A Mouse Model with Ablated Asparaginase and Isoaspartyl Peptidase 1 (Asrgl1) Develops Early Onset Retinal Degeneration (RD) Recapitulating the Human Phenotype
Abstract
1. Introduction
2. Methodology
3. Results
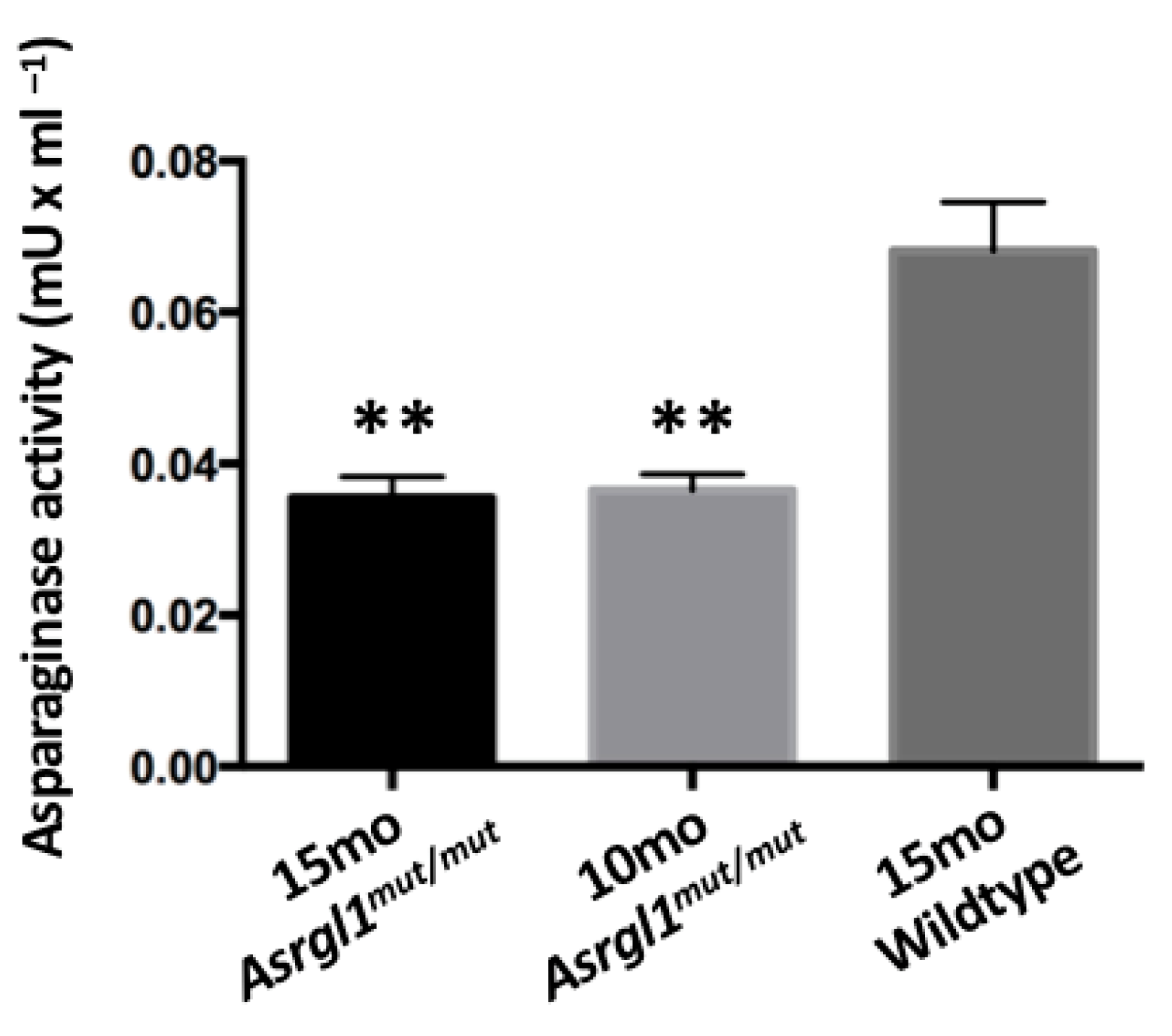
3.1. Generation of Asrgl1mut/mut Mouse Model
3.2. Ophthalmic Evaluation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- RetNet. Available online: http://www.sph.uth.tmc.edu/Retnet/ (accessed on 1 January 2022).
- Hamel, C. Retinitis pigmentosa. Orphanet J. Rare Dis. 2006, 1, 40. [Google Scholar] [CrossRef] [PubMed]
- Lev, S. Molecular aspects of retinal degenerative diseases. Cell Mol. Neurobiol. 2001, 21, 575–589. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, S.; Di Iorio, E.; Barbaro, V.; Ponzin, D.; Sorrentino, F.S.; Parmeggiani, F. Retinitis pigmentosa: Genes and disease mechanisms. Curr. Genom. 2011, 12, 238–249. [Google Scholar]
- Daiger, S.P.; Sullivan, L.S.; Bowne, S.J. Genes and mutations causing retinitis pigmentosa. Clin. Genet. 2013, 84, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Collin, G.B.; Gogna, N.; Chang, B.; Damkham, N.; Pinkney, J.; Hyde, L.F.; Stone, L.; Naggert, J.K.; Nishina, P.M.; Krebs, M.P. Mouse Models of Inherited Retinal Degeneration with Photoreceptor Cell Loss. Cells 2020, 9, 931. [Google Scholar] [CrossRef]
- Beryozkin, A.; Matsevich, C.; Obolensky, A.; Kostic, C.; Arsenijevic, Y.; Wolfrum, U.; Banin, E.; Sharon, D. A new mouse model for retinal degeneration due to Fam161a deficiency. Sci. Rep. 2021, 11, 2030. [Google Scholar] [CrossRef]
- Qin, W.; Kutny, P.M.; Maser, R.S.; Dion, S.L.; Lamont, J.D.; Zhang, Y.; Perry, G.A.; Wang, H. Generating Mouse Models Using CRISPR-Cas9-Mediated Genome Editing. Curr. Protoc. Mouse Biol. 2016, 6, 39–66. [Google Scholar] [CrossRef]
- Fauser, S.; Luberichs, J.; Schuttauf, F. Genetic animal models for retinal degeneration. Surv. Ophthalmol. 2002, 47, 357–367. [Google Scholar] [CrossRef]
- Petersen-Jones, S.M. Animal models of human retinal dystrophies. Eye 1998, 12 Pt 3b, 566–570. [Google Scholar] [CrossRef]
- Biswas, P.; Chavali, V.R.M.; Agnello, G.; Stone, E.; Chakarova, C.; Duncan, J.L.; Kannabiran, C.; Homsher, M.; Bhattacharya, S.S.; Naeem, M.A.; et al. A missense mutation in ASRGL1 is involved in causing autosomal recessive retinal degeneration. Hum. Mol. Genet. 2016, 25, 2483–2497. [Google Scholar]
- Cantor, J.R.; Stone, E.M.; Chantranupong, L.; Georgiou, G. The human asparaginase-like protein 1 hASRGL1 is an Ntn hydrolase with β-aspartyl peptidase activity. Biochemistry 2009, 48, 11026–11031. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, H.; Shivalila, C.S.; Dawlaty, M.M.; Cheng, A.W.; Zhang, F.; Jaenisch, R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013, 153, 910–918. [Google Scholar] [CrossRef] [PubMed]
- Inui, M.; Miyado, M.; Igarashi, M.; Tamano, M.; Kubo, A.; Yamashita, S.; Asahara, H.; Fukami, M.; Takada, S. Rapid generation of mouse models with defined point mutations by the CRISPR/Cas9 system. Sci. Rep. 2014, 4, 5396. [Google Scholar] [CrossRef]
- Chavali, V.R.; Khan, N.W.; Cukras, C.A.; Bartsch, D.U.; Jablonski, M.M.; Ayyagari, R. A CTRP5 gene S163R mutation knock-in mouse model for late-onset retinal degeneration. Hum. Mol. Genet. 2011, 20, 2000–2014. [Google Scholar] [CrossRef] [PubMed]
- Mandal, N.A.; Vasireddy, V.; Jablonski, M.M.; Wang, X.; Heckenlively, J.R.; Hughes, B.A.; Reddy, G.B.; Ayyagari, R. Spatial and temporal expression of MFRP and its interaction with CTRP5. Investig. Ophthalmol. Vis. Sci. 2006, 47, 5514–5521. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Cantor, J.R.; Yogesha, S.D.; Yang, S.; Chantranupong, L.; Liu, J.Q.; Agnello, G.; Georgiou, G.; Stone, E.M.; Zhang, Y. Uncoupling intramolecular processing and substrate hydrolysis in the N-terminal nucleophile hydrolase hASRGL1 by circular permutation. ACS Chem. Biol. 2012, 7, 1840–1847. [Google Scholar] [CrossRef][Green Version]
- Li, W.; Irani, S.; Crutchfield, A.; Hodge, K.; Matthews, W.; Patel, P.; Zhang, Y.J.; Stone, E. Intramolecular Cleavage of the hASRGL1 Homodimer Occurs in Two Stages. Biochemistry 2016, 55, 960–969. [Google Scholar] [CrossRef]
- Edqvist, P.H.D.; Huvila, J.; Forsström, B.; Talve, L.; Carpén, O.; Salvesen, H.B.; Krakstad, C.; Grénman, S.; Johannesson, H.; Ljungqvist, O.; et al. Loss of ASRGL1 expression is an independent biomarker for disease-specific survival in endometrioid endometrial carcinoma. Gynecol. Oncol. 2015, 137, 529–537. [Google Scholar] [CrossRef]
- Radadiya, A.; Zhu, W.; Coricello, A.; Alcaro, S.; Richards, N.G.J. Improving the Treatment of Acute Lymphoblastic Leukemia. Biochemistry 2020, 59, 3193–3200. [Google Scholar] [CrossRef]
- Kumar, K.; Kaur, J.; Walia, S.; Pathak, T.; Aggarwal, D. L-asparaginase: An effective agent in the treatment of acute lymphoblastic leukemia. Leuk Lymphoma 2014, 55, 256–262. [Google Scholar] [CrossRef]
- Zhou, Y.; Tian, W.; Jiang, X.; Yang, H.; Jiang, Z.; Li, X.; Jiang, D.; Sun, K.; Yang, Y.; Liu, W.; et al. Deletion of Asrgl1 Leads to Photoreceptor Degeneration in Mice. Front. Cell Dev. Biol. 2021, 9, 783547. [Google Scholar] [CrossRef] [PubMed]
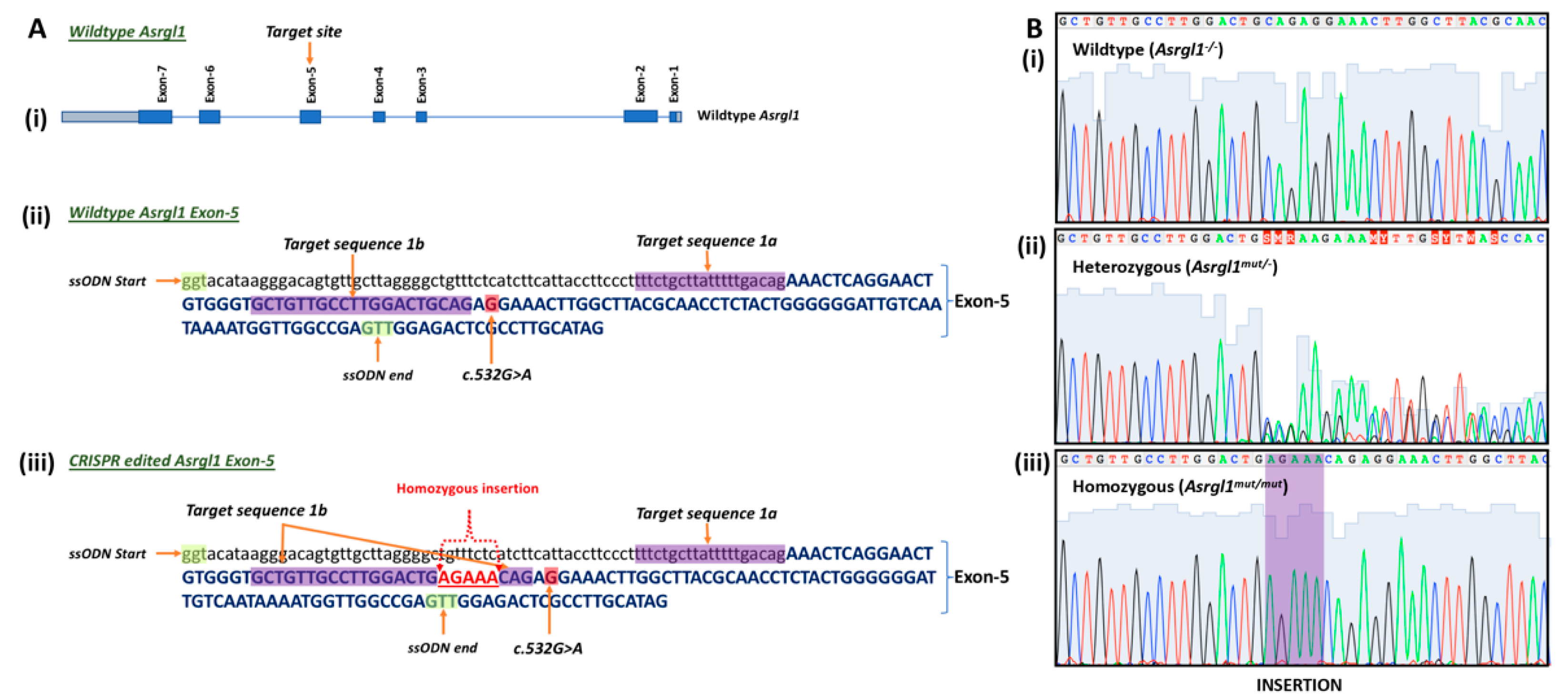
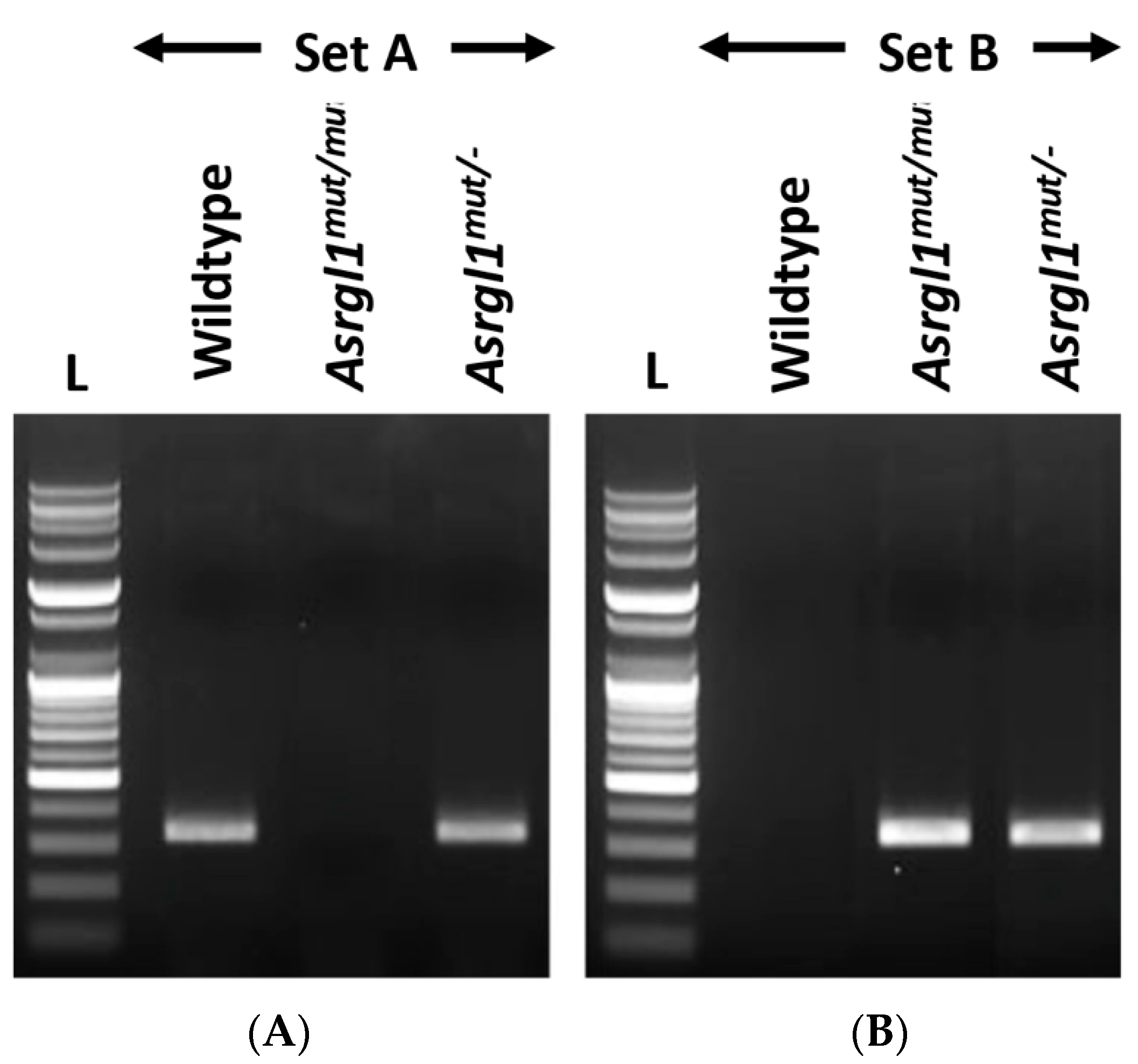

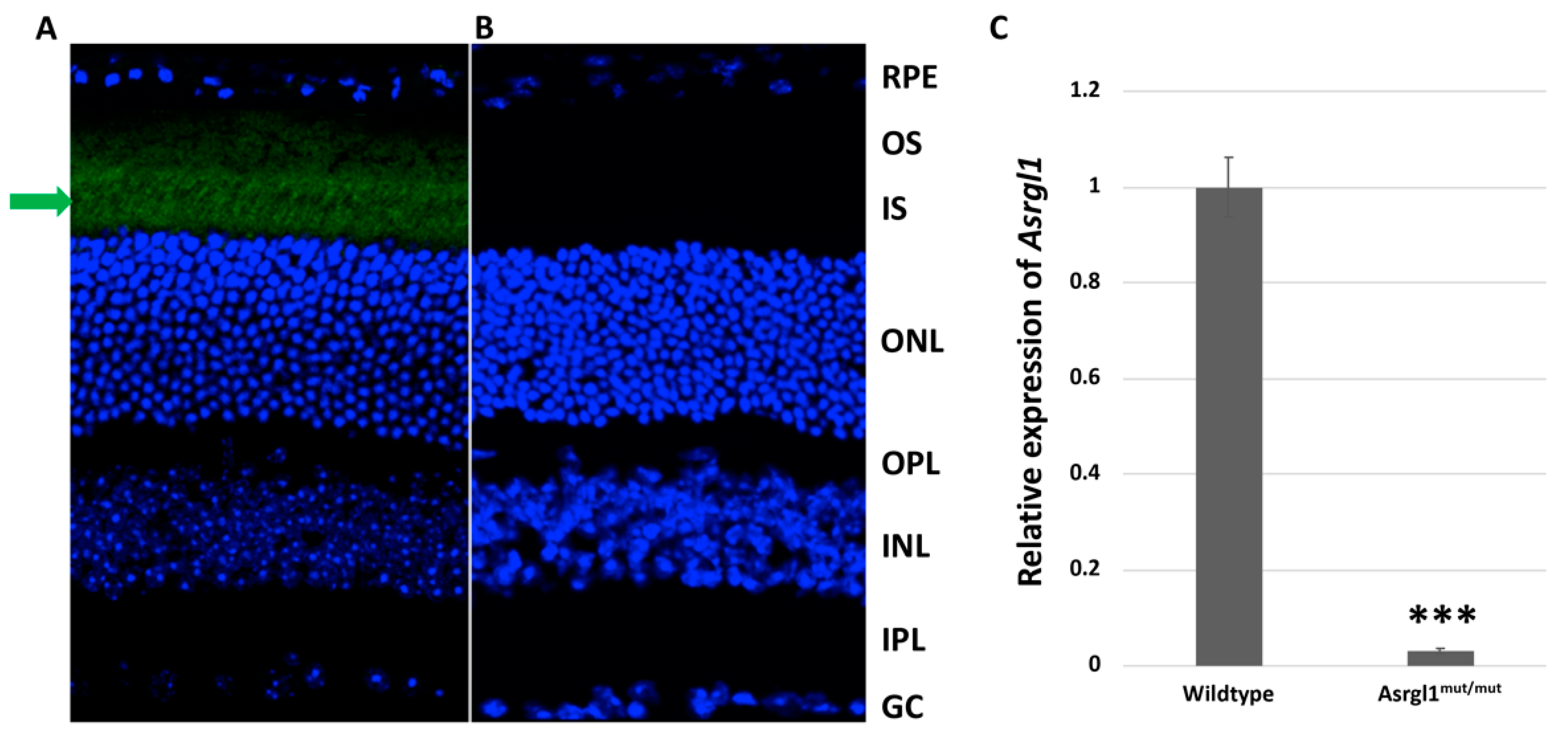
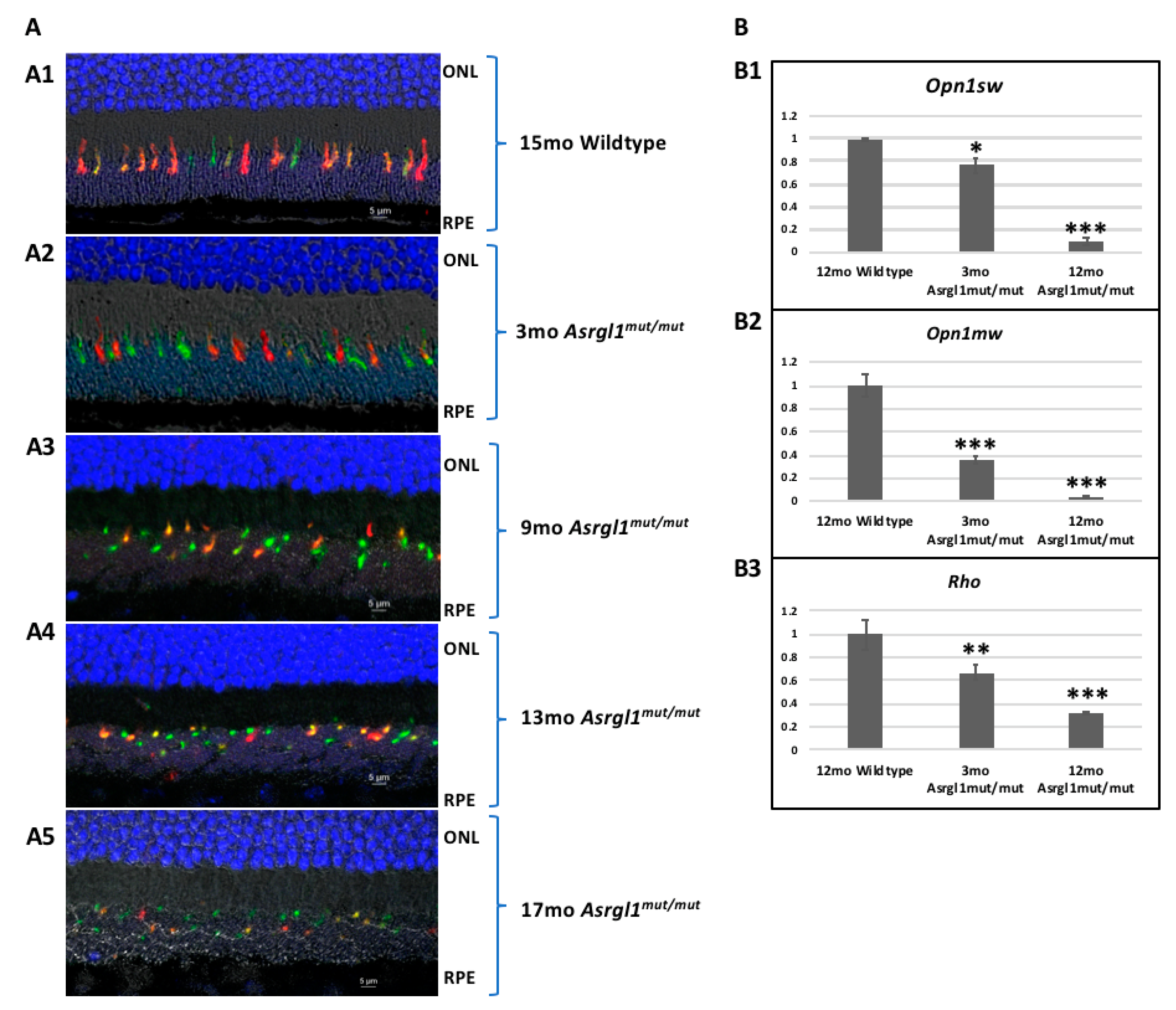
| Primer ID | Sequence | Length | Tm | GC% | Amplicon |
|---|---|---|---|---|---|
| 1. mAsrgl1 wt F | AAGCCAAGTTTCCTCTGAAGTC | 22 | 60.3 | 45.5 | 328 bp |
| 2. mAsrgl1 mut F | AAGCCAAGTTTCCTCTGGTTCT | 22 | 60.3 | 45.5 | 333 bp |
| 3. mAsrgl1 uni R | ACCAAGCCAACCTCAGACTC | 20 | 60.5 | 55.0 |
| Sample | Set A (Wildtype Allele Specific) | Set B (Mutant Allele Specific) |
|---|---|---|
| Wildtype (Asrgl1−/−) | 328 bp product | None |
| Homozygous (Asrgl1mut/mut) | None | 333 bp product |
| Heterozygous (Asrgl1mut/−) | 328 bp product | 333 bp product |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biswas, P.; Berry, A.M.; Zawaydeh, Q.; Bartsch, D.-U.G.; Raghavendra, P.B.; Hejtmancik, J.F.; Khan, N.W.; Riazuddin, S.A.; Ayyagari, R. A Mouse Model with Ablated Asparaginase and Isoaspartyl Peptidase 1 (Asrgl1) Develops Early Onset Retinal Degeneration (RD) Recapitulating the Human Phenotype. Genes 2022, 13, 1461. https://doi.org/10.3390/genes13081461
Biswas P, Berry AM, Zawaydeh Q, Bartsch D-UG, Raghavendra PB, Hejtmancik JF, Khan NW, Riazuddin SA, Ayyagari R. A Mouse Model with Ablated Asparaginase and Isoaspartyl Peptidase 1 (Asrgl1) Develops Early Onset Retinal Degeneration (RD) Recapitulating the Human Phenotype. Genes. 2022; 13(8):1461. https://doi.org/10.3390/genes13081461
Chicago/Turabian StyleBiswas, Pooja, Anne Marie Berry, Qais Zawaydeh, Dirk-Uwe G. Bartsch, Pongali B. Raghavendra, J. Fielding Hejtmancik, Naheed W. Khan, S. Amer Riazuddin, and Radha Ayyagari. 2022. "A Mouse Model with Ablated Asparaginase and Isoaspartyl Peptidase 1 (Asrgl1) Develops Early Onset Retinal Degeneration (RD) Recapitulating the Human Phenotype" Genes 13, no. 8: 1461. https://doi.org/10.3390/genes13081461
APA StyleBiswas, P., Berry, A. M., Zawaydeh, Q., Bartsch, D.-U. G., Raghavendra, P. B., Hejtmancik, J. F., Khan, N. W., Riazuddin, S. A., & Ayyagari, R. (2022). A Mouse Model with Ablated Asparaginase and Isoaspartyl Peptidase 1 (Asrgl1) Develops Early Onset Retinal Degeneration (RD) Recapitulating the Human Phenotype. Genes, 13(8), 1461. https://doi.org/10.3390/genes13081461








