The Rm1 and Rm2 Resistance Genes to Green Peach Aphid (Myzus persicae) Encode the Same TNL Proteins in Peach (Prunus persica L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
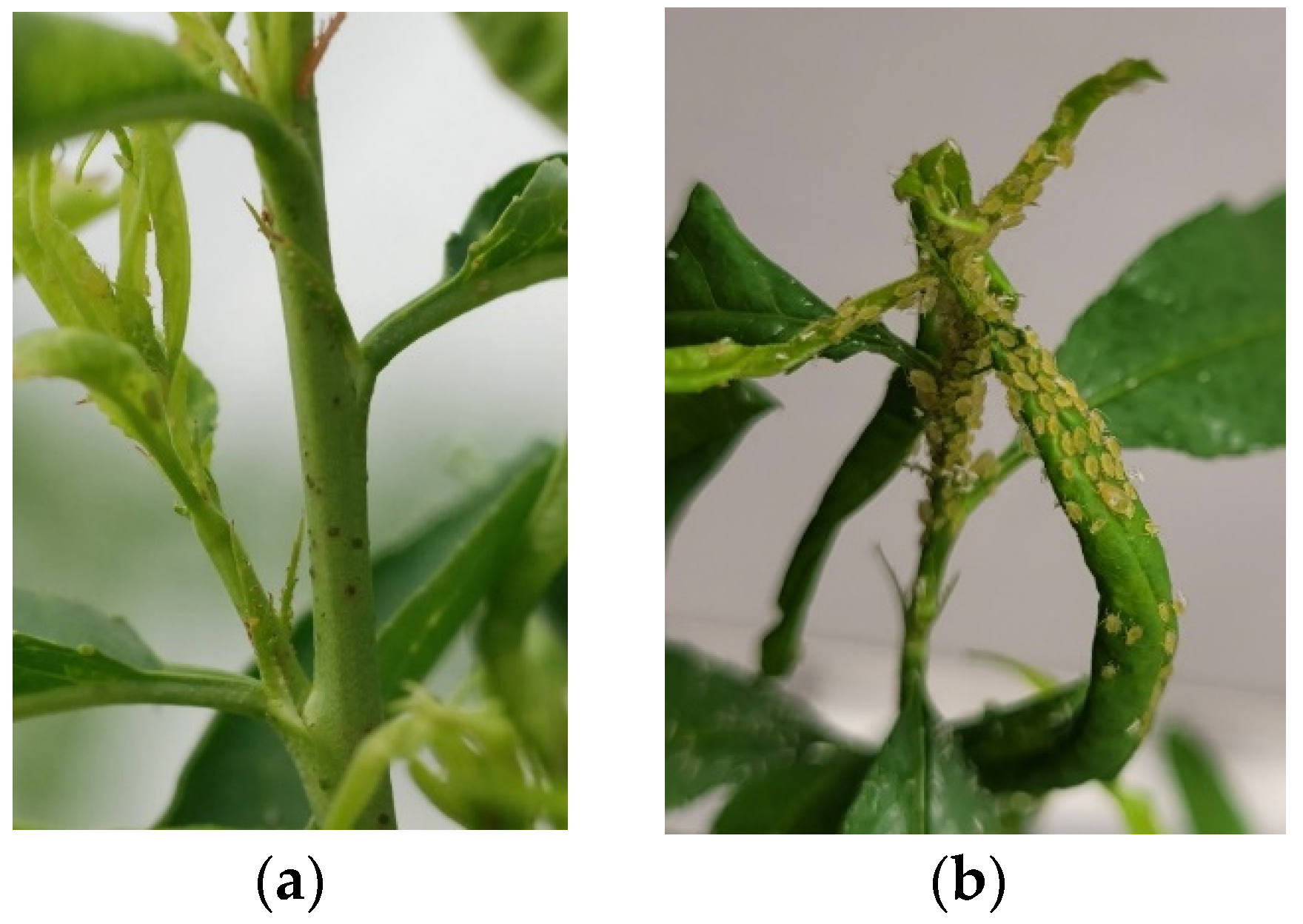
2.2. Phenotyping for Aphid Resistance
2.3. Nucleic Acid Extraction and Genotyping
2.4. NGS Genome Sequencing of the Peach Genotypes
2.5. Genomic Library and Bacterial Artificial Chromosome (BAC) Sequencing
2.6. Construction and Sequencing Full-Length RNA-Seq Library
2.7. Gene Expression (qPCR)
2.8. NGS and Gene Sequence Analysis
3. Results
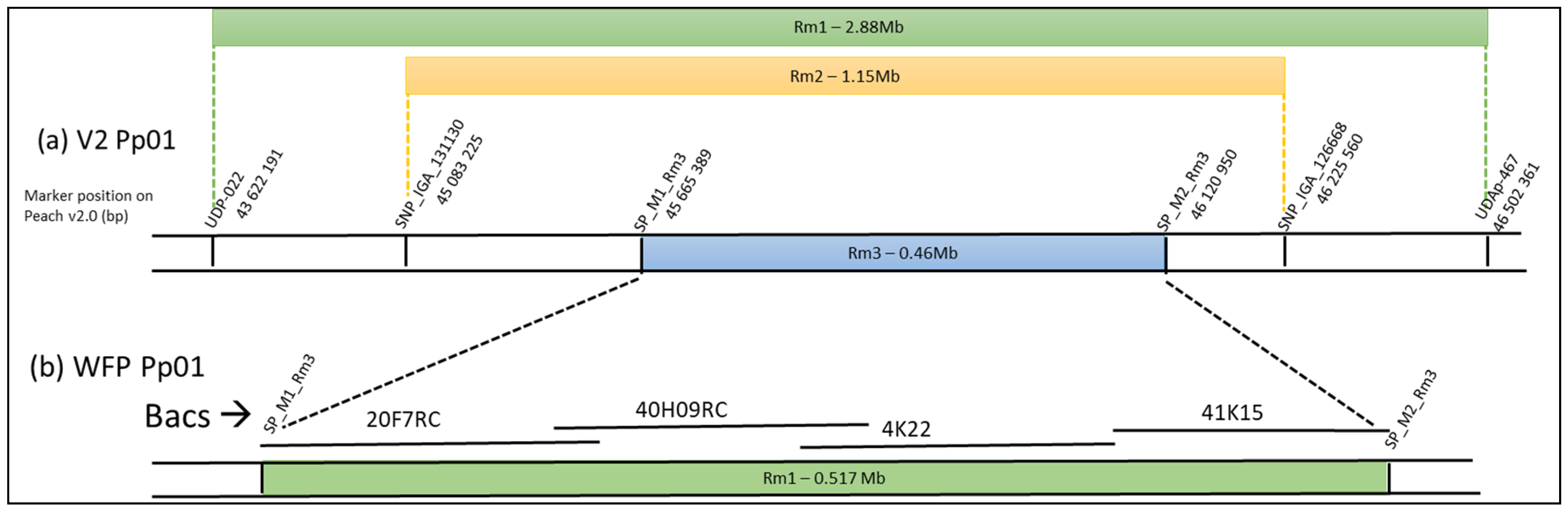
3.1. Reconstruction of the Genomic Region Encompassing Rm1 in WFP
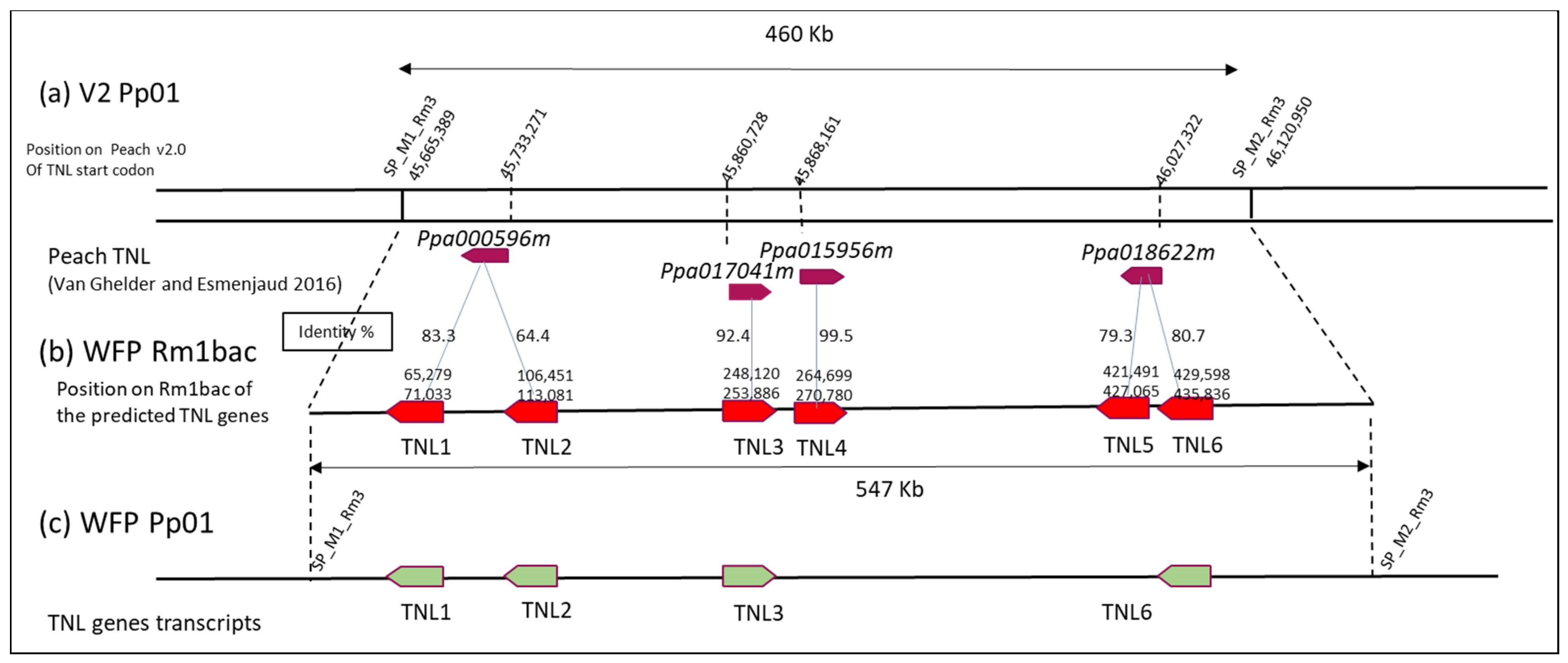
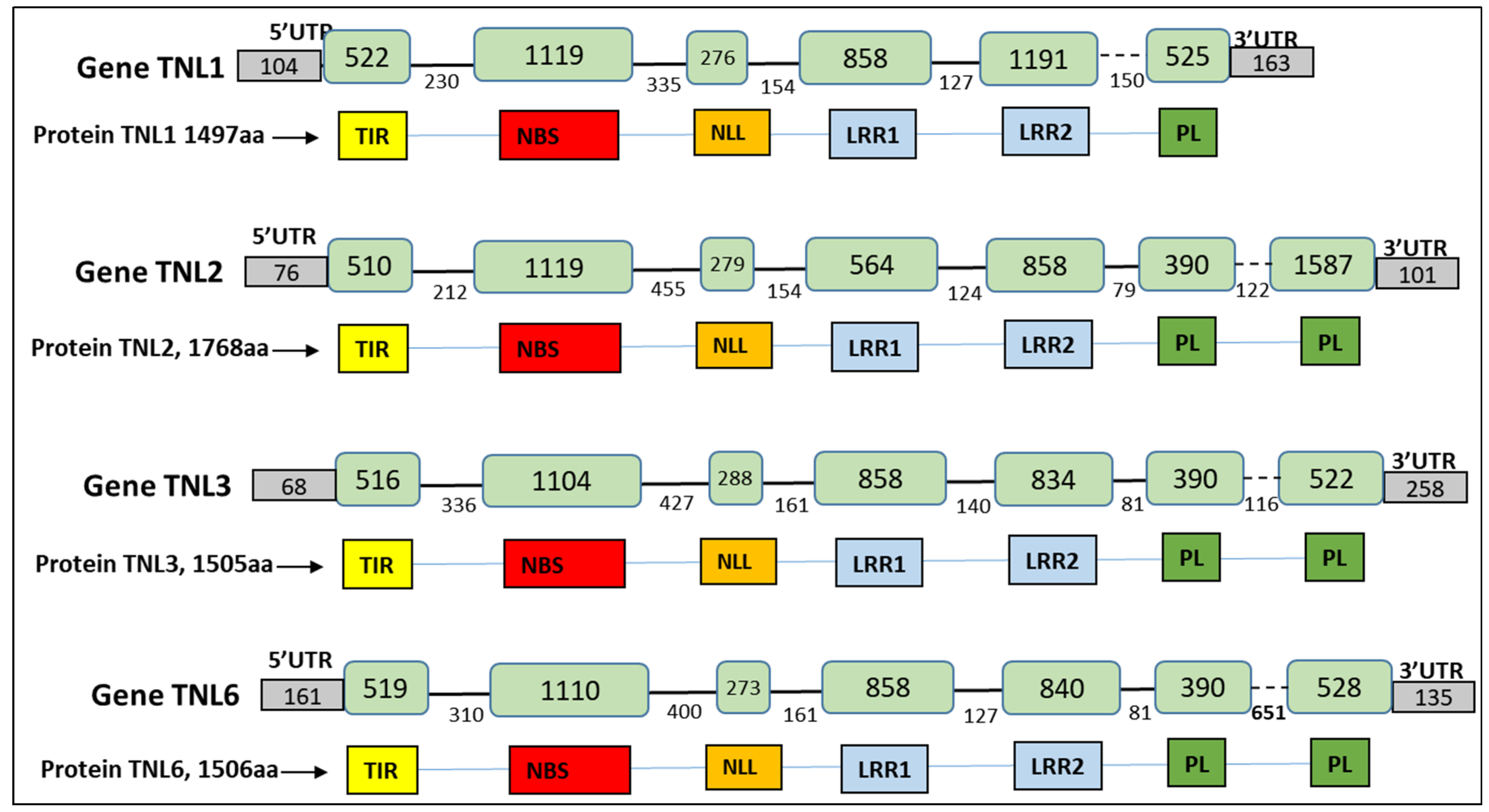
3.2. Structural Annotation of the TNL Genes in the Novel Rm1Bac Sequence from WFP
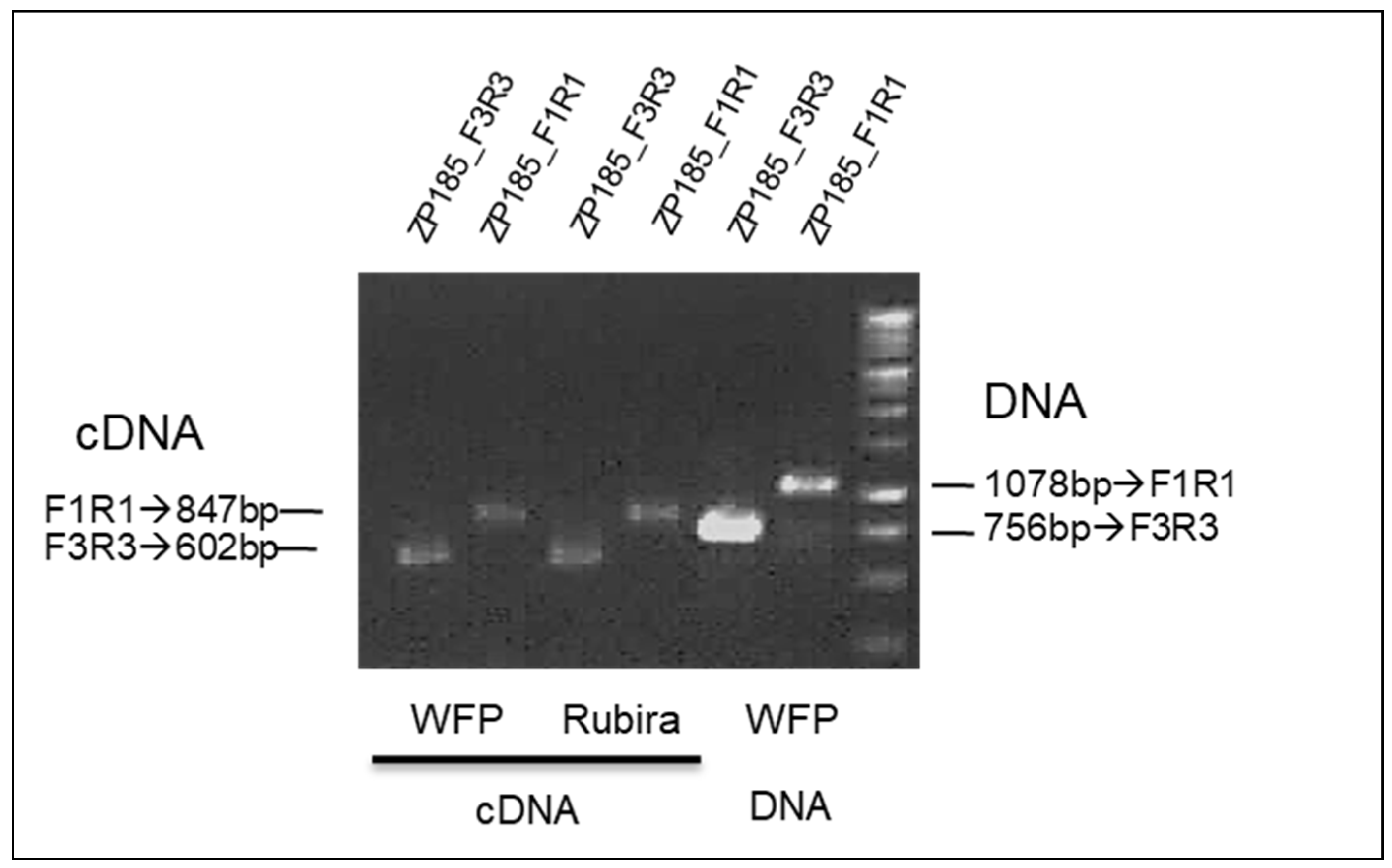
3.3. Quantitative Expression Analysis of the Transcript TNL Genes
3.4. Genomic Sequence of Rm2 in Rubira
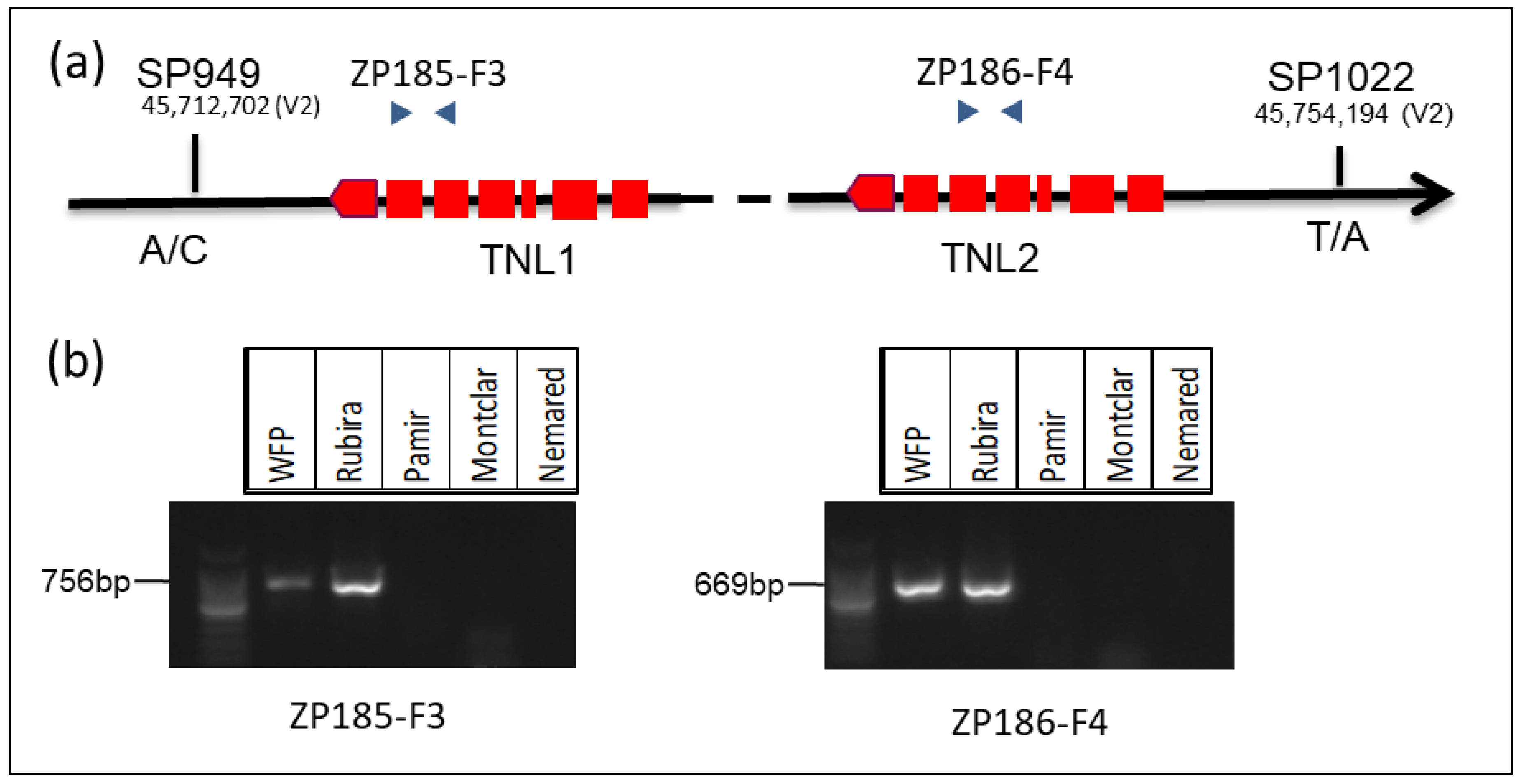
3.5. Marker Development for Marker Assisted Selection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aranzana, M.J.; Decroocq, V.; Dirlewanger, E.; Eduardo, I.; Gao, Z.S.; Gasic, K.; Iezzoni, A.; Jung, S.; Peace, C.; Prieto, H.; et al. Prunus genetics and applications after de novo genome sequencing: Achievements and prospects. Hortic. Res. 2019, 6, 58. [Google Scholar] [CrossRef] [PubMed]
- Gradziel, T.M. Exotic genes for solving emerging peach production challenges. Sci. Hortic. 2022, 295, 110801. [Google Scholar] [CrossRef]
- Claverie, M.; Dirlewanger, E.; Bosselut, N.; Van Ghelder, C.; Voisin, R.; Kleinhentz, M.; Lafargue, B.; Abad, P.; Rosso, M.-N.; Chalhoub, B.; et al. The Ma Gene for Complete-Spectrum Resistance to Meloidogyne Species in Prunus Is a TNL with a Huge Repeated C-Terminal Post-LRR Region. Plant Physiol. 2011, 156, 779–792. [Google Scholar] [CrossRef] [PubMed]
- Van Ghelder, C.; Esmenjaud, D.; Callot, C.; Dubois, E.; Mazier, M.; Duval, H. Ma Orthologous Genes in Prunus spp. Shed Light on a Noteworthy NBS-LRR Cluster Conferring Differential Resistance to Root-Knot Nematodes. Front. Plant Sci. 2018, 9, 1269. [Google Scholar] [CrossRef]
- Marimon, N.; Luque, J.; Arús, P.; Eduardo, I. Fine mapping and identification of candidate genes for the peach powdery mildew resistance gene Vr3. Hortic. Res. 2020, 7, 175. [Google Scholar] [CrossRef]
- Verde, I.; Abbott, A.; Scalabrin, S.; Jung, S.; Shu, S.; Marroni, F.; Zhebentyayeva, T.; Dettori, M.; Grimwood, J.; Cattonaro, F.; et al. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 2013, 45, 487–494. [Google Scholar] [CrossRef]
- Verde, I.; Jenkins, J.; Dondini, L.; Micali, S.; Pagliarani, G.; Vendramin, E.; Paris, R.; Aramini, V.; Gazza, L.; Rossini, L.; et al. The Peach v2.0 release: High-resolution linkage mapping and deep resequencing improve chromosome-scale assembly and contiguity. BMC Genom. 2017, 18, 225. [Google Scholar] [CrossRef]
- Massonié, G.; Maison, P.; Monet, R.; Grasselly, C. Resistance to green peach aphid Myzus-Persicae Sulzer in Prunus persica (L) Batsch and others Prunus. Agronomie 1982, 2, 63–70. [Google Scholar] [CrossRef]
- Massonie, G.; Maison, P. Preliminary-Results on the Study of the Resistance of Prunus-Persica L (batsch) Varieties to Myzus-Persicae Sulz and Myzus-Varians Davids. Ann. Zool. Ecol. Anim. 1979, 11, 479–485. [Google Scholar]
- Monet, R.; Massonie, G. Genetic Determinism of Resistance to the Green Aphid (myzus-Persicae) in the Peach—Further Results. Agronomie 1994, 14, 177–182. [Google Scholar] [CrossRef][Green Version]
- Pascal, T.; Aberlenc, R.; Confolent, C.; Hoerter, M.; Lecerf, E.; Tuero, C.; Lambert, P. Mapping of new resistance (Vr2, Rm1) and ornamental (Di2, pl) Mendelian trait loci in peach. Euphytica 2017, 213, 132. [Google Scholar] [CrossRef]
- Lambert, P.; Pascal, T. Mapping Rm2 gene conferring resistance to the green peach aphid (Myzus persicae Sulzer) in the peach cultivar “Rubira (R)”. Tree Genet. Genomes 2011, 7, 1057–1068. [Google Scholar] [CrossRef]
- Sauge, M.H.; Kervella, J.; Pascal, T. Settling behaviour and reproductive potential of the green peach aphid Myzus persicae on peach varieties and a related wild Prunus. Entomol. Exp. Appl. 1998, 89, 233–242. [Google Scholar] [CrossRef]
- Niu, L.; Lu, Z.; Zeng, W.; Cui, G.; Pan, L.; Xu, Q.; Li, G.; Wang, Z. Inheritance analysis of resistance to green peach aphids(Myzus persicae Sulzer)for peach cultivar‘Fen Shouxing’(Prunus persica var. densa). J. Fruit Sci. 2016, 33, 578–584. [Google Scholar]
- Pan, L.; Lu, Z.; Yan, L.; Zeng, W.; Shen, Z.; Yu, M.; Bu, L.; Cui, G.; Niu, L.; Wang, Z. NLR1 is a strong candidate for the Rm3 dominant green peach aphid (Myzus persicae) resistance trait in peach. J. Exp. Bot. 2022, 73, 1357–1369. [Google Scholar] [CrossRef]
- Lambert, P.; Campoy, J.A.; Pacheco, I.; Mauroux, J.-B.; Linge, C.D.S.; Micheletti, D.; Bassi, D.; Rossini, L.; Dirlewanger, E.; Pascal, T.; et al. Identifying SNP markers tightly associated with six major genes in peach [Prunus persica (L.) Batsch] using a high-density SNP array with an objective of marker-assisted selection (MAS). Tree Genet. Genomes 2016, 12, 121. [Google Scholar] [CrossRef]
- Zhang, N.; Lu, Z.; Cui, G.; Pan, L.; Zeng, W.; Niu, L.; Wang, Z. Gene mapping of aphid-resistant for peach using SNP markers. Sci. Agric. Sin. 2017, 50, 4613–4621. [Google Scholar]
- Duval, H.; Hoerter, M.; Polidori, J.; Confolent, C.; Masse, M.; Moretti, A.; Van Ghelder, C.; Esmenjaud, D. High-resolution mapping of the RMia gene for resistance to root-knot nematodes in peach. Tree Genet. Genomes 2014, 10, 297–306. [Google Scholar] [CrossRef]
- Peterson, D.G.; Tomkins, J.P.; Frisch, D.A.; Paterson, A.H. Construction of plant bacterial artificial chromosome (bac) libraries: An illustrated guide. J. Agric. Genom. 2000, 5, 1–100. [Google Scholar]
- Gonthier, L.; Bellec, A.; Blassiau, C.; Prat, E.; Helmstetter, N.; Rambaud, C.; Huss, B.; Hendriks, T.; Berges, H.; Quillet, M.-C. Construction and characterization of two BAC libraries representing a deep-coverage of the genome of chicory (Cichorium intybus L., Asteraceae). BMC Res. Notes 2010, 3, 225. [Google Scholar] [CrossRef]
- Chalhoub, B.; Belcram, H.; Caboche, M. Efficient cloning of plant genomes into bacterial artificial chromosome (BAC) libraries with larger and more uniform insert size. Plant Biotechnol. J. 2004, 2, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.; Alexander, D.; Marks, P.; Klammer, A.; Drake, J.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.; et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative Genomics Viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed]
- Solovyev, V.; Kosarev, P.; Seledsov, I.; Vorobyev, D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 2006, 7, S10. [Google Scholar] [CrossRef]
- Van Ghelder, C.; Esmenjaud, D. TNL genes in peach: Insights into the post-LRR domain. BMC Genom. 2016, 17, 317. [Google Scholar] [CrossRef]
- Meyers, B.; Kozik, A.; Griego, A.; Kuang, H.; Michelmore, R. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 2003, 15, 809–834. [Google Scholar] [CrossRef]
- Xun, H.; Yang, X.; He, H.; Wang, M.; Guo, P.; Wang, Y.; Pang, J.; Dong, Y.; Feng, X.; Wang, S.; et al. Over-expression of GmKR3, a TIR–NBS–LRR type R gene, confers resistance to multiple viruses in soybean. Plant Mol. Biol 2019, 99, 95–111. [Google Scholar] [CrossRef]
- Sauge, M.; Mus, F.; Lacroze, J.; Pascal, T.; Kervella, J.; Poessel, J. Genotypic variation in induced resistance and induced susceptibility in the peach—Myzus persicae aphid system. Oikos 2006, 113, 305–313. [Google Scholar] [CrossRef]
- Sauge, M.H.; Lacroze, J.P.; Poessel, J.L.; Pascal, T.; Kervella, J. Induced resistance by Myzus persicae in the peach cultivar “Rubira. ” Entomol. Exp. Appl. 2002, 102, 29–37. [Google Scholar] [CrossRef]
- Eitas, T.K.; Dangl, J.L. NB-LRR proteins: Pairs, pieces, perception, partners, and pathways. Curr. Opin. Plant Biol. 2010, 13, 472–477. [Google Scholar] [CrossRef]
- Gallois, J.; Moury, B.; German-Retana, S. Role of the Genetic Background in Resistance to Plant Viruses. Int. J. Mol. Sci. 2018, 19, 2856. [Google Scholar] [CrossRef] [PubMed]
- Dogimont, C.; Chovelon, V.; Pauquet, J.; Boualem, A.; Bendahmane, A. The Vat locus encodes for a CC-NBS-LRR protein that confers resistance to Aphis gossypii infestation and A. gossypii-mediated virus resistance. Plant J. 2014, 80, 993–1004. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Goggin, F.; Milligan, S.; Kaloshian, I.; Ullman, D.; Williamson, V. The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc. Natl. Acad. Sci. USA 1998, 95, 9750–9754. [Google Scholar] [CrossRef] [PubMed]
- Boualem, A.; Dogimont, C.; Bendahmane, A. The battle for survival between viruses and their host plants. Curr. Opin. Virol. 2016, 17, 32–38. [Google Scholar] [CrossRef]
- Milligan, S.; Bodeau, J.; Yaghoobi, J.; Kaloshian, I.; Zabel, P.; Williamson, V. The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 1998, 10, 1307–1319. [Google Scholar] [CrossRef] [PubMed]
- Nombela, G.; Williamson, V.; Muniz, M. The root-knot nematode resistance gene Mi-1.2 of tomato is responsible for resistance against the whitefly Bemisia tabaci. Mol. Plant-Microbe Interact. 2003, 16, 645–649. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.; Meyers, B.; Chen, J.; Tian, D.; Yang, S. Tracing the origin and evolutionary history of plant nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes. New Phytol. 2012, 193, 1049–1063. [Google Scholar] [CrossRef]
- Gassmann, W.; Hinsch, M.; Staskawicz, B. The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 1999, 20, 265–277. [Google Scholar] [CrossRef]
- Reighard, G.L.; Newall, W.C.; Beckman, T.G.; Okie, W.R.; Zehr, E.I.; Nyczepir, A.P. Field performance of Prunus rootstock cultivars and selections on replant soils in South Carolina. In Proceedings of the International Symposium on Integrating Canopy, Rootstocks and Environmental Physiology in Orchard Systems, Proceedings—Vols 1 and 2, Stellenbosch, South Africa, 3–6 December 2012; Barritt, B.H., Kappel, F., Eds.; International Society Horticultural Science: Leuven, Belgium, 1997; pp. 243–249. [Google Scholar]
- Kadasa, N.M.; Metwali, E.M.R.; Soliman, H.I.A.; Alshehri, W.A. Creation of borer pests resistance genetically engineering peach (Prunus persica L.) plants by constitutively overexpressing the cry1Ab gene. Plant Cell Tiss Organ Cult. 2022, 148, 465–477. [Google Scholar] [CrossRef]
- Sauge, M.-H.; Lambert, P.; Pascal, T. Co-localisation of host plant resistance QTLs affecting the performance and feeding behaviour of the aphid Myzus persicae in the peach tree. Heredity 2012, 108, 292–301. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duval, H.; Heurtevin, L.; Dlalah, N.; Callot, C.; Lagnel, J. The Rm1 and Rm2 Resistance Genes to Green Peach Aphid (Myzus persicae) Encode the Same TNL Proteins in Peach (Prunus persica L.). Genes 2022, 13, 1489. https://doi.org/10.3390/genes13081489
Duval H, Heurtevin L, Dlalah N, Callot C, Lagnel J. The Rm1 and Rm2 Resistance Genes to Green Peach Aphid (Myzus persicae) Encode the Same TNL Proteins in Peach (Prunus persica L.). Genes. 2022; 13(8):1489. https://doi.org/10.3390/genes13081489
Chicago/Turabian StyleDuval, Henri, Laure Heurtevin, Naïma Dlalah, Caroline Callot, and Jacques Lagnel. 2022. "The Rm1 and Rm2 Resistance Genes to Green Peach Aphid (Myzus persicae) Encode the Same TNL Proteins in Peach (Prunus persica L.)" Genes 13, no. 8: 1489. https://doi.org/10.3390/genes13081489
APA StyleDuval, H., Heurtevin, L., Dlalah, N., Callot, C., & Lagnel, J. (2022). The Rm1 and Rm2 Resistance Genes to Green Peach Aphid (Myzus persicae) Encode the Same TNL Proteins in Peach (Prunus persica L.). Genes, 13(8), 1489. https://doi.org/10.3390/genes13081489





