Cathepsin D Plays a Vital Role in Macrobrachium nipponense of Ovary Maturation: Identification, Characterization, and Function Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Animals
2.2. Nucleotide Sequence and Bioinformatics Analysis of Mn-CTSD
2.3. In Situ Hybridization
2.4. Mn-CTSD RNA Interference
2.5. Statistical Analysis
3. Results
3.1. Characteristics of Mn-CTSD Sequence
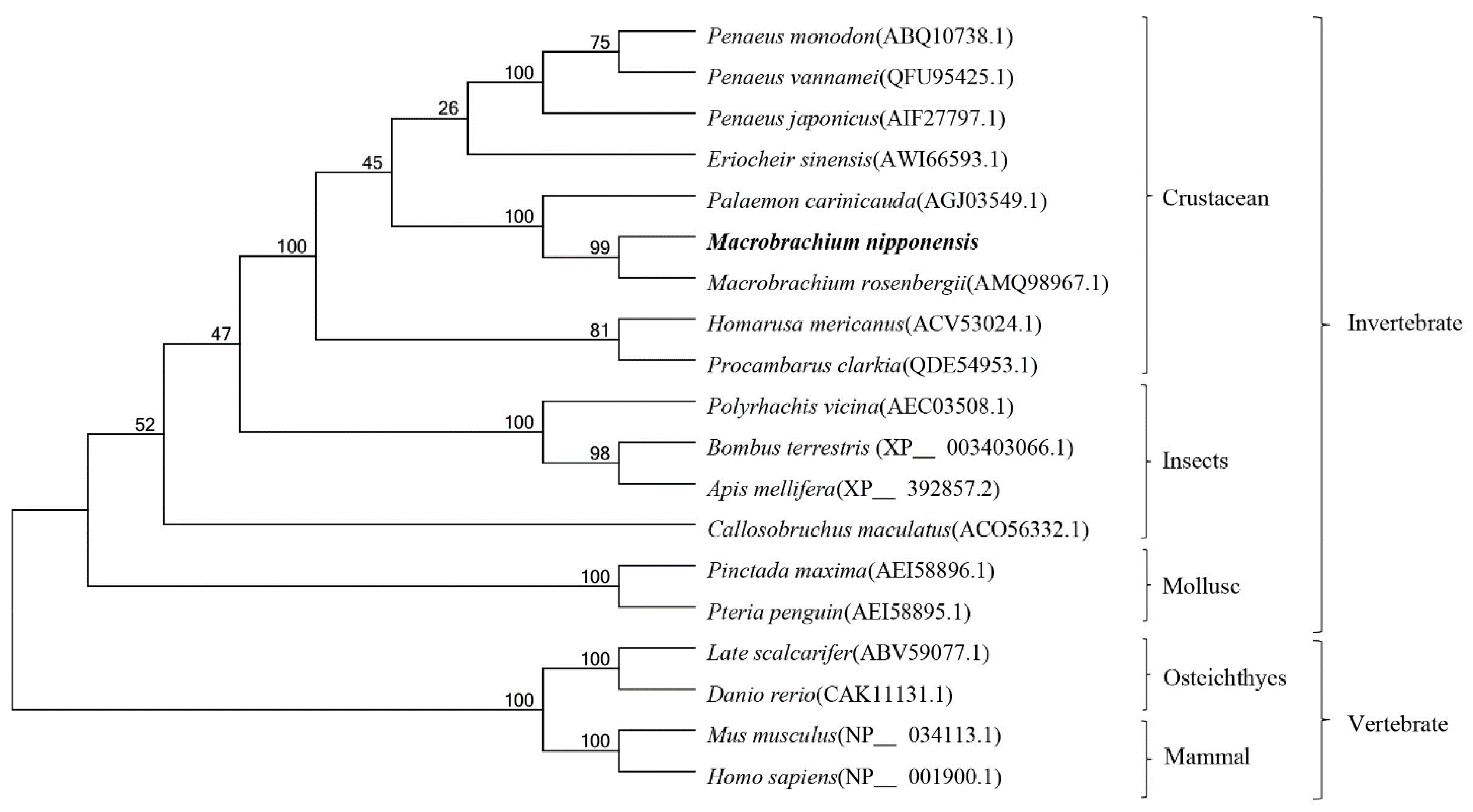
3.2. Phylogenetic and Sequence Alignment Analysis of Mn-CTSD
3.3. Spatiotemporal Expression of Mn-CTSD
3.4. Mn-CTSD Localization in Five Ovarian Stages by ISH
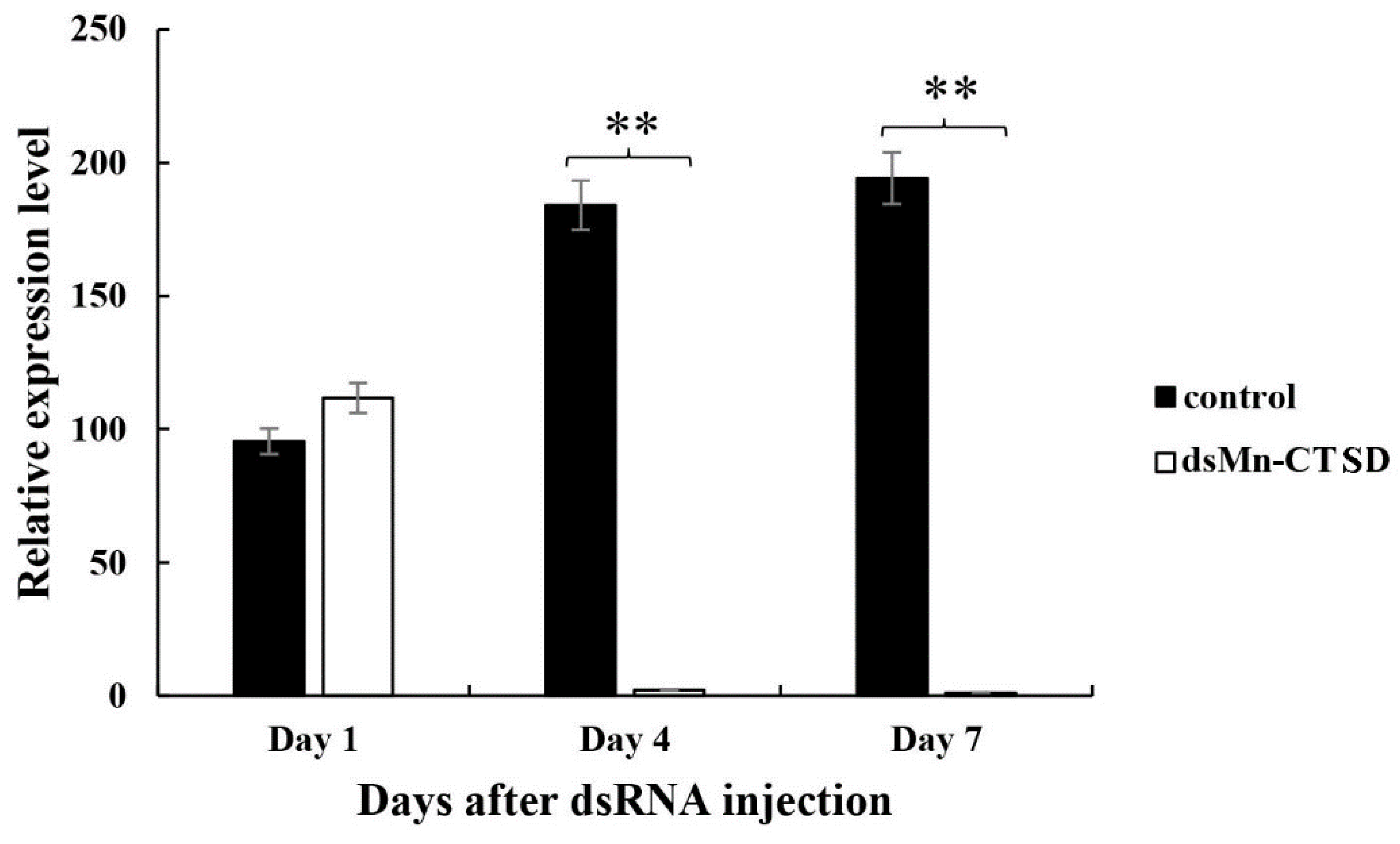
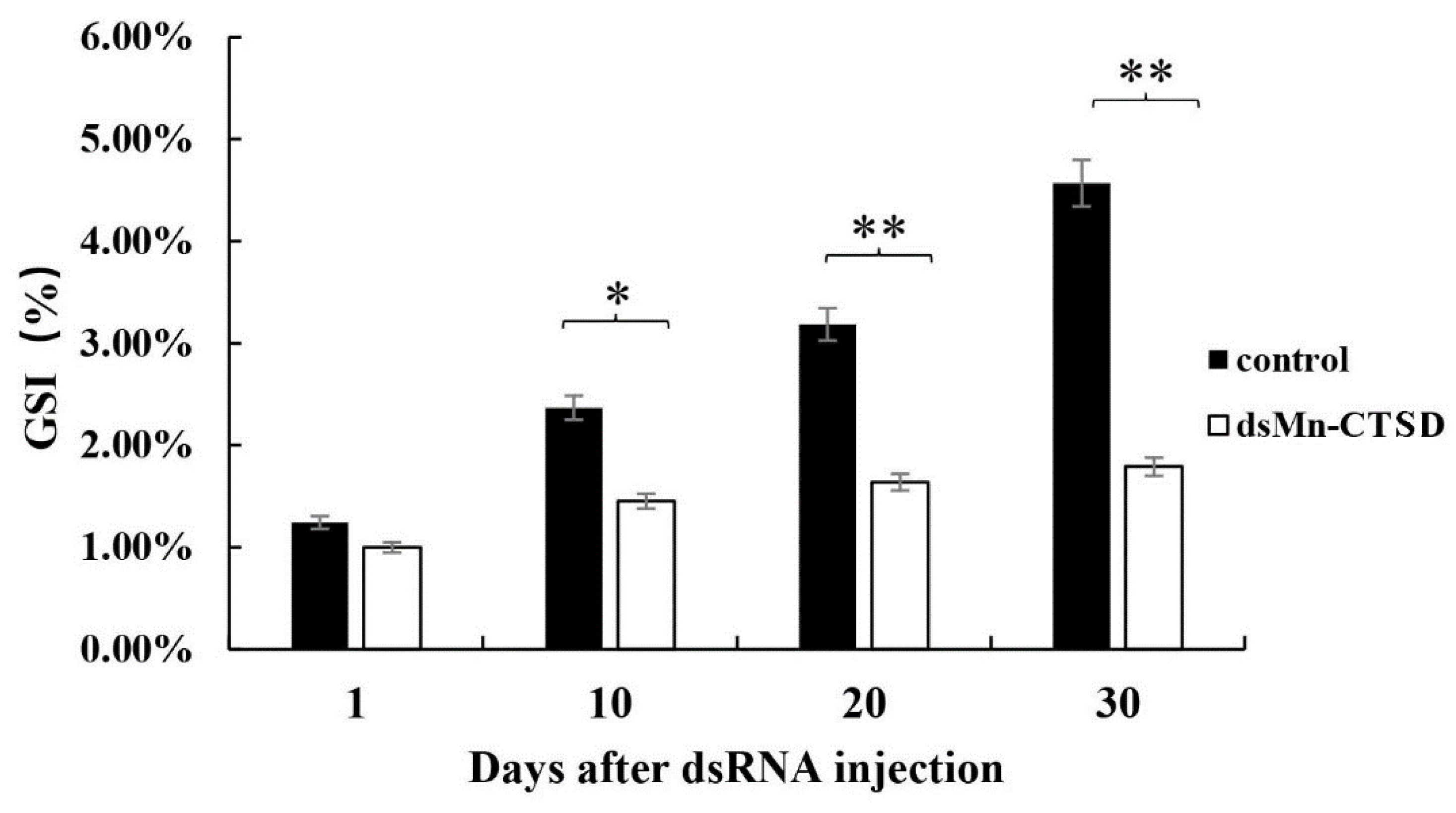
3.5. Effect of Mn-CTSD on Ovarian Maturation Was Knocked Out by RNAi
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, F.; Zhang, S.; Fu, C.; Li, T.; Cui, X. Molecular and functional analysis of the insulin-like peptides gene in the oriental river prawn Macrobrachium nipponense. Gen. Comp. Endocrinol. 2019, 280, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H.; Fu, H.; Xiong, Y.; Jiang, S.; Zhang, W.; Sun, S.; Jin, S.; Gong, Y.; Wang, Y.; Shan, D. Molecular insights into reproduction regulation of female Oriental River prawns Macrobrachium nipponense through comparative transcriptomic analysis. Sci. Rep. 2017, 7, 12161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Jiang, S.; Qiao, H.; Xiong, Y.; Wu, Y. Transcriptome analysis of five ovarian stages reveals gonad maturation in female Macrobrachium nipponense. BMC Genom. 2021, 22, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H.; Fu, H.; Jin, S.; Wu, Y.; Jiang, S.; Gong, Y.; Xiong, Y. Constructing and random sequencing analysis of normalized cDNA library of testis tissue from oriental river prawn (Macrobrachium nipponense). Comp. Biochem. Physiol. Part D Genom. Proteom. 2012, 7, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fu, Y.; Jiang, S.; Qiao, H.; Wu, Y. Comparative metabolomics analysis of ovarian developmental stages in Macrobrachium nipponense. Comp. Biochem. Physiol. Part D Genom. Proteom. 2020, 34, 100648. [Google Scholar] [CrossRef]
- Abreu, L.A.; Valle, D.; Manso, P.; Façanha, A.R.; Pelajo-Machado, M.; Masuda, H.; Masuda, A.; Vaz, I.; Lenzi, H.; Oliveira, P.L. Proteolytic activity of Boophilus microplus Yolk pro-Cathepsin D (BYC) is coincident with cortical acidification during embryogenesis. Insect Biochem. Mol. Biol. 2004, 34, 443–449. [Google Scholar] [CrossRef]
- Choi, K.M.; Sang, H.S.; An, C.M.; Nam, B.H.; Kim, Y.O.; Kim, J.W.; Park, C.I. Cloning, characterisation, and expression analysis of the cathepsin D gene from rock bream (Oplegnathus fasciatus). Fish Shellfish Immunol. 2014, 40, 253–258. [Google Scholar] [CrossRef]
- Fialho, E.; Nakamura, A.; Juliano, L.; Masuda, H.; Silva-Neto, M. Cathepsin D-mediated yolk protein degradation is blocked by acid phosphatase inhibitors. Arch. Biochem. Biophys. 2005, 436, 246–253. [Google Scholar] [CrossRef]
- Roberg, K.; Kågedal, K.; Ollinger, K. Microinjection of Cathepsin D Induces Caspase-Dependent Apoptosis in Fibroblasts. Am. J. Pathol. 2002, 161, 89–96. [Google Scholar] [CrossRef] [Green Version]
- Bańkowska, A.; Gacko, M.; Chyczewska, E.; Worowska, A. Biological and diagnostic role of cathepsin D. Rocz. Akad. Med. W Białymstoku 1997, 42, 79–85. [Google Scholar]
- Cha, I.S.; Kwon, J.; Mun, J.Y.; Park, S.B.; Jang, H.B.; Nho, S.W.; del Castillo, C.S.; Hikima, J.-I.; Aoki, T.; Jung, T.S. Cathepsins in the kidney of olive flounder, Paralichthys olivaceus, and their responses to bacterial infection. Dev. Comp. Immunol. 2012, 38, 538–544. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.D.; Jiao, Z.; Ji, X.S.; Zhou, F.N.; Fu, Y.; Chen, W.; Zeng, Y.Q.; Li, T.M.; Wang, H. Molecular cloning, characterization and expression of cathepsin D from grass carp (Ctenopharyngodon idella). Fish Shellfish Immunol. 2012, 33, 1207–1214. [Google Scholar] [CrossRef] [PubMed]
- Feng, T.; Zhang, H.; Liu, H.; Zhou, Z.; Niu, D.; Wong, L.; Kucuktas, H.; Liu, X.; Peatman, E.; Liu, Z. Molecular characterization and expression analysis of the channel catfish cathepsin D genes. Fish Shellfish Immunol. 2011, 31, 164–169. [Google Scholar] [CrossRef]
- Yu, X.M.; Chen, J.L.; Abbas, M.N.; Gul, I.; Kausar, S.; Dai, L.S. Characterization of the cathepsin D in Procambarus clarkii and its biological role in innate immune responses. Dev. Comp. Immunol. 2020, 111, 103766. [Google Scholar] [CrossRef]
- Yang, C.; Zhang, J.; Li, F.; Ma, H.; Zhang, Q.; Jose Priya, T.A.; Zhang, X.; Xiang, J. A Toll receptor from Chinese shrimp Fenneropenaeus chinensis is responsive toVibrio anguillarum infection. Fish Shellfish Immunol. 2008, 24, 564–574. [Google Scholar] [CrossRef]
- Li, C.S.; Kausar, S.; Gul, I.; Yao, X.X.; Dai, L.S. Heat shock protein 20 from Procambarus clarkii is involved in the innate immune responses against microbial infection. Dev. Comp. Immunol. 2020, 106, 103638. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H.; Sun, S.; Zhang, W.; Jin, H.; Xiong, S. Characterization, expression, and function analysis of gonad-inhibiting hormone in Oriental River prawn, Macrobrachium nipponense and its induced expression by temperature. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2015, 185, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jin, S.; Fu, H.; Qiao, H.; Sun, S.; Zhang, W.; Jiang, S.; Gong, Y.; Xiong, Y.; Wu, Y. Identification and Characterization of the DMRT11E Gene in the Oriental River Prawn Macrobrachium nipponense. Int. J. Mol. Sci. 2019, 20, 1734. [Google Scholar] [CrossRef] [Green Version]
- Lei, X.; Ming, Y.; Fu, H.; Sun, S.; Yan, W. Molecular Cloning and Expression of MnGST-1 and MnGST-2 from Oriental River Prawn, Macrobrachium nipponense, in Response to Hypoxia and Reoxygenation. Int. J. Mol. Sci. 2018, 19, 3102. [Google Scholar]
- Hu, Y.; Fu, H.; Hui, Q.; Sun, S.; Zhang, W.; Jin, S.; Jiang, S.; Gong, Y.; Xiong, Y.; Wu, Y. Validation and Evaluation of Reference Genes for Quantitative Real-Time PCR in Macrobrachium Nipponense. Int. J. Mol. Sci. 2018, 19, 2258. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Qiao, H.; Fu, H.; Sun, S.; Zhang, W.; Jin, S.; Jiang, S.; Gong, Y.; Xiong, Y.; Wu, Y. Identification and characterization of opsin gene and its role in ovarian maturation in the oriental river prawn Macrobrachium nipponense. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2018, 218, S1096495917302129. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H.; Jiang, F.; Xiong, Y.; Jiang, S.; Fu, H.; Li, F.; Zhang, W.; Sun, S.; Jin, S.; Gong, Y. characterization, expression patterns of molt- inhibiting hormone gene of macrobrachium nipponense and its roles in molting and growth. PLoS ONE 2018, 13, e0198861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2-DDCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Rojo, L.; Muhlia-Almazan, A.; Saborowski, R.; García-Carreo, F. Aspartic Cathepsin D Endopeptidase Contributes to Extracellular Digestion in Clawed Lobsters Homarus americanus and Homarus gammarus. Mar. Biotechnol. 2010, 12, 696–707. [Google Scholar] [CrossRef] [Green Version]
- Xiao, R.; Zhang, Z.; Wang, H.; Han, Y.; Gou, M.; Li, B.; Duan, D.; Wang, J.; Liu, X.; Li, Q. Identification and characterization of a cathepsin D homologue from lampreys (Lampetra japonica). Dev. Comp. Immunol. 2015, 49, 149–156. [Google Scholar] [CrossRef]
- Padilha, M.; Pimentel, A.C.; Ribeiro, A.F.; Terra, W.R. Sequence and function of lysosomal and digestive cathepsin D-like proteinases of Musca domestica midgut. Insect Biochem. Mol. Biol. 2009, 39, 782–791. [Google Scholar] [CrossRef]
- Mei, Y.; Chen, Y.; Li, J.; Gao, P.; Zhang, G. Sequence Identification, Tissue Distribution and Polymorphism of the Porcine Cathepsin D ( CTSD ) Gene. Anim. Biotechnol. 2008, 19, 144–158. [Google Scholar] [CrossRef]
- Brooks, S.; Tyler, C.R.; Carnevali, O.; Coward, K.; Sumpter, J.P. Molecular characterisation of ovarian cathepsin D in the rainbow trout, Oncorhynchus mykiss. Gene 1997, 201, 45–54. [Google Scholar] [CrossRef]
- Garcia, M.; Platet, N.; Liaudet, E.; Laurent, V.; Derocq, D.; Brouillet, J.P.; Rochefort, H. Biological and Clinical Significance of Cathepsin D in Breast Cancer Metastasis. Stem Cells 2010, 14, 642–650. [Google Scholar] [CrossRef]
- Khalkhali-Ellis, Z.; Zhang, M.; Hendrix, M.J.C. Maspin and cathepsin d partnership in regulating mammary gland development and breast cancer. In Breast Cancer: Causes, Diagnosis and Treatment; Romero, M.R., Ed.; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2011; pp. 161–176. [Google Scholar]
- Masson, O.; Prébois, C.; Derocq, D.; Meulle, A.; Liaudet-Coopman, E. Cathepsin-D, A Key Protease in Breast Cancer, Is Up-Regulated in Obese Mouse and Human Adipose Tissue, and Controls Adipogenesis. PLoS ONE 2011, 6, e16452. [Google Scholar] [CrossRef] [Green Version]
- Delbarre-Ladrat, C.; Lamballerie-Anton, M.; Verrez-Bagnis, V. Calpain and cathepsin activities in post mortem fish and meat muscles. Food Chem. 2007, 101, 1474–1479. [Google Scholar]
- Jia, A.; Zhang, X.-H. Molecular cloning, characterization and expression analysis of cathepsin D gene from turbot Scophthalmus maximus. Fish Shellfish Immunol. 2009, 26, 606–613. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, L.B.; Nielsen, H.H. Purification and characterization of cathepsin D from herring muscle (Clupea harengus). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2001, 128, 351–363. [Google Scholar] [CrossRef]
- Follo, C.; Ozzano, M.; Montalenti, C.; Santoro, M.M.; Isidoro, C. Knockdown of cathepsin D in zebrafish fertilized eggs determines congenital myopathy. Biosci. Rep. 2013, 33, e00034. [Google Scholar] [CrossRef] [PubMed]
- Yoshizaki, N.; Yonezawa, S. Cysteine proteinase plays a key role for the initiation of yolk digestion during development of Xenopus laevis. Dev. Growth Differ. 2010, 40, 659–667. [Google Scholar] [CrossRef]
- Palomino, J.; Herrera, G.; Torres-Fuentes, J.; Dettleff, P.; Patel, A.; Martínez, V. Assessment of cathepsin mRNA expression and enzymatic activity during early embryonic development in the yellowtail kingfish Seriola lalandi. Anim. Reprod. Sci. 2017, 180, 23–29. [Google Scholar] [CrossRef]
- Kwon, J.Y.; Prat, F.; Randall, C.; Tyler, C.R. Molecular Characterization of Putative Yolk Processing Enzymes and Their Expression during Oogenesis and Embryogenesis in Rainbow Trout (Oncorhynchus mykiss). Biol. Reprod. 2001, 65, 1701–1709. [Google Scholar] [CrossRef] [Green Version]
- Haendeler, J.; Popp, R.; Goy, C.; Tischler, V.; Zeiher, A.M.; Dimmeler, S. Cathepsin D and H2O2 stimulate degradation of thioredoxin-1: Implication for endothelial cell apoptosis. J. Biol. Chem. 2005, 280, 42945–42951. [Google Scholar] [CrossRef] [Green Version]
- Cheung, M.; Seres, T.; Cleveland, J.; Kounis, N.G. Kounis syndrome, a coronary hypersensitivity disorder: A rare case of amiodarone-induced coronary vasospasm and simultaneous peripheral vasodilation intraoperatively. Int. J. Cardiol. 2016, 218, 267–268. [Google Scholar] [CrossRef]
- Bonavida, B.; Mencia-Huerta, J.M.; Braquet, P. Effects of platelet-activating factor on peripheral blood monocytes: Induction and priming for TNF secretion. J. Lipid Mediat. 1990, 2, S65–S76. [Google Scholar]




| Ovarian Stage | Ovarian Maturation Stage | Ovarian Appearance Color |
|---|---|---|
| O-I | undeveloped stage | transparent |
| O-II | developing stage | yellow |
| O-III | nearly-ripe stage | light green |
| O-IV | ripe stage | dark green |
| O-V | worn out stage | gray |
| Different Embryonic and Larva Development Stages | |
|---|---|
| CS | cleavage stage |
| BS | blastula stage |
| GS | gastrulation stage |
| NS | nauplius stage |
| ZS | zoea stage |
| L1 | the 1st day larvae after hatching |
| L5 | the 5th day larvae after hatching |
| L10 | the 10th day larvae after hatching |
| L15 | the 15th day after hatching |
| PL1 | the 1st day after metamorphosis |
| PL5 | the 5th day after metamorphosis |
| PL10 | the 10th day after metamorphosis |
| PL15 | the 15th day after metamorphosis |
| PL25 | the 25th day after metamorphosis |
| Primer Name | Sequence (5′-3′) | Usage |
|---|---|---|
| Mn-CTSD 3′F1 | TGAAGGTGTTGATTTTGCTGGC | 3′RACE |
| Mn-CTSD 3′F2 | CTCCATAAAACTTACACGCCGC | 3′RACE |
| Mn-CTSD 3′F3 | TCATCCAATCTCTGGGTTCCTTC | 3′RACE |
| Mn-CTSD qF | GACGTTGTCTTCCCAAACTGG | RT-PCR |
| Mn-CTSD qR | GCTTGGAGGTTCTGACCCAAA | RT-PCR |
| EIF-F | CATGGATGTACCTGTGGTGAAAC | RT-PCR |
| EIF-R | CTGTCAGCAGAAGGTCCTCATTA | RT-PCR |
| Mn-CTSD iF | TAATACGACTCACTATAGGGGACGTTGTCTTCCCAAACTGG | CTSD dsRNA F |
| Mn-CTSD iR | TAATACGACTCACTATAGGGGCTTGGAGGTTCTGACCCAAA | CTSD dsRNA R |
| GFP iF | GATCACTAATACGACTCACTATAGGGTCCTGGTCGAGCTGGACGG | GFP F |
| GFP iR | GATCACTAATACGACTCACTATAGGGCGCTTCTCGTTGGGGTCTTTG | GFP R |
| probe | CTCGTCTTATCAACAAGAAGATTGGTGCTAAGCCTATTGTTGGAGGAGAGTGGATGGTTGACTGTGGTCTTA | ISH |
| anti-probe | TAAGACCACAGTCAACCATCCACTCTCCTCCAACAATAGGCTTAGCACCAATCTTCTTGTTGATAAGACGAG | ISH |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, D.; Zhang, W.; Jiang, S.; Xiong, Y.; Jin, S.; Pan, F.; Zhu, J.; Gong, Y.; Wu, Y.; Qiao, H.; et al. Cathepsin D Plays a Vital Role in Macrobrachium nipponense of Ovary Maturation: Identification, Characterization, and Function Analysis. Genes 2022, 13, 1495. https://doi.org/10.3390/genes13081495
Cheng D, Zhang W, Jiang S, Xiong Y, Jin S, Pan F, Zhu J, Gong Y, Wu Y, Qiao H, et al. Cathepsin D Plays a Vital Role in Macrobrachium nipponense of Ovary Maturation: Identification, Characterization, and Function Analysis. Genes. 2022; 13(8):1495. https://doi.org/10.3390/genes13081495
Chicago/Turabian StyleCheng, Dan, Wenyi Zhang, Sufei Jiang, Yiwei Xiong, Shubo Jin, Fangyan Pan, Junpeng Zhu, Yongsheng Gong, Yan Wu, Hui Qiao, and et al. 2022. "Cathepsin D Plays a Vital Role in Macrobrachium nipponense of Ovary Maturation: Identification, Characterization, and Function Analysis" Genes 13, no. 8: 1495. https://doi.org/10.3390/genes13081495
APA StyleCheng, D., Zhang, W., Jiang, S., Xiong, Y., Jin, S., Pan, F., Zhu, J., Gong, Y., Wu, Y., Qiao, H., & Fu, H. (2022). Cathepsin D Plays a Vital Role in Macrobrachium nipponense of Ovary Maturation: Identification, Characterization, and Function Analysis. Genes, 13(8), 1495. https://doi.org/10.3390/genes13081495







