Wang, Q.; Tu, X.; Zhang, J.; Chen, X.; Rao, L. Heat stress-induced BBX18 negatively regulates the thermotolerance in Arabidopsis. Mol. Biol. Rep. 2013, 40, 2679–2688. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, S.; Liu, Q.; Dai, X.; Wang, X. Transcriptomic Analysis Reveals the Correlation between End-of-Day Far Red Light and Chilling Stress in Setaria viridis. Genes 2022, 13, 1565. https://doi.org/10.3390/genes13091565
Sun S, Liu Q, Dai X, Wang X. Transcriptomic Analysis Reveals the Correlation between End-of-Day Far Red Light and Chilling Stress in Setaria viridis. Genes. 2022; 13(9):1565. https://doi.org/10.3390/genes13091565
Chicago/Turabian StyleSun, Shilei, Qingjia Liu, Xiuru Dai, and Xianglan Wang. 2022. "Transcriptomic Analysis Reveals the Correlation between End-of-Day Far Red Light and Chilling Stress in Setaria viridis" Genes 13, no. 9: 1565. https://doi.org/10.3390/genes13091565
APA StyleSun, S., Liu, Q., Dai, X., & Wang, X. (2022). Transcriptomic Analysis Reveals the Correlation between End-of-Day Far Red Light and Chilling Stress in Setaria viridis. Genes, 13(9), 1565. https://doi.org/10.3390/genes13091565




